Abstract
Arabinogalactan proteins are abundant cell-surface proteoglycans in plants and are involved in many cellular processes including somatic embryogenesis, cell-cell interactions, and cell elongation. We reported a glucuronosyltransferase encoded by Arabidopsis AtGlcAT14A, which catalyzes an addition of glucuronic acid residues to β-1,3- and β-1,6-linked galactans of arabinogalactan (Knoch et al. 2013). The knockout mutant of this gene resulted in the enhanced growth rate of hypocotyls and roots of seedlings, suggesting an involvement of AtGlcAT14A in cell elongation. AtGlcAt14A belongs to the family GT14 in the Carbohydrate Active Enzyme database (CAZy; www.cazy.org), in which a total of 11 proteins, including AtGLCAT14A, are classified from Arabidopsis thaliana. In this paper, we report the enzyme activities for the rest of the Arabidopsis GT14 isoforms, analyzed in the same way as for AtGlcAT14A. Evidently, two other Arabidopsis GT14 isoforms, At5g15050 and At2g37585, also possess the glucuronosyltransferase activity adding glucuronic acid residues to β-1,3- and β-1,6-linked galactans. Therefore, we named At5g15050 and At2g37585 as AtGlcAT14B and AtGlcAT14C, respectively.
Keywords: Arabidopsis thaliana, CAZy GT14, arabinogalactan, glucuronic acid, glucuronosyltransferase, glycosyltransferase, plant cell walls, proteoglycan
Arabinogalactan proteins (AGPs, AG proteins) are abundant proteoglycans on plant cell surfaces, commonly found in many species and involved in many cellular processes.1 AGPs consist of proteins, AG glycans, and some also contain lipids (glycosyl-phosphatidylinositol anchor) of which AG glycans are the major component, often occupying more than 90% of the molecule. The AG glycans are synthesized on proteins by post-translational modifications catalyzed by glycosyltransferases (GTs) in the secretory pathway. AG glycans are highly heterogeneous but commonly consist of β-1,3-linked galactans as the main chains, which are substituted with β-1,6-linked galactan side chains decorated by many arabinose and other minor sugars, such as glucuronic acid (GlcA), rhamnose, and fucose.
We recently reported an Arabidopsis AtGLCAT14A, which possesses GlcA transferase (GlcAT) activity, adding GlcA to β-1,3 and β-1,6-linked galactans of AG glycans.2 GlcA residues in AG side chains may play a role in the binding and release of extracellular calcium,3 and AG GlcAT activities may be important to guarantee GlcA residues in AGPs. AtGlcAT14A belongs to the Carbohydrate Active Enzyme family GT14, and 11 proteins from Arabidopsis thaliana are classified to this family. In this paper, we report redundant activities of AG GlcAT investigated in all 11 Arabidopsis GT14 isoforms. We basically followed the same procedure of heterologous expression of the isoforms in Pichia pastoris and in vitro enzyme assay using UDP-14[C]-GlcA as the donor-substrate described in Knoch et al.2 In the enzyme assay, we used polysaccharide acceptors representative of AG-derived polysaccharides, GAGP8 or β-1,3-galactan.4 GAGP8 acceptor is the microsomes after expression of recombinant AGP having 8 repetitive core motif of gum Arabic (GAGP8)5 in N. benthamiana and it consists of β-1,6-galactans of degree of polymerization from 1 to 8 which are partially decorated with arabinose.4 β-1,3-Galactan is prepared by three times Smith degradation of gum Arabic and it consists of β-1,3-galactan of average 25 kDa with average degree of polymerization of 154.6 We first verified these acceptors in the reaction of AtGLCAT14A.
The recombinant AtGLCAT14A transferred 14[C]-GlcA from UDP-14[C]-GlcA to the GAGP8 acceptor, and the 14[C]-GlcA incorporated products were susceptible to the treatment with endo-β-1,6-galactanase (Fig. 1A),4 which released small oligosaccharides with a degree of polymerization (DP) of 2–3, analyzed by size exclusion chromatography (Fig. 1A). Co-treatment with β-glucuronidase further cleaved 14[C]-GlcA from the above materials with DP2–3 (Fig. 1A). Because endo-β-1,6-galactanase cleaves unsubstituted β-1,6-galactans longer than DP3,7 the results indicate that AtGlcAT14A transferred 14[C]-GlcA to the β-1,6-galactans longer than DP3 in the GAGP8 acceptor.
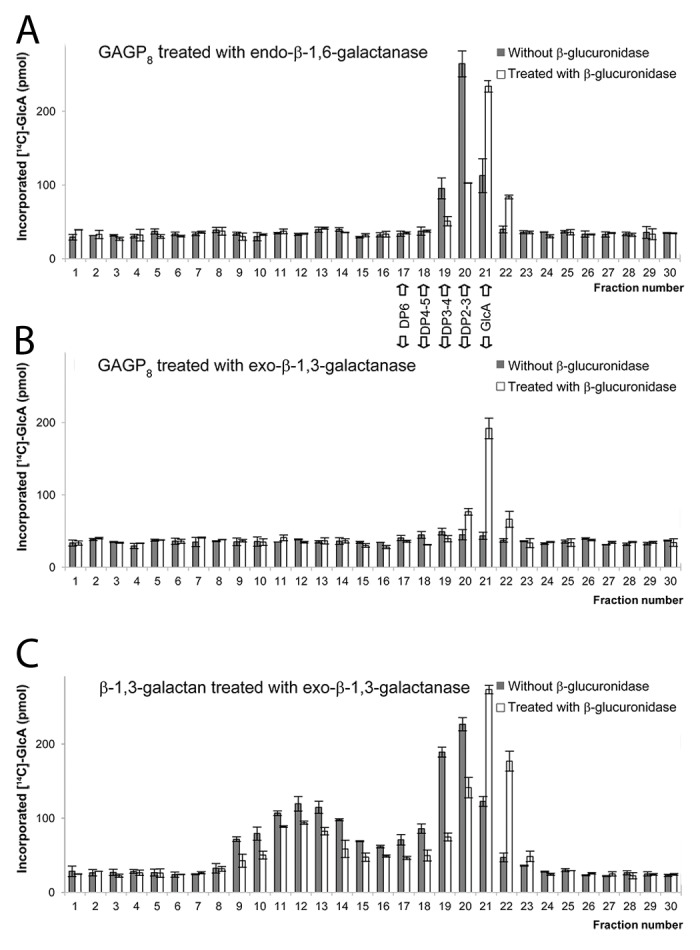
Figure 1. Evaluation of acceptors used in this study. GAGP8 and β-1,3-galactan acceptors were incubated with recombinant AtGLCAT14A and a UDP-14[C]-GlcA donor-substrate. The 14[C]-GlcA incorporated products were treated with specific hydrolases and analyzed by size exclusion chromatography. The [14C]-sugars in the fractions were analyzed by scintillation counting. (A) GAGP8 products treated with endo-β-1,6-galactanase with or without co-treatment with β-glucuronidase; (B) GAGP8 products treated with exo-β-1,3-galactanase with or without co-treatment with β-glucuronidase; (C) products made on β-1,3-galactan treated with exo-β-1,3-galactanase with or without co-treatment with β-glucuronidase. Control sample prepared from Pichia pastoris harboring an empty vector does not produce 14[C]-GlcA incorporated products with these acceptors.
In contrast, 14[C]-GlcA incorporated products transferred to the GAGP8 acceptor were not susceptible to the exo-β-1,3-galactanase treatment (Fig. 1B). Exo-β-1,3-galactanase cleaves β-1,3-linked galactan regardless of the presence or absence of the substituted side chains.8 The incorporation of GlcA into β-1,3-linked galactan should be detected as GlcA-Gal disaccharide after exo-β-1,3-galactanase treatment; however, the treatment did not release detectable oligosaccharides of DP2 (Fig. 1B). Treatment of the 14[C]-GlcA-products using β-glucuronidase released 14[C]-GlcA at a level similar to that released by the treatment of endo-β-1,6-galactanase (compare Fig. 1B and 1A), indicating that the GAGP8 serves primarily β-1,6-galactans, not β-1,3-galactans, as acceptors for AtGLCAT14A activity.
The recombinant AtGLCAT14A also transferred 14[C]-GlcA to the second acceptor, β-1,3-galactan, and the 14[C]-GlcA incorporated products were susceptible to the exo-β-1,3-galactanase treatment, which released various sizes of polysaccharides/oligosaccharides, including DP2 (Fig. 1C). The larger 14[C]-GlcA products (fraction 9–14) are most likely large β-1,3-galactans substituted with 14[C]-GlcA but left from exo-β-1,3-galactanase digestion. Similar incomplete digestion by Phanerochaete chrysosporium exo-β-1,3-galactanase has previously been observed.4 The materials in fraction 20 are likely 14[C]-GlcA-Gal and further degraded to 14[C]-GlcA by β-glucuronidase treatment (Fig. 1C), indicating that this acceptor worked as a β-1,3-galactan acceptor for AtGLCAT14A.
Thus, we used GAGP8 and β-1,3-galactan acceptors as representative of β-1,6- and β-1,3-galactans, respectively, for the enzyme characterization of the Arabidopsis GT14 isoforms.
We expressed soluble catalytic domains with an N-terminal FLAG tag in Pichia pastoris and used immunoprecipitated materials as an enzyme source, as described previously.2 The Western blot analysis revealed various levels of expression among different isoforms; specifically, At4g03340 and At3g24040 were highly expressed; At4g27480, At1g03520, and At5g39990 (AtGlcAT14A) were also expressed; At1g53100, At5g15050, and At2g37585 were barely detected; and At3g03690, At1g71070, and At3g15350 were not detected (Fig. 2A). When we tested GlcAT activity, At5g15050, At2g37585 as well as At5g39990 (AtGLCAT14A) demonstrated transfer of 14[C]-GlcA to both GAGP8 and β-1,3-galactan, whereas other isoforms did not exhibit this activity (Fig. 2B). Based on the result, we named At5g15050 and At2g37585 as AtGlcAT14B and AtGlcAT14C, respectively. Because the expression levels of AtGlcAT14B and AtGlcAT14C were much lower than that of AtGLCAT14A (only approximately 5% of the level of AtGLCAT14A), relative enzyme activity normalized by the protein concentration is 20–30 times higher in AtGlcAT14C compared with AtGLCAT14A and AtGlcAT14B (Fig. 2B, inset).

Figure 2. Western blot analysis of the recombinant proteins and enzyme assays. (A) The recombinant proteins were prepared from a Pichia culture broth by concentration and subsequent collection on agarose conjugated with anti-FLAG antibody as described previously.2 The protein samples collected on the anti-FLAG agarose were subjected to SDS-PAGE and analyzed by Western blot using anti-FLAG antibody. The two images are from the same blot, but the bottom image is a longer exposure after excision of lane 5. Lane 1, At4g27480; lane 2, At4g03340; lane 3, At3g03690; lane 4, At1g71070; lane 5, At3g24040; lane 6, At3g15350; lane 7, At1g03520; lane 8, At1g53100; lane 9, At5g15050; lane 10, At2g37585; lane 11, At5g39990 ( = AtGlcAT14A). A relative protein concentration is estimated as 0.05 for At5g15050 and At2g37585 when the level of AtGLCAT14A is set as 1. (B) Enzyme activities among different Arabidopsis GT14 isoforms. Transferase activity of 14[C]-GlcA was tested using GAGP8 (black bar) and β-1,3-galactan (gray bar) as acceptors. At5g15050 (AtGLCAT14B) and At2g37585 (AtGLCAT14C) in addition to AtGLCAT14A showed GlcAT activity with both acceptors. Specific activity based on a relative protein concentration (AtGlcAT14A as 1, AtGLCAT14B and C as 0.05) is shown in inset.
Because AtGlcAT14B is the closest homolog of AtGlcAt14A (see Fig. 1 in Knoch et al.2), the same GlcAT activity was expected, whereas AtGlcAT14C is rather distantly related to AtGlcAT14A and AtGlcAT14B and therefore its possession of GlcAT activity in spite of several isoforms present in between AtGlcAT14C and AtGlcAT14A/14B was rather unexpected. The transcript-levels for AtGlcAT14A, B, and C are low to medium throughout development of Arabidopsis and they have rather similar expression profiles (Genevestigator: http://www.genevestigator.com/gv/). However, AtGlcAT14A and B (especially B) are very highly expressed in micropylar endosperm and suspensor of embryo at preglobular and globular stages, while AtGlcAT14C is not expressed in those tissues (eFP browser9); AtGlcAT14A is expressed 10–30 fold more than B and C in seeds after imbibition; AtGlcAT4C is expressed specifically in guard cells and pollen.9 In roots, all three isoforms are expressed in xylem, but AtGlcAt14A and B are expressed ~7-fold more than C in the meristematic zone, while AtGlcAT14C is expressed 4–6-fold more than A and B in the root tip.9 The result shown in Figure 2B indicates that AtGlcAT14A and B prefers β-1,6-galactan while AtGlcAT14C prefers β-1,3-galactan as substrate. We do not know whether GlcA substituted β-1,3 and β-1,6-galactans possess different functions during development, but the tissue specific expression of AtGlcAT14A and B (e.g., micropylar endosperm and suspensor) vs. C (e.g., guard cells and pollen) may represent a specific function of GlcA substituted β-1,6- and β-1,3-galactans in the respective tissues.
Altogether, we report AG GlcAT activity for AtGlcAT14B and AtGlcAT14C in addition to AtGlcAT14A2 within the Arabidopsis isoforms classified to the CAZy GT14 family. All three recombinant enzymes possess the same GlcAT activity, transferring GlcA to β-1,3 and β-1,6-galactans of AG; therefore, these most likely serve as redundant activities in plants.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by funding provided by the Danish Agency for Science, Technology and Innovation’s Strategic Research for Health, Food and Welfare (DSF:09-067059) and Technology and Production (FTP:274-09-0113) programs to NG.
References
- 1.Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2007;58:137–61. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- 2.Knoch E, Dilokpimol A, Tryfona T, Poulsen CP, Xiong G, Harholt J, Petersen BL, Ulvskov P, Hadi MZ, Kotake T, et al. A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 2013;76:1016–29. doi: 10.1111/tpj.12353. [DOI] [PubMed] [Google Scholar]
- 3.Lamport DT, Várnai P. Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 2013;197:58–64. doi: 10.1111/nph.12005. [DOI] [PubMed] [Google Scholar]
- 4.Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T, et al. A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 2013;76:128–37. doi: 10.1111/tpj.12281. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Shpak E, Gu T, Moo-Young M, Kieliszewski M. Production of recombinant plant gum with tobacco cell culture in bioreactor and gum characterization. Biotechnol Bioeng. 2005;90:578–88. doi: 10.1002/bit.20441. [DOI] [PubMed] [Google Scholar]
- 6.Sekimata M, Ogura K, Tsumuraya Y, Hashimoto Y, Yamamoto S. A beta-Galactosidase from Radish (Raphanus sativus L.) Seeds. Plant Physiol. 1989;90:567–74. doi: 10.1104/pp.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichinose H, Kotake T, Tsumuraya Y, Kaneko S. Characterization of an endo-beta-1,6-Galactanase from Streptomyces avermitilis NBRC14893. Appl Environ Microbiol. 2008;74:2379–83. doi: 10.1128/AEM.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichinose H, Yoshida M, Kotake T, Kuno A, Igarashi K, Tsumuraya Y, Samejima M, Hirabayashi J, Kobayashi H, Kaneko S. An exo-beta-1,3-galactanase having a novel beta-1,3-galactan-binding module from Phanerochaete chrysosporium. J Biol Chem. 2005;280:25820–9. doi: 10.1074/jbc.M501024200. [DOI] [PubMed] [Google Scholar]
- 9.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


