Abstract
Virus-induced gene complementation (VIGC), a plant virus technology based on Potato virus X for transient overexpression of endogenous genes complemented tomato mutants, resulting in non-ripening fruits to ripen. This efficient “gain-of-function” approach involves no stable transformation, and reveals a fruit-specific transcriptional network that may exist among key transcription factors in modulating tomato ripening. Thus, VIGC represents a novel and feasible strategy for gene functional analysis in plants.
Keywords: plant virus technology, functional complementation, VIGC, ripening inhibitor, colourless non-ripening, Potato virus X
Plant virus technology including small interfering RNA-mediated virus induced transcriptional and posttranscriptional gene silencing (Sir VIGS), microRNA-mediated VIGS (Mr VIGS), virus-based RNA mobility assays, and virus-based ectopic gene expression, is an important forward and reverse genetic approach to delineate gene function in plants. In particular, Sir VIGS has been widely used in many plant species, owing to its capability of effective and rapid gene “knockdown.”1,2 A drawback for Sir VIGS is the off-target silencing, and it can cause nonspecific mRNA degradation or translation repression.3-5 This disadvantage can be overcome to a certain degree by Mr VIGS in which only one known mature microRNA can be expressed from a specifically designed microRNA precursor by a virus vector, leading to highly specific silencing of target genes.6 On the other hand, solid evidence for illuminating biological significance of a given gene often involves “gain-of-function” analysis, especially phenotypic complementation in mutant background. This is usually performed through labor-intensive and time-consuming stable plant transformation.
However, considerable progress has been made to test the feasibility of plant virus technology as a gene overexpression system. For instance, expression of heterologous genes from recombinant viral vectors results in a high level of foreign proteins in plants.7-9 This implies that a viral transient expression system could be applied to express endogenous genes for rapid induction of specific phenotypes and phenotypic complementation, thus as a “gain of function” tool to define plant gene functions and bridge knowledge gaps between genotypes and phenotypes.
To exploit whether virus technology could be indeed used in a “gain-of-function” assay, we have developed a virus-induced gene complementation (VIGC) system from a modified Potato virus X (PVX) vector. Viral ectopic overexpression of the MADS-box transcription factor (TF) gene SlMADS-RIN, involving no stable tomato transformation, was able to complement non-ripening phenotype in tomato Ripening inhibitor (rin) mutant.10 To further test the usefulness of the VIGC technique, we expressed the SBP-box SlSPL-CNR TF in the tomato Colorless non-ripening (Cnr) mutant fruit, and found that viral delivery of a functional SlSPL-CNR gene caused complementation of the Cnr mutant (Fig. 1). Moreover, comparative gene expression analysis of ripening and non-ripening tissues at the precisely equivalent growing stage collected from same individual fruits, revealed that SlMADS-RIN is a master TF in a transcriptional network in modulating tomato fruit ripening.10 This model is supported by the fact that SlMADS-RIN directly interacts with promoters of several key ripening-associated TFs.11,12
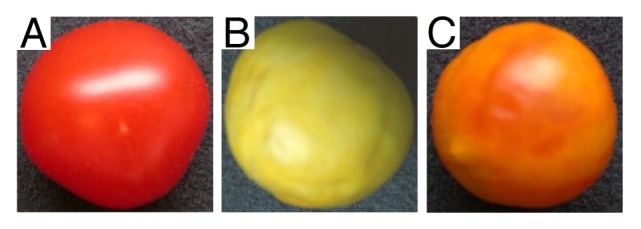
Figure 1. VIGC-mediated complementation of ripening in Cnr mutant fruit. (A) Wild-type tomato (Solanum lycopersicum Ailsa Craig) ripened normally. (B) Cnr mutant fruit in Ailsa Craig background remained colorless non-ripening. (C) Complementation of ripening occurred in Cnr mutant fruit treated with a VIGC vector PVX carrying a functional SlSPL-CNR gene.
Taken together, our results indicate that the efficient VIGC technology represents a novel strategy to reveal gene functions in tomato. Considering the availability of a wild range of plant virus-based gene expression vectors, VIGC technique based on different plant viruses can be developed for functional genomics in diverse plant species including many economically important crops.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by an innovative R&D grant from Hangzhou Normal University, a grant from National Natural Science Foundation of China (NSFC31370180), and a BBSRC-Warwick HRI core grant (BBS/E/H/00YH0271) to Hong Y, and grants (LQ13C020004, LQ13C060003, LQ12C02005) from the Natural Science Foundation of Zhejiang province, China.
References
- 1.Becker A, Lange M. VIGS--genomics goes functional. Trends Plant Sci. 2010;15:1–4. doi: 10.1016/j.tplants.2009.09.002. http://dx.dor.org/10.1016/j.tplants.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 2.Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16:656–65. doi: 10.1016/j.tplants.2011.08.006. http://dx.dor.org/ 10.1016/j.tplants.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 3.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. http://dx.dor.org/ doi:10.1038/nbt831 [DOI] [PubMed] [Google Scholar]
- 4.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–4. doi: 10.1016/j.tig.2004.08.006. http://dx.dor.org/ 10.1016/j.tig.2004.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 2006;142:429–40. doi: 10.1104/pp.106.083295. http://dx.dor.org/10.1104/pp.106.083295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y. Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol. 2010;153:632–41. doi: 10.1104/pp.110.155796. http://dx.dor.org/10.1104/pp.110.155796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takamatsu N, Ishikawa M, Meshi T, Okada Y. Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants mediated by TMV-RNA. EMBO J. 1987;6:307–11. doi: 10.1002/j.1460-2075.1987.tb04755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donson J, Kearney CM, Hilf ME, Dawson WO. Systemic expression of a bacterial gene by a tobacco mosaic virus-based vector. Proc Natl Acad Sci U S A. 1991;88:7204–8. doi: 10.1073/pnas.88.16.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman S, Kavanagh T, Baulcombe D. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–57. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou T, Zhang H, Lai T, Qin C, Shi N, Jin M, Zhong S, Fan Z, Liu Y, Wu Z, et al. Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening. Sci Rep. 2012;2:836. doi: 10.1038/srep00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011;157:1568–79. doi: 10.1104/pp.111.181107. http://dx.dor.org/ 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, Ito Y. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta. 2012;235:1107–22. doi: 10.1007/s00425-011-1561-2. http://dx.dor.org/ 10.1007/s00425-011-1561-2 [DOI] [PubMed] [Google Scholar]


