Abstract
The plant Glutamate-Like Receptors (GLRs) are homologs of animal ionotropic glutamate receptors (iGluRs), and are hypothesized to be potential amino acid sensors in plants. Genetic studies of proteins from this family implicate individual GLRs in a diversity of physiological roles in plants. Recently, amino-acid gated channel activities have been proven for a few plant GLRs, suggesting that at least some of the functional mechanisms are conserved between plant GLRs and animal iGluRs. Animal iGluRs generally form heterotetramers, and the ligand-binding specificity and channel functionality is determined by interaction between the subunits. In order to investigate whether plant GLRs interact with each other, a modified yeast-2-hybrid system (mbSUS) approach was taken on 15 of the 20 Arabidopsis GLRs to identify potential interaction partners. Using this approach, we have successfully identified GLR subunits that are capable of interacting with multiple other GLRs. Unlike iGluRs, sequence similarity between the subunit was not correlated with the likelihood of interaction among 2 given subunits. Interactions between selected GLRs (GLR1.1, 2.9, 3.2, and 3.4) were further tested in another heterologous expression system, mammalian HEK293 cells, using Förster resonance energy transfer (FRET). Two separate approaches (sensitized FRET and acceptor photobleaching) indicated that GLRs 1.1 and 3.4 are capable of forming homomers, whereas other combinations did not result in detectable FRET between the subunits.
Keywords: glutamate receptor, glutamate, ligand-gated channel, amino acids, membrane protein, calcium ion, protein-protein interaction
Introduction
Due to their sessile nature, plants need to sense the availability of minerals to modulate development and maximize fitness. Nitrogen (N) is quantitatively the most important mineral nutrient, and its availability has a profound short- and long-term effect on plant growth and development.1 For most plants, nitrate and ammonia are the primary sources of available N, which are converted to amino acids. Amino acids are the predominant form of transported organic N in the plant and are thus considered to be the N currency of the plant body. Considering the critical roles amino acids play in N assimilation and transport, it is not surprising that the change in amino acid levels induce a rapid change in transcriptome and in the activities of key enzymes in the N assimilation pathway.2,3 In fact, amino acids have long been suggested as signals that regulate genes involved in N transport and metabolism in plants, but the exact mechanism of amino acid sensing in plants is not well understood.4,5
Some mechanisms for amino acid sensing have been documented in bacteria, yeast, and mammals, and potential homologs of such amino acid sensors from other organisms do exist in plants. One example is Glb1, an Arabidopsis homolog of bacterial PII proteins that are considered N “master regulators,”6-9 although the role of Glb1 seems to be constrained compared with the bacterial counterparts.10 Likewise, Arabidopsis homolog of Target of Rapamycin (TOR) plays a role in integrating nutrition signals including nitrogen in a similar manner as its yeast and animal counterparts.11 Lastly, the plant homologs of amino acid gated channels from mammals has recently been established as amino acid gated channels,12,13 representing a potential mechanism for amino acid sensing in plants.
Plant GLRs exist in all tracheophytes (vascular plants) and bryophytes (non-vascular plants) sequenced so far. Phylogenetic analyses have revealed that plant GLRs have diverged from metazoan and bacterial iGluRs, which probably happened as early as the last universal common ancestor.14,15 Despite a large evolutionary distance, plant GLRs and mammalian iGluRs share structural similarities with regards to the signature “3 + 1” trans- membrane domains M1 to M4 as well as the ligand binding domain (LBD) and N-terminal domain (ATD), which show high amino acid sequence identity (63– 16%), particularly with the M3 domain (63%) of animal NMDA receptor iGluRs. In animal glutamate receptors, N-terminal domain (ATD) and ligand binding domain (LBD) are responsible for interdomain interaction and ligand-gated conformational change, respectively, and the composition of subunits determines the channel properties.16 The predicted membrane topology and orientation of the protein as a tetramer, with the ATD and LBD exposed to the external side of the membrane, are considered to be conserved in plant GLRs.17
Understanding the function of plant GLRs has previously been hampered by gene redundancy and toxicity although genetic, pharmacological, and electrophysiological approaches suggested diverse physiological roles such as carbon/nitrogen balance regulation,18,19 stomatal opening,20 pollen tube growth,21 plant-pathogen interaction,22-24 responses to wound,25,26 and lateral root formation.12 Recently, 2 of the Arabidopsis GLRs, AtGLR3.4 and 1.4, were shown to function as amino acid gated channels when expressed in heterologous systems,13,27 further confirming the functional commonality of plant and mammalian glutamate receptors.
So far, most approaches to investigate the roles of GLRs have been centered on understanding the function of one protein through genetic or electrophysiological approaches. We hypothesize that formation of homo- or hetero-tetramers would be required for the formation of the ion-conducting pore (therefore for full function) of plant GLRs, similar to their animal and bacterial counterparts. This level of organization may result in modulation of sensing capabilities in response to multiple ligands, stress, or developmental situations in which plant GLRs may play a role. Recently, it was reported that AtGLRs 3.2 and 3.4 are capable of physically interacting with one another, and to a lesser extent, with themselves and AtGLR3.3 in mammalian HEK293 cells as well as Nicotiana benthamiana cells.12 Despite this, a comprehensive study of inter-domain interaction between plant GLRs has not been reported so far. Such information will allow us to develop hypotheses about possible composition of the channels in a given cell, with particular focus on importance in signaling in different plant tissues and during development.
In this study, we studied 2-way interactions between 15 different GLR subunits from Arabidopsis. A modified yeast 2-hybrid approach was performed to identify potential interactors in yeast. In this heterologous expression system, members of all 3 clades (AtGLR1, 2, and 3s) interact with each other, which is in contrast with the mammalian iGluRs which only interact with subunits within their own clade. Putative interactors were then confirmed using Förster resonance energy transfer (FRET) in mammalian HEK293. Data suggests that AtGLR1.1 and 3.4 are capable of forming homomeric channels.
Results
Split ubiquitin system
A yeast mating-based split ubiquitin system (mbSUS) was used to allow for systematic analyses of GLR protein interactions.28 Unlike standard yeast 2-hybrid, the method does not require the fusion proteins to travel to the nucleus, therefore is suitable in detecting interactions between membrane proteins.29,30 Fifteen out of 20 GLR cDNAs were successfully isolated, and fused to either a modified N-terminal (NubG) or C-terminal (Cub) half of ubiquitin. Both interaction-dependent auxotrophy and enzymatic activity assays were used to evaluate the interactions between the subunits.
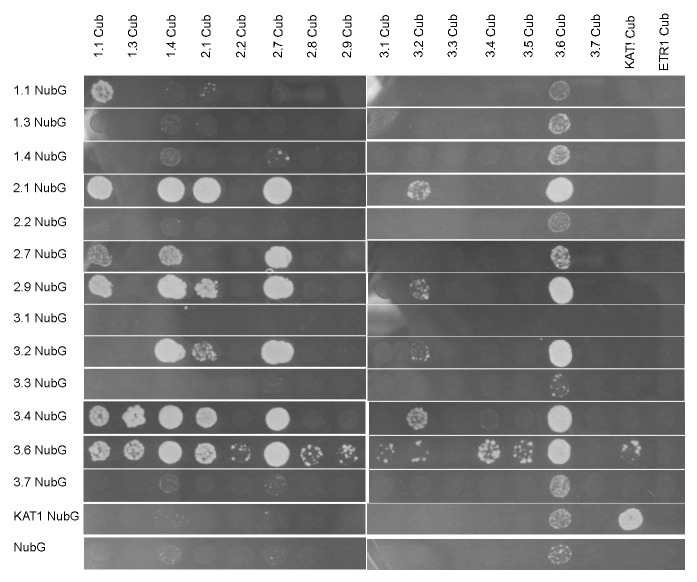
Mating-based SUS of 15 AtGLRs (listed in Table S1) revealed both putative homo- and hetero- GLR subunit interactions. At least 3 independent experiments were performed for each combination (the results are summarized in Table S1). One representative experiment of mbSUS is shown in Figure 1. Homomeric interactions were consistently observed for GLR1.1NubG/GLR1.1Cub and GLR2.1NubG/GLR2.1Cub (Table S1). Interestingly, homomeric interaction for GLR1.4NubG/GLR1.4Cub and GLR3.4NubG/GLR3.4Cub, the subunits capable of acting as homomeric channels in heterologous expression systems,13,27 were not among the strongest interactions observed (Fig. 1; Table S1). Heteromeric interactions were also observed between a number of subunits (Fig. 1; Table S1). In many cases reciprocal interaction (the same interaction partners with the half-ubiquitin tags swapped) was not observed.
Figure 1. Potential GLR interactors identified using mbSUS in yeast. Representation of one mbSUS experiment demonstrating a number of putative interactors, where the growth on interaction-selective media indicates a potential interaction. KAT1 serves as a positive control for homomeric interaction of membrane protein and ETR1 serves as a negative control.
Our experiments identified several subunits (1.1, 2.1, 2.9, 3.2, and 3.4) that consistently interact with multiple other subunits. Both NubG- and Cub- tagged GLR3.6 proteins were capable of auto-activating the transcription of reporters, hence the results were disregarded. None of the other GLR-NubG/Cub fusions interacted with the negative controls NubG or KAT1-NubG, nor promoted growth of yeast on its own (Fig. 1). These results were supported using β-galactosidase assays to confirm activity of the LacZ reporter gene in interacting yeast (Fig. 2 and data not shown).
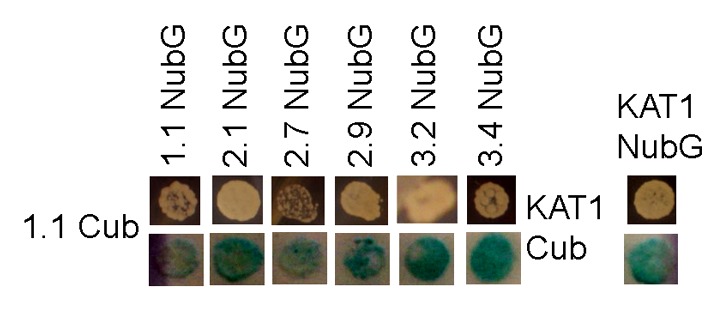
Figure 2. GLR 1.1 interacting partners identified from mbSUS. Yeast growth on auxotrophy-selective media indicates protein-protein interaction. Blue coloring indicates activation of β-galactosidase reporter gene and interaction between proteins. KAT1 serves as a positive control for membrane protein homomer formation.
Intra-molecular Förster Resonance Energy Transfer (FRET)
Intra-molecular Förster Resonance Energy Transfer (FRET) was used to analyze interactions between selected GLR subunits found through mbSUS using human epithelial kidney (HEK293) cell culture.31 Two techniques of FRET measurement were applied for determining the FRET efficiency between co-localizing subunits: sensitized emission FRET32 and acceptor photobleaching.31
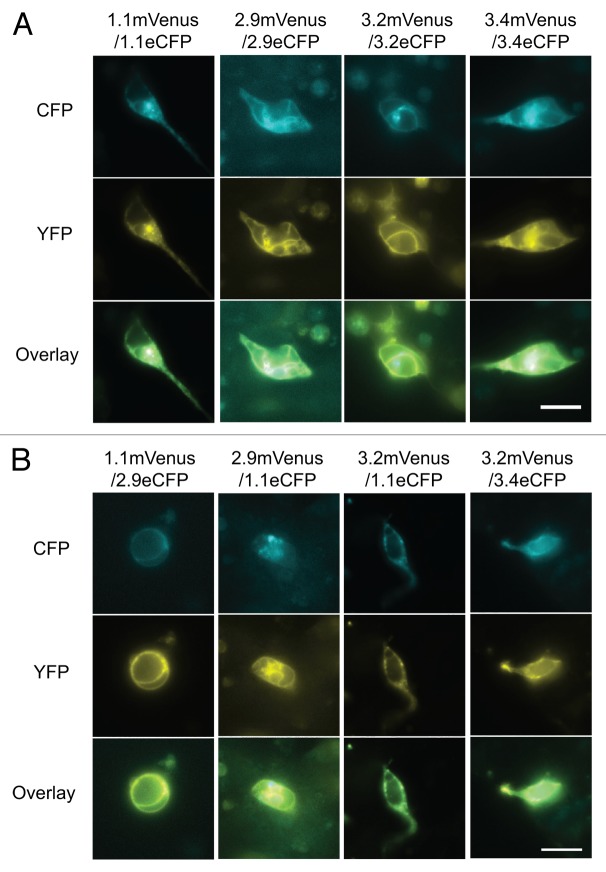
Among the GLRs that interacted with multiple other GLRs in the mbSUS experiments, AtGLRs 1.1, 2.9, 3.2, and 3.4 were successfully expressed in mammalian HEK293 cells. Each construct was fused to either monomeric Venus (mVenus)33 or enhanced CFP (ECFP, Clontech Laboratories), and FRET efficiency between the 2 subunits were measured using either epifluorescent or confocal microscopy. The cells singly expressing eCFP- or mVenus-tagged GLR subunits are shown in Figure 3. All of the constructs localized mainly to the plasma membrane, although in many cases internal membranes were also visible. When eCFP- and mVenus- tagged subunits are co-transfected into HEK293 cells, many combinations resulted in co-localization on the plasma membrane as expected (Fig. 4). Co-expression of 3.2eCFP with other subunits tagged with mVenus, except for 3.2mVenus, resulted in minimal or no cells co-expressing both fluorescent tags (Fig. S1 and data not shown). Likewise, co-expression of 2.9mVenus/3.4eCFP in single cell was not observed consistently (data not shown).
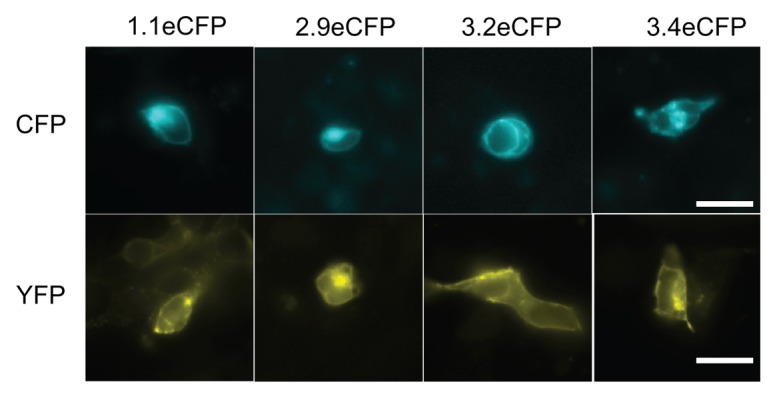
Figure 3. Expression of individual GLR-fluorescent protein fusions in HEK293 cells. Expression of GLR potential subunits identified from mbSUS C-terminally fused with eCFP (top panels) or mVenus (bottom pannels) in mammalian HEK293. Scale bar: 20µm.
Figure 4. Co-expression of GLR-fluorescent protein fusions in HEK293 cells. (A) Homomeric and (B) heteromeric co-expression of GLR subunits that are C-terminally fused with eCFP or mVenus in mammalian HEK293. Scale bar: 20µm.
Next, FRET efficiencies between eCFP- and mVenus-tagged GLR subunits co-expressed in HEK293 were examined. Combinations that did not produce consistent double-labeling of the cells (1.1mVenus/3.2eCFP, 2.9mVenus/3.2eCFP, 3.4mVenus/3.2eCFP, 2.9 mVenus/3.4eCFP) were excluded from this study. FRET efficiencies were successfully determined for all 12 combinations. Among the combinations tested, homomeric interactions were identified between GLRs 1.1 and 3.4 (Fig. 5). However, no other homomeric or heteromeric combinations resulted in detectable FRET, regardless of apparent co-localization on the plasma membrane (Fig. 3B). Homomeric interaction of GLR1.1 and 3.4 were further confirmed using a YFP acceptor photobleaching strategy (Fig. 6). Both of the homomeric interactions produced positive FRET values with 77.8 ± 30.0 and 47.6 ± 29.5 for GLR1.1 and 26.2 ± 16.1 and 40.4 ± 23.9 for GLR3.4 using sensitized emission and YFP acceptor photobleaching, respectively, whereas no detectable FRET was observed with GLR1.1/3.4 combinations. From these results, we concluded that GLR1.1 and 3.4 are capable of forming monomeric channels.
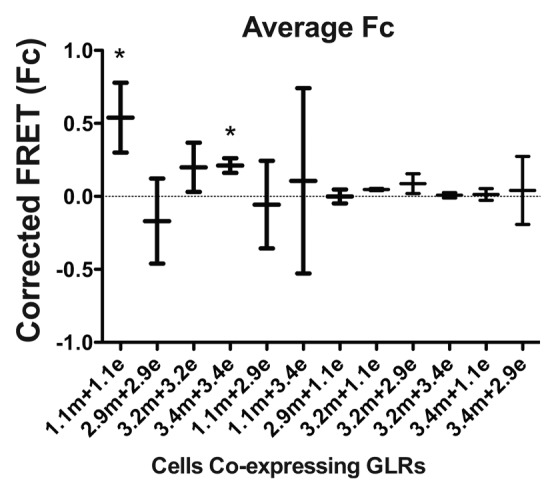
Figure 5. Corrected FRET values of interactors identified from mbSUS. Corrected FRET (sRET) values were obtained from HEK cells co-expressing 2 GLR subunits. Error bars indicate standard deviation. Combinations with statistically significant positive values are indicated with asterisks.
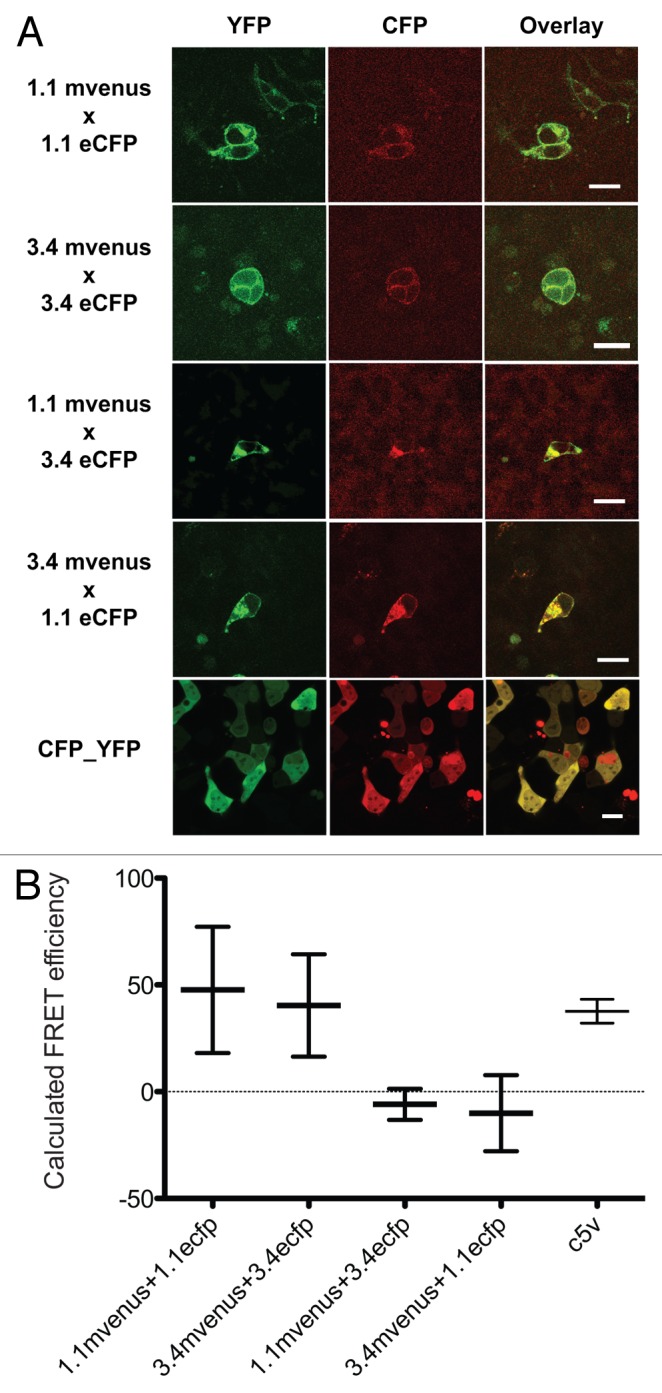
Figure 6. Acceptor photobleaching FRET between GLR1.1 and 3.4 subunits. (A) Confocal images of HEK293 cells co-expressing different combinations of GLR1.1 and 3.4. Cells shown in the bottom panels are expressing positive control for FRET, C5V (Chen et al., 2007). (B) FRET efficiencies calculated using acceptor photobleaching techinique. Values are obtained from at least 6 independent cells. Error bars indicate standard deviation.
Discussion
Understanding the stoichiometry and assembly of plant GLR subunits can give insight into the overall function of the plant glutamate receptor homologs. Systematic analysis of 15 GLR cDNA clones in yeast revealed both homo- and hetero-meric GLR subunit interactions (Fig. 1). These interactions were also confirmed by using β-galactosidase assays (Fig. 2). Interestingly, interaction between the subunits did not correlate with the similarity between the 2 sequences. Rather, the studies identified several subunits (GLRs 1.1, 2.1, 2.9, 3.2, and 3.4) that more frequently interacted with other subunits. In contrast, animal glutamate receptors form homo-/hetero-tetramers only within their own group (e.g., NMDA receptors only interacts with subunits that also belong to the NMDA family).34 In animals, inter-subunit interaction is mainly determined by the N-terminal domains.35,36 Although the N-terminal domains are conserved in plant glutamate receptors, the mechanism through which the subunit composition is determined can be different in plants.
Among the subunits identified through yeast mbSUS assay, interactions among GLRs 1.1, 2.9, 3.2, and 3.4 were further studied using heterologous expression in mammalian cell line HEK293. Two separate approaches using Förster Resonance Energy Transfer (FRET) identified homomeric interactions for GLR1.1 and GLR3.4. GLR3.4 has been shown to function as a Ca2+-conducting channel when expressed in HEK293 cells in a previous work,27 corroborating the finding in this study. In contrast to the results published from the same group, however, we did not observe consistent interaction between GLR3.2 and GLR3.4.12 The difference could be due to a subtle variation in the constructs used, since FRET is sensitive to both the distance and relative dipole orientations of the fluorophores, and the linker regions between the subunit and the fluorescent proteins could have a large effect on FRET efficiency.37 Therefore, the results need to be interpreted with caution; the lack of apparent energy transfer does not exclude the inter-subunit interactions, nor can FRET efficiency be used as a measure of the strength of interactions. Both of the techniques used in this study employed heterologous expression systems. Ideally another approach such as co-immunoprecipitation should be applied to examine the interactions in plant cells. However, we were not successful in expressing epitope-tagged GLRs in plant tissue at the level necessary for such experiments (data not shown), potentially due to the toxicity of GLRs when overexpressed in plant cells. Therefore, further examination of individual interactions might require other approaches such as electrophysiological studies of cells expressing multiple subunits, or examining genetic interactions.12
In conclusion, our study identified multiple potentially interacting AtGLRs in heterologous expression systems. Utilization of high-resolution transcriptome and proteome data38-40 can be used to further examine co-expression of GLR subunits. Such analyses along with this data will guide identification of potential GLR subunits that could comprise a channel in a given cell type.
Methods and Materials
Split ubiquitin system
GLR cDNA clones 1.1 (AT3G04110), 1.3 (AT5G48410), 2.2 (AT2G24720), 2.9 (AT2G29100), 3.2 (AT4G35290), 3.3 (AT1G42540), 3.4 (AT1G05200), and 3.7 (AT2G32400) were isolated through a high-throughput membrane protein interactome project (NSF 0618402, http://www.associomics.org/index.php) and were amplified from Arabidopsis cDNA and cloned into either pDONR221 (GLR1.1, 2.2, and 3.7) or pCR8/GW/TOPO (GLR1.3, 2.2, 3.2, 3.3, 3.4, and 3.7) (courtesy of Dr Wolf Frommer, Carnegie Institution). GLRs 1.4 (AT3G07520), 2.1 (AT5G27100), 2.7 (AT2G29120), 3.1 (AT2G17260), and 3.6 (AT3G51480) genes were successfully amplified using reverse transcriptase PCR from Arabidopsis thaliana ecotype Columbia leaf-derived cDNA and cloned into pENTR1A vector (Invitrogen), using the primers listed in Table S2. GLR3.5 (AT2G32390) was amplified from a pENTR clone generously provided by Dr June Kwak (University of Maryland). Multiple attempts to amplify GLRs 1.2 (AT5G48400), 2.3 (AT2G24710), 2.4 (AT4G31710), 2.5 (AT5G11210), 2.6 (AT5G11180), and 2.8 (AT2G29110) were unsuccessful. A series of controls including WT Nub, KAT1-NubG, KAT1-Cub, KEA3-Cub, NIP7.1-Cub, and ETR1-Cub were kindly provided by Dr Guillaume Pilot (Virginia Tech).
GLR cDNA entry clones were mobilized into the vectors pXN22_GW and pMetYC_GW, carrying NubG and Cub sequences respectively,41 using GATEWAY cloning system. The resulting fusion constructs in pXN22_GW and pMetYC_GW were transformed into yeast mating strain THY.AP5 (MATα leu2–3, 112 trp1–289 his3-Δ1 loxP::ade2 URA3) and THY.AP4 (MATa leu2–3, 112 ura3–52 lexA::HIS3 lexA::ADE2 lexA::lacZ::trp1) respectively using a modified lithium acetate method.42
Transformants of AP4 and AP5 expressing Cub and Nub constructs, respectively, were mated by mixing 10µL of each cell suspension in micro-titer plates then spotted on YPAD media and allowed to grow at 28 °C. Once visible growth was observed (overnight to 20 h) cells were scraped and re-suspended in water, and each suspension was spotted on SC –leu –trp +ade +his +met for selection of mated yeasts. Interaction between Nub and Cub fusion proteins were observed by growing diploids on SC –leu –trp –ade –his, and growth of yeast was recorded as a positive indicator of interaction between Nub and Cub fusion constructs. In order to exclude growth effect independent of the interaction between the 2 constructs, cell suspensions were spotted on SC –leu –trp –ade –his +met (125µM and 250µM) to increase the repression of the Cub expression cassette, which is repressed by methionine. In addition, Arabidopsis KAT1 NubG/Cub and ETR1/Cub fusions served as positive and negative controls for interaction.43
β-Galactosidase assay in yeast
β-galactosidase activity of yeast interactors was measured following the manufacturer’s protocol (Clontech, manual no. PT3024–1) to confirm interaction between NubG and Cub GLR fusion constructs. Briefly, diploid yeast were spotted onto Hybond C-Extra nitrocellulose membrane (Amersham Biosciences) atop SC –leu –trp –ade –his agar media and grown for 2 d at 28 °C. The nitrocellulose sheet was freeze-thawed 3 times in liquid nitrogen and warmed to room temperature to lyse the cells. Β-galactosidase activity was monitored by placing the nitrocellulose membrane on Whatman 3M filter paper soaked with Z-buffer (100 mM Na2HPO4, pH 7, 10 mM KCl, 1 mM MgSO4, and 5 mM β-mercaptoethanol) and incubated at 37 °C until blue color developed.
Transfection of HEK293 cells
GLR 1.1, 2.9, 3.2, and 3.4 cDNA clones were restriction digested with SalI and SmaI and ligated to SalI/AfeI site of pENTR-mVenus or pENTR-ECFP (Altaf Ahmad, unpublished results) to produce entry clones carrying either mVenus or ECFP at the C-termini. The mVenus and ECFP fusion constructs were then Gateway recombined into the mammalian expression vector pCDNA 3.2 V5 DEST (Invitrogen). HEK293 cells were grown to 80% confluency in Dulbecco’s modified eagle medium containing 10% cosmic calf serum and 100U penicillin/streptomycin (DMEM, HyClone, ThermoScientific), trypsinated and split into glass-bottom 8-well chamber (Lab-Tek). At 50% confluency, cells were transfected using Opti-MEM I Reduced serum media (Invitrogen) containing 1µL Lipofectamine 2000 (Invitrogen) and 400ng of plasmid and allowed to transfect 24 h and then exchanged back to DMEM media for one day prior to imaging, cells approximately 80–100% confluency.
Sensitized emission FRET using wide-field microscopy
Cells were imaged using an epi-fluorescence microscope (IX81, Olympus) with appropriate filters for excitation: YFP 500/20 nm and CFP 430/24 nm; dichroic beam splitter: YFP 535/30 nm and CFP 470/24 nm; and emission: YFP 535/30 nm and CFP 470/24 nm (Chroma). Images were taken with a microscope equipped with a 40x oil-immersion objective and a Lamda-XL filter wheel (Sutter) that allows for rapid exchange of filters and captured with an EMCCD Rolera-MGI FAST 1394 monochromatic camera (Olympus). Regions of cells were selected and the pixels quantified for each channel using the appropriate software (Slidebook, 3I) and then corrected for spectral cross-talk and relative abundance of fluorophores to give a normalized FRET efficiency, NFRET, using the following equation:44
| (Equation | 1) |
Where IFRET, ICFP, and IYFP are the intensities under FRET, CFP, and YFP filter sets, respectively, in each region of interest (ROI), and a and b are the normalization of percentage of CFP- or YFP-bleed-through, respectively, under the FRET filter set. The normalizations for a and b varied between experiments and thus were calculated for each individual experiment by analyzing images of cells expressing only CFP or YFP and quantifying the relative intensity ratio under the FRET/CFP or FRET/YFP filter sets.45
YFP acceptor photobleaching using confocal microscopy
Cells were imaged on a Zeiss LSM510 confocal microscope using a plan-apo 63x oil immersion objective. CFP and YFP fluorophores were excited using 458 nm and 514 nm light, respectively. Colocalization was determined by using the RG2B colocalization plugin with autothresholding (Christopher Mauer, Northwestern University) in Image J software (NIH). For FRET the cells were imaged and bleached for 16 iterations of 100% 514 nm laser intensity on selected ROIs to 30% or less of initial YFP intensity. CFP fluorescence intensity was determined using fluorescence intensity (FI) NIH Image J software. FRET efficiency was calculated as follows:
| (Equation | 2) |
Where FRET efficiency, EFRET, is determine by correcting fluorescence intensity for the donor (YFP) after YFP bleaching for nonspecific bleaching using an ROI outside of the bleached area to determine amount of fluorescence change over time. A minimum of 16 cells were analyzed for each construct from at least 2 separate transfections.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work in this publication was supported by Jeffress Memorial Trust (J-908).
References
- 1.Marschner H. Functions of Mineral Nutrients: Macronutrients. In: Mineral Nutrition of Higher Plants. San Diego CA: Academic Press Inc, 1997:231-54. [Google Scholar]
- 2.Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci U S A. 2008;105:4939–44. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz C, Mueller C, Matt P, Feil R, Stitt M. Impact of the C-N status on the amino acid profile in tobacco source leaves. Plant Cell Environ. 2006;29:2055–76. doi: 10.1111/j.1365-3040.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 4.Vincentz M, Moureaux T, Leydecker MT, Vaucheret H, Caboche M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 1993;3:315–24. doi: 10.1111/j.1365-313X.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 5.Forde BG, Lea PJ. Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot. 2007;58:2339–58. doi: 10.1093/jxb/erm121. [DOI] [PubMed] [Google Scholar]
- 6.Arcondéguy T, Jack R, Merrick M. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutts G, Thomas G, Blakey D, Merrick M. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 2002;21:536–45. doi: 10.1093/emboj/21.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev. 2004;28:319–33. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Osanai T, Tanaka K. Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria. Plant Cell Physiol. 2007;48:908–14. doi: 10.1093/pcp/pcm072. [DOI] [PubMed] [Google Scholar]
- 10.Feria Bourrellier AB, Valot B, Guillot A, Ambard-Bretteville F, Vidal J, Hodges M. Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci U S A. 2010;107:502–7. doi: 10.1073/pnas.0910097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24:4850–74. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincill ED, Clarin AE, Molenda JN, Spalding EP. Interacting glutamate receptor-like proteins in Phloem regulate lateral root initiation in Arabidopsis. Plant Cell. 2013;25:1304–13. doi: 10.1105/tpc.113.110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapken D, Anschütz U, Liu LH, Huelsken T, Seebohm G, Becker D, Hollmann M. A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci Signal. 2013;6:ra47. doi: 10.1126/scisignal.2003762. [DOI] [PubMed] [Google Scholar]
- 14.Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol Biol Evol. 2002;19:1066–82. doi: 10.1093/oxfordjournals.molbev.a004165. [DOI] [PubMed] [Google Scholar]
- 15.Price MB, Jelesko J, Okumoto S. Glutamate receptor homologs in plants: functions and evolutionary origins. Front Plant Sci. 2012;3:235. doi: 10.3389/fpls.2012.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–56. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G. Glutamate-receptor genes in plants. Nature. 1998;396:125–6. doi: 10.1038/24066. [DOI] [PubMed] [Google Scholar]
- 18.Kang J, Turano FJ. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2003;100:6872–7. doi: 10.1073/pnas.1030961100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J, Mehta S, Turano FJ. The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004;45:1380–9. doi: 10.1093/pcp/pch159. [DOI] [PubMed] [Google Scholar]
- 20.Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM. De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J. 2009;58:437–49. doi: 10.1111/j.1365-313X.2009.03789.x. [DOI] [PubMed] [Google Scholar]
- 21.Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science. 2011;332:434–7. doi: 10.1126/science.1201101. [DOI] [PubMed] [Google Scholar]
- 22.Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J. 2011;440:355–65. doi: 10.1042/BJ20111112. [DOI] [PubMed] [Google Scholar]
- 23.Li F, Wang J, Ma C, Zhao Y, Wang Y, Hasi A, Qi Z. Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol. 2013;162:1497–509. doi: 10.1104/pp.113.217208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwaaitaal M, Maintz J, Cavdar M, Panstruga R. On the ligand binding profile and desensitization of plant ionotropic glutamate receptor (iGluR)-like channels functioning in MAMP-triggered Ca²⁺ influx. Plant Signal Behav. 2012;7:1373–7. doi: 10.4161/psb.21761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature. 2013;500:422–6. doi: 10.1038/nature12478. [DOI] [PubMed] [Google Scholar]
- 26.Meyerhoff O, Müller K, Roelfsema MR, Latz A, Lacombe B, Hedrich R, Dietrich P, Becker D. AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta. 2005;222:418–27. doi: 10.1007/s00425-005-1551-3. [DOI] [PubMed] [Google Scholar]
- 27.Vincill ED, Bieck AM, Spalding EP. Ca(2+) conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol. 2012;159:40–6. doi: 10.1104/pp.112.197509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci U S A. 2004;101:12242–7. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehming N. Analysis of protein-protein proximities using the split-ubiquitin system. Brief Funct Genomic Proteomic. 2002;1:230–8. doi: 10.1093/bfgp/1.3.230. [DOI] [PubMed] [Google Scholar]
- 30.Lalonde S, Ehrhardt DW, Loqué D, Chen J, Rhee SY, Frommer WB. Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 2008;53:610–35. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- 31.Van Munster EB, Kremers GJ, Adjobo-Hermans MJ, Gadella TW., Jr. Fluorescence resonance energy transfer (FRET) measurement by gradual acceptor photobleaching. J Microsc. 2005;218:253–62. doi: 10.1111/j.1365-2818.2005.01483.x. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP. Advanced fluorescence microscopy techniques--FRAP, FLIP, FLAP, FRET and FLIM. Molecules. 2012;17:4047–132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 34.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 35.Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–13. doi: 10.1016/S0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 36.Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem. 2005;280:15053–60. doi: 10.1074/jbc.M408413200. [DOI] [PubMed] [Google Scholar]
- 37.Okumoto S, Jones A, Frommer WB. Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol. 2012;63:663–706. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- 38.Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al. The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci U S A. 2012;109:6811–8. doi: 10.1073/pnas.1202546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105:803–8. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 41.Lalonde S, Sero A, Pratelli R, Pilot G, Chen J, Sardi MI, Parsa SA, Kim DY, Acharya BR, Stein EV, et al. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol. 2010;1:24. doi: 10.3389/fphys.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–20. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 43.Schachtman DP. Molecular insights into the structure and function of plant K(+) transport mechanisms. Biochim Biophys Acta. 2000;1465:127–39. doi: 10.1016/S0005-2736(00)00134-6. [DOI] [PubMed] [Google Scholar]
- 44.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophysical Journal. 2001;81:2395–402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.