Abstract
Autophagy, especially selective autophagy, is poorly characterized in plants compared with mammals and yeasts, where numerous factors required for the proper regulation of autophagy have been identified. The evidence for the importance of the cytoskeleton (both actin filaments and microtubules) in various aspects of autophagy comes mostly from work on yeasts and mammals, while in plant cells these links are poorly explored. In this report we demonstrate that tobacco protein Joka2, a member of a family of selective autophagy cargo receptors closely related to mammalian NBR1 and p62 colocalizes with both major cytoskeletal components, microtubules and microfilaments and, additionally, resides in close proximity of the ER.
Keywords: actin, autophagy, cytoskeleton, ER, Joka2, microfilament, microtubule, NBR1-like protein, selective autophagy cargo receptor, tobacco, tubulin
The plant cytoskeleton is involved in numerous fundamental processes such as mitosis, cytokinesis, cell polarity or intracellular trafficking. It also modulates the plant response to environmental stimuli and affects cell morphogenesis by controlling the arrangement of cellulose microfibrils, the cell wall assembly and expansion.1 The cytoskeleton comprises of 2 distinct networks of fibrous polymers, microtubules (MTs) and microfilaments (MFs), composed of tubulin and actin respectively. The MTs are polar tubular structures built by α-β tubulin heterodimers. They are mobile and highly dynamic structures characterized by rapid polymerization at the plus end and slower depolymerization at the minus end.2 The minus ends of MTs are anchored in the central part of the cell while the plus ends directed mainly toward the cell perimeter. The MFs are polar as well but are shorter and thinner than the MTs, and are distributed more randomly in the cell. In plants, in contrast to animals, the intracellular traffic seems to rely mostly on the MFs, while the MTs mainly position (stabilize) the organelles (mitochondria, chloroplasts).3,4 The intracellular transport depends on motor proteins carrying the cargo along the cytoskeletal filaments, myosins along the MFs, and kinesins and dyneins along the MTs.5 Interestingly, different groups of organisms have different subsets of molecular motors, suggesting a diversity of strategies for intracellular transport. The MTs and MFs co-operate with each other owing to their direct physical interactions and indirect interactions mediated by various bifunctional proteins or multiprotein complexes.6 The structure and dynamics of the cytoskeleton can be studied by fluorescence microscopy with the use of diverse specific microtubule-associated proteins (MAPs) or actin-interacting proteins with actin binding domains (ABD).7-9
Autophagy is an evolutionarily conserved process of degradation of unwanted cell components in UPS-independent manner with over 30 genes (ATG-autophagy-related genes encoding Atg proteins) involved.10,11 The best characterized type of autophagy is macroautophagy (here referred to simply as autophagy) where double-membrane structures (autophagosomes) are created for cargo uptake and sequestration to the lysosomes or vacuole(s) for degradation.12 For a long time autophagy was considered as a bulk process bereft selectivity. However, an increasing numbers of evidences that specific cargos are selectively carried for degradation have revealed recently that selective autophagy cargo receptors exist and are involved in cargo recognition.13 The tobacco Joka2 protein14 and its homolog from Arabidopsis thaliana AtNBR115 are so far the only examples of the selective autophagy receptors described in plants. Despite the increasing interest in autophagy, the recognition of proteins connecting it with other cellular components, such as the cytoskeleton, is limited and mostly refers to yeasts and mammals.16,17
In yeasts, the MTs are not essential for bulk autophagy despite the fact that in vitro the Atg8 protein binds to tubulin1 and 2 via Atg4.18 Unlike the MTs, actin filaments seem to play a crucial role in the biogenesis of the Cvt (cytoplasm-to-vacuole targeting) pathway.19-22 Actin, in complex with Atg11 and Arp2/3 (actin-related protein2/3 complex, an actin nucleation factor), is involved in the trafficking of the Cvt complex and Atg9-containing membranes to the phagophore assembly site (PAS).20,22-25 As mentioned above, actin is required in yeast for cargo selection in the Cvt pathway and also for pexophagy or specific removal of the ER.19,20,22,23,25 Little is known about the relationships between actin and autophagy in mammals or plants. It has only been shown that in mammals GABARAP (a member of the ATG8 family) co-localizes with MFs.26 However, it is known that autophagosomes move along microtubules and dynein is involved, since lysosomes are mainly located near the MT organizing center (MTOC), while new autophagosomes are formed peripherally.27-29 Additionally, the LC3 protein from the ATG8 family binds to MTs directly or via MAP1A and MAP1B but the significance of this binding is unknown.30,31 The participation of MTs in autophagosome formation and fusion with late endosomes/lysososmes in mammals has been postulated as well.32 Nevertheless, it is still unknown how the ubiquitin-recognizing selective autophagy receptors are moved close to the polyubiquitinated components or how they are transported to the PAS.
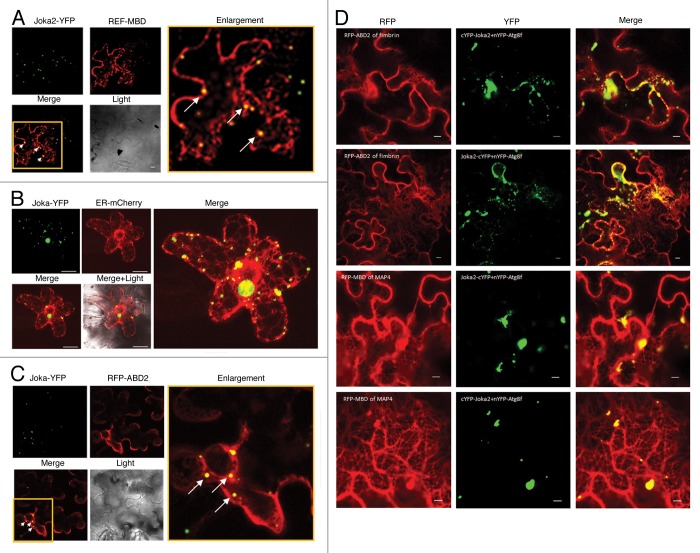
The Arabidopsis Atg8s, similarly to the mammalian GABARAP/LC3 protein family, bind MTs33 and probably use them to relocate autophagosomes to the vacuole. On the other hand, it has been shown recently that the mammalian selective autophagy receptor NBR1 binds to the light chain of the microtubule-associated protein (MAP1B-LC1) through the NBR1/FW domain conserved in all NBR1-like proteins.34 A recently characterized NBR1-like protein from tobacco, Joka2, interacts with ATG8s,14,35 but no evidence of its interaction with MTs has been presented so far. Here we show that Joka2-YFP transiently co-expressed in planta with microtubule-binding domain (MBD) of MAP4 fused to RFP (used as MTs marker) (Fig. 1A) co-localizes, suggesting a Joka2-MTs interaction. This result also argues for the Joka2 involvement in autophagy by showing its resemblance to the well-characterized mammalian selective autophagy receptor NBR1.34 Furthermore, the co-localization of Joka2-YFP with ER-targeted mCherry protein (Fig. 1B) suggest association of Joka2 with newly formed autophagosomes since the phagophores are formed from the ER membrane.39 It could be also hypothesized that Joka2 receives cargo from the ER and docks it into autophagosomes during phagophore maturation, thereby preventing the ER from overloading. Since the ER controls protein folding, posttranslational modifications and subsequently initial steps of their delivery through the secretory pathway to their destination a mechanism is required to prevent defects in ER functioning caused by the accumulation of aberrant and/or misfolded proteins within the ER lumen. Perhaps bulk autophagy and selective autophagy mediated by the selective autophagy cargo receptors such as Joka2 recognizing polyubiquitinated proteins could be a mechanism of misfolded or unwanted protein clearance from the ER supplementing the UPS system. Concluding, the presented co-localization of Joka2 with MTs is the first evidence of a connection between the cytoskeleton and selective autophagy in planta and lets us hypothesize that also in plants the MTs facilitate autophagosome trafficking, as they do in mammals. However, it is also possible that MTs could be involved in the formation of early autophagosomes or in their fusion with the vacuole since such a role of MTs in mammals has been under extensive debate.
Figure 1. Co-localization of Joka2-YFP with MTs (A), ER (B), MFs (C), and co-localization of the Joka2-Atg8 complex with MTs and MFs (D). (A) Confocal microscope image of co-localization of fluorescent signals from fusion proteins Joka2-YFP and RFP-MBD (for microtubules labeling) in N. benthamiana epidermal cells transfected by Agrobacterium containing plasmids pK7WGR2-MBD of MAP436 and pJ4.14 Observation under Nicon confocal microscope Eclipse TE2000-E was done 3 days post agroinfiltration.35 Selected region of the Merge panel has been enlarged to expose yellow spots (showed by white arrows) indicating the Joka2-YFP signals co-localized with microtubules. (B) Confocal microscope image of Joka2-YFP and ER structures labeled with mCherry protein in N. benthamiana epidermis one day post particle bombardment. For the biolistic gene transformation (bombardment) of N. benthamiana leaves cultivated in non-stressed conditions used tungsten-M10 as a microcarrier. Tungsten 0.7µm particles (325 µg) were coated with 2 µg of mixture of plasmids pJ414 and ER-mCherry.37 BIO-RAD PDS-1000/He Biolistic Particle Delivery System was used at 1300psi helium pressure. After that leaves were placed on solid 0.5xMS medium with 3% sucrose and kept in darkness for 24h at room temperature. Yellow spots in the Merge panels indicate the Joka2-YFP signals co-localized with ER structures. (C) Confocal microscope image of co-localization of fluorescent signals from fusion proteins Joka2-YFP and RFP-ABD2 (for microfilaments labeling) 3 days post agroinfiltration of N. benthamiana epidermal cells with Agrobacterium containing plasmids pK7WGR2- ABD2 of fimbrin38 and pJ4.14 Selected region of the Merge panel has been enlarged to expose yellow spots (showed by white arrows) indicating the Joka2-YFP signals co-localized with microfilaments. (D) Confocal microscope images of co-localization of fluorescent signals from YFP formed by BiFC from YC-Joka2 or Joka2-YC and YN-ATG8f35 and RFP-ABD2 or RFP-MBD fusion proteins (labeled microtubules and microfilaments). Yellow spots in the Merge panels indicate the BiFC signals resulting from Joka2-ATG8f interactions (visualized in YFP panels) which is co-localized with microfilaments or microtubules in 2 upper rows or 2 lower rows, respectively (2 BiFC combinations were used).
On the other hand, we also observed a co-localization of Joka2-YFP and actin-binding domain 2 (ABD2) of fimbrin fused to RFP serving as an MFs marker transiently co-expressed in planta, suggesting a Joka2 association with actin (Fig. 1C). Notably, we found that not only Joka2 alone but also the Joka2-Atg8f complex (complex formation was monitored by BiFC) co-localizes with fluorescently marked MTs and MFs (Fig. 1D). Subsequent bioinformatic analysis of Joka2 sequence has revealed the existence of a WH2 actin-binding bipartite motif in the first interdomain region (between PB1 domain and NES motif). The N-terminus of this motif forms a short α-helix rich in hydrophobic acids and binds to the hydrophobic cleft formed by subdomains 1 and 3 of actin, while the C-terminus of this motif forms a disordered region, stabilized upon actin binding. Interestingly, the WH2 motif is conserved only in plant NBR1-like proteins but not in the mammalian NBR1s (not shown). The Joka2-Atg8f interaction with actin could either promote or inhibit filament assembly and regulate filament nucleation.40 One could speculate that by promoting actin polymerization Joka2 recruits polyubiquitinated cargo into phagophore or that it is moved there with other protein(s). Interestingly, it is known that p62 (another mammalian homolog of Joka2 besides NBR1, also called SQSTM1) requires self-oligomerization but not LC3 binding to be loaded to the PAS and this step is accelerated by starvation.41 However, it is still unknown how p62 is targeted to the autophagosome formation site. An involvement of actin filaments in this process cannot be excluded. On the other hand, it has been shown that the proteasome physically associates with actin filaments.42 Since p62 can deliver substrates to the proteasome in mammals.43,44 and Joka2 could have a similar function in plants (due to the high similarity of their PB1 domains) an involvement of the actin cytoskeleton in transporting Joka2 to proteasome should also be considered. The 2 hypotheses about the involvement of actin in autophagosome formation and selective docking, and in the recruitment of selective autophagy receptors to the proteasome require further verification and are not mutually exclusive.
To conclude, the presented results shed new light on the role of the cytoskeleton in selective autophagy in plants by showing that a tobacco NBR1-like protein, Joka2, either alone or as in complex with Atg8 (possibly along with an ubiquitinated cargo), co-localizes with both types of the cytoskeleton components: microtubules and actin microfilaments. The functional significance of this co-localization remains to be determined.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Polish Ministry of Science and Higher Education (grant nr W16/7.PR/2011).
References
- 1.Ivakov A, Persson S. Plant cell shape: modulators and measurements. Front Plant Sci. 2013;4:439. doi: 10.3389/fpls.2013.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nogales E. A structural view of microtubule dynamics. Cell Mol Life Sci. 1999;56:133–42. doi: 10.1007/s000180050012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Gestel K, Köhler RH, Verbelen JP. Plant mitochondria move on F-actin, but their positioning in the cortical cytoplasm depends on both F-actin and microtubules. J Exp Bot. 2002;53:659–67. doi: 10.1093/jexbot/53.369.659. [DOI] [PubMed] [Google Scholar]
- 4.Romagnoli S, Cai G, Faleri C, Yokota E, Shimmen T, Cresti M. Microtubule- and actin filament-dependent motors are distributed on pollen tube mitochondria and contribute differently to their movement. Plant Cell Physiol. 2007;48:345–61. doi: 10.1093/pcp/pcm001. [DOI] [PubMed] [Google Scholar]
- 5.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–80. doi: 10.1016/S0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 6.Petrásek J, Schwarzerová K. Actin and microtubule cytoskeleton interactions. Curr Opin Plant Biol. 2009;12:728–34. doi: 10.1016/j.pbi.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Blanchoin L, Boujemaa-Paterski R, Henty JL, Khurana P, Staiger CJ. Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Curr Opin Plant Biol. 2010;13:714–23. doi: 10.1016/j.pbi.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Higaki T, Sano T, Hasezawa S. Actin microfilament dynamics and actin side-binding proteins in plants. Curr Opin Plant Biol. 2007;10:549–56. doi: 10.1016/j.pbi.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner J, Marc J. Arabidopsis thaliana, a plant model organism for the neuronal microtubule cytoskeleton? J Exp Bot. 2011;62:89–97. doi: 10.1093/jxb/erq278. [DOI] [PubMed] [Google Scholar]
- 10.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fimia GM, Kroemer G, Piacentini M. Molecular mechanisms of selective autophagy. Cell Death Differ. 2013;20:1–2. doi: 10.1038/cdd.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reggiori F, Komatsu M, Finley K, Simonsen A. Selective types of autophagy. Int J Cell Biol. 2012;2012:156272. doi: 10.1155/2012/156272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzyńska A, Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011;7:1145–58. doi: 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009;84:431–48. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackeh R, Perdiz D, Lorin S, Codogno P, Poüs C. Autophagy and microtubules - new story, old players. J Cell Sci. 2013;126:1071–80. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 18.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamasaki M, Noda T, Baba M, Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 20.He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–35. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monastyrska I, Shintani T, Klionsky DJ, Reggiori F. Atg11 directs autophagosome cargoes to the PAS along actin cables. Autophagy. 2006;2:119–21. doi: 10.4161/auto.2.2.2298. [DOI] [PubMed] [Google Scholar]
- 22.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–56. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–9. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell. 2008;19:1962–75. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Olsen RW. Binding of the GABA(A) receptor-associated protein (GABARAP) to microtubules and microfilaments suggests involvement of the cytoskeleton in GABARAPGABA(A) receptor interaction. J Neurochem. 2000;75:644–55. doi: 10.1046/j.1471-4159.2000.0750644.x. [DOI] [PubMed] [Google Scholar]
- 27.Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–6. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 28.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–87. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–22. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 30.Kouno T, Mizuguchi M, Tanida I, Ueno T, Kanematsu T, Mori Y, Shinoda H, Hirata M, Kominami E, Kawano K. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J Biol Chem. 2005;280:24610–7. doi: 10.1074/jbc.M413565200. [DOI] [PubMed] [Google Scholar]
- 31.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–7. [PubMed] [Google Scholar]
- 32.Köchl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–45. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 33.Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004;567:302–6. doi: 10.1016/j.febslet.2004.04.088. [DOI] [PubMed] [Google Scholar]
- 34.Marchbank K, Waters S, Roberts RG, Solomon E, Whitehouse CA. MAP1B Interaction with the FW Domain of the Autophagic Receptor Nbr1 Facilitates Its Association to the Microtubule Network. Int J Cell Biol. 2012;2012:208014. doi: 10.1155/2012/208014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zientara-Rytter K, Sirko A. Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front Plant Sci. 2014;5:13. doi: 10.3389/fpls.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D. In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 2004;136:3956–67. doi: 10.1104/pp.104.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–36. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 38.Vanstraelen M, Van Damme D, De Rycke R, Mylle E, Inzé D, Geelen D. Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr Biol. 2006;16:308–14. doi: 10.1016/j.cub.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 39.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 40.Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–7. doi: 10.1016/S0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- 41.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haarer B, Aggeli D, Viggiano S, Burke DJ, Amberg DC. Novel interactions between actin and the proteasome revealed by complex haploinsufficiency. PLoS Genet. 2011;7:e1002288. doi: 10.1371/journal.pgen.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 44.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–8. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]