Abstract
CnAIP2 (Callitropsis nootkatensis ABI3-Interacting Protein 2) was previously identified as a protein that interacts with the yellow-cedar ABI3 protein. CnAIP2 plays important roles during several key transitions of the plant lifecycle and acts as a global regulator with functions opposite to those of ABI3 proteins. Here we report that the CnAIP2 gene promoter is strongly upregulated by all of the major plant hormones. Young Arabidopsis seedlings expressing a chimeric CnAIP2pro-GUS construct were subjected to exogenously applied hormones; the maximum fold-enhancement of GUS activity was as high as 47-fold, and each hormone showed a distinctive cell/tissue-specific pattern of GUS induction. By far the greatest response was elicited by the synthetic auxin 2,4-D (47-fold induction); the other hormones tested stimulated GUS activities by 8- to 21-fold. The CnAIP2 promoter also responded to glucose and salt (NaCl), albeit to a lesser extent (2- to 3-fold induction). As well as acting in an antagonistic way to the global regulator ABI3, CnAIP2 appears to participate in multiple hormonal crosstalk pathways to carry out its functions.
Keywords: ABI3-interacting protein, plant hormones, CnAIP2, yellow-cedar, hormonal crosstalk
Seeds of yellow-cedar (Callitropsis nootkatensis, formerly known as Chamaecyparis nootkatensis) exhibit deep dormancy at maturity and require several months of cool moist conditions to break dormancy. As a high elevation species whose seeds are dispersed in the fall, seed dormancy represents an adaptive trait that prevents germination during the cold winter months. We have isolated the ortholog of the ABI3 gene from yellow-cedar, CnABI3 (Callitropsis nootkatensis ABscisic Acid Insensitive 3). Characterization of the gymnosperm ABI3 in both yellow-cedar, and in heterologous (angiosperm) hosts, shows that the CnABI3 protein has similar roles as its angiosperm counterparts.1-3 As reported in a recent publication, using a yeast 2-hybrid approach,4 we further identified a new protein of yellow-cedar, CnAIP2, that interacts with CnABI3. CnAIP2 plays important roles during several key transitions of the plant lifecycle, and this protein appears to act as a global regulator with functions opposite to those of ABI3 proteins. Four processes or transitions are impacted by CnAIP2: 1) early-to-mid seed development, in which overexpression of CnAIP2 decreases seed viability, 2) lateral root initiation, 3) dormancy to germination, and 4) vegetative growth to reproductive initiation (flowering). Expression of CnAIP2pro in roots and seeds is greatly enhanced by auxin.
Here we report that besides auxin, all of the major plant hormones significantly increased the expression of a GUS reporter gene driven by the CnAIP2 gene promoter. Glucose and salt were able to induce expression by 2- to 3-fold. Multiple hormone induction of gene expression is a unique characteristic of a plant promoter.
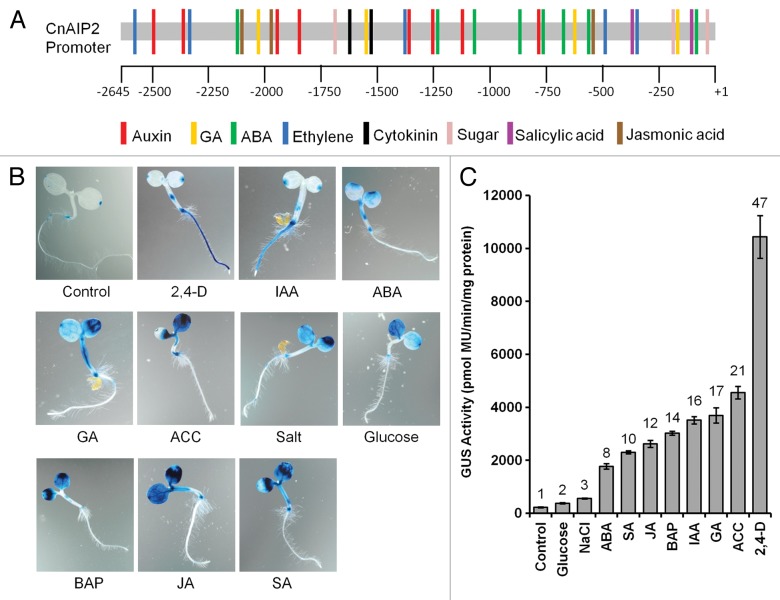
We first conducted analyses to identify putative responsive cis-elements on the 2.6-kb CnAIP2 gene promoter fragment that we had isolated previously4 using the online software PLACE (http://www.dna.affrc.go.jp/PLACE/,5) and PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/6). This uncovered responsive elements to all the major plant hormones including abscisic acid (ABA), gibberellins (GA), auxins, cytokinins, and ethylene, and to the defense-response-hormones, jasmonic acid (JA) and salicylic acid (SA) (Fig. 1A). Sugar-responsive elements were also evident. Although this analysis is only based on sequence similarities to the known responsive elements (some of which are indicated in Figure 1), obviously they may not represent bonafide response elements. Yet clearly there were sufficient numbers of candidate sequences to warrant further testing to determine their authenticity. This is verified below by the hormonal responses when transgenic Arabidopsis seedlings expressing CnAIP2pro-GUS were treated with various hormones.
Figure 1.CnAIP2 promoter putative cis-elements and hormonal responses. (A) Putative cis-elements of the CnAIP2 gene promoter analyzed using the online software PLACE (http://www.dna.affrc.go.jp/PLACE/) and PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Each colored bar represents the approximate position along the promoter of a putative cis-element conferring responsiveness to the indicated hormone. (B) Expression of GUS driven by the CnAIP2 gene promoter as indicated by the GUS histochemical assay using X-gluc as substrate. Staining was conducted over 16 h on 3-d-old seedlings that had been treated for 24 h in a half-strength liquid MS medium containing 5 µM of hormone, 200 mM NaCl, or 6% glucose. The control is shown for 3-d-old seedlings incubated in half-strength MS medium (alone) for 24 h. (C) Enhancement of GUS expression driven by the CnAIP2 gene promoter in seedlings treated as described in (B) as determined by the flurometric activity assay.7 Numbers on bars indicate the fold-increase of GUS activity elicited by each treatment as compared with that of the control (seedlings incubated in half-strength MS medium alone).
In our recent report we showed that the CnAIP2 promoter was strongly upregulated by auxin, especially by the synthetic auxin 2,4-D.4 Here our further investigations indicated that all of the major plant hormones were able to upregulate the expression of this promoter. As shown in Figure 1 (B, C), ABA, GA, the ethylene precursor ACC, BAP (cytokinin), jasmonic acid, and salicylic acid all significantly increased GUS expression driven by the CnAIP2 promoter. The treatments were performed for 24 h on 3-d-old seedlings using 5 µM of each hormone in half-strength liquid Murashige and Skoog (MS) media. For the salt/high sugar treatments, the young seedlings were treated for 24 h with 200 mM NaCl and 6% of glucose, respectively, in half-strength liquid MS media. In the control seedlings incubated in the medium lacking hormone, GUS expression was mostly localized within the roots, especially in the top part of the root adjacent to the hypocotyl, and at sites of lateral root initiation (Fig. 1B;4). The cell-/tissue-characteristics of expression induced or enhanced by the other hormones differed greatly from that induced by the auxin treatments (Fig. 1B). In auxin-treated seedlings, there was a huge increase in GUS expression in roots, while almost no effect was detected in cotyledons (and leaves), except at the very tips (Fig. 1B;4). In sharp contrast, when seedlings were treated with all of the other hormones, there were minimal increases of GUS expression in roots, but significant upregulation in cotyledons (Fig. 1B). We also detected upregulation in the leaves of older seedlings (data not shown).
The GUS expression driven by the CnAIP2 gene promoter was also measured using fluorometric activity assays.7 All of the hormones upregulated GUS expression significantly, with activities increasing by several-fold, as compared with those of control seedlings (Fig. 1C). The synthetic auxin 2,4-D had the strongest effect, enhancing GUS activity by 47-fold. Treatment with high concentrations of salt (200 mM NaCl) and sugar (6% glucose) increased GUS activities by 2- to 3-fold. Although these latter treatments enhanced the GUS activity to a lesser extent than did the hormone treatments, the fold-enhancement (2- to 3-fold) was nonetheless significant. It is unclear if the upregulation was direct, or if these stresses mediated an enhancement of GUS activity through increasing the levels of specific hormones (e.g., ABA).
Our recent report showed that CnAIP2 plays important roles in several key transitions of the plant lifecycle, such as during seed development, flowering, and root development.4 The functions of CnAIP2 appear to be the opposite of those of ABI3/CnABI3. Intriguingly, CnAIP2 expression appears to be greatly affected by all the major hormones, including JA and SA, which play major roles in plant defense responses. Even sugar and salt at high concentrations also increased the expression of CnAIP2. It is evident that CnAIP2 is involved in multiple hormonal crosstalk, and the regulatory characteristics of the CnAIP2 gene promoter are unique in this respect. Crosstalk between plant hormones in controlling a given developmental process is very complex; it may involve 2 hormones, sometimes with antagonistic or synergistic actions, or it may involve simultaneous or sequential interactions between multiple hormones.8 As an example of the former scenario, ABA can have antagonistic interactions with SA and MeJA; yet the effect of simultaneous hormone treatments on gene expression reveals that ABA interacts with SA and MeJA cooperatively as well.9 To respond to ever-changing environmental conditions, plants have developed extraordinary flexibility and adaptability. Plant hormones act in an interconnected complex network of interactions and feedback circuits that determines the final outcome of the individual hormone actions.10 The responses of the CnAIP2 gene promoter to all of the hormones tested indicate that this gene is uniquely positioned at nodes of hormonal crosstalk. Further mechanistic investigations on how CnAIP2 participates in hormonal regulation during the different lifecycle transitions will be of great significance in understanding the functions of both CnAIP2 and CnABI3/ABI3 genes.
Acknowledgments
This research is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant awarded to ARK.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zeng Y, Raimondi N, Kermode AR. Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol Biol. 2003;51:39–49. doi: 10.1023/A:1020762304937. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Y, Kermode AR. A gymnosperm ABI3 gene functions in a severe abscisic acid-insensitive mutant of Arabidopsis (abi3-6) to restore the wild-type phenotype and demonstrates a strong synergistic effect with sugar in the inhibition of post-germinative growth. Plant Mol Biol. 2004;56:731–46. doi: 10.1007/s11103-004-4952-y. [DOI] [PubMed] [Google Scholar]
- 3.Kermode AR, Zeng Y, Hu X, Lauson S, Abrams SR, He X. Ectopic expression of a conifer Abscisic Acid Insensitive3 transcription factor induces high-level synthesis of recombinant human alpha-L-iduronidase in transgenic tobacco leaves. Plant Mol Biol. 2007;63:763–76. doi: 10.1007/s11103-006-9122-y. [DOI] [PubMed] [Google Scholar]
- 4.Zeng Y, Zhao T, Kermode AR. A conifer ABI3-interacting protein plays important roles during key transitions of the plant life cycle. Plant Physiol. 2013;161:179–95. doi: 10.1104/pp.112.206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rombauts S, Déhais P, Van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27:295–6. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–7. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–73. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto M, Tsuboi Y, Goda H, Yoshizumi T, Shimada Y, Hirayama T. Multiple hormone treatment revealed novel cooperative relationships between abscisic acid and biotic stress hormones in cultured cells. Plant Biotechnol. 2012;29:19–34. doi: 10.5511/plantbiotechnology.11.1130a. [DOI] [Google Scholar]
- 10.Vanstraelen M, Benková E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012;28:463–87. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]