Abstract
Lately we have published on the characterization of the upstream of SamDC gene from rice and investigated the involvement of various cis-elements present in the promoter region in its transcriptional regulation. Analysis of SamDC expression showed that it was inducible by abiotic stresses like salinity, drought, and cold as well as by light and ABA treatment. Furthermore, DNA protein interaction studies have identified transacting actors responsible for its expression after abiotic stresses or light inducibility. Here we have further discussed on the possible role of these cis-elements in modulating the transcriptional network and comment on their function in relation to polyamine biosynthesis during periods of abiotic stress in rice.
Keywords: ABA, Drought, Light, Low Temperature, Polyamines, Salinity, SamDC
Introduction
Poyamines are low molecular weight multi-cationic organic compounds found in plants. Polyamines are distributed in all the compartments of plant cell including the nucleus thereby clearly suggesting a multi-functional role for these polycations along with abiotic stress tolerance. Polyamine biosynthesis in plants is regulated by arginine decarboxylase (ADC), orninthine decarboxylase (ODC) and S-adenosylmethionine decarboxylase (SamDC).2 Rice shows enormous variation in their susceptibility to abiotic as well as biotic stresses. SamDC is a key enzyme in polyamine biosynthesis3 and abiotic stress tolerance in rice catalyzing the biosynthesis of spermine and spermidine by providing decarboxylated S-adenosyl methionine as aminopropyl donor.4
Regulation of SamDC Expression in Plants
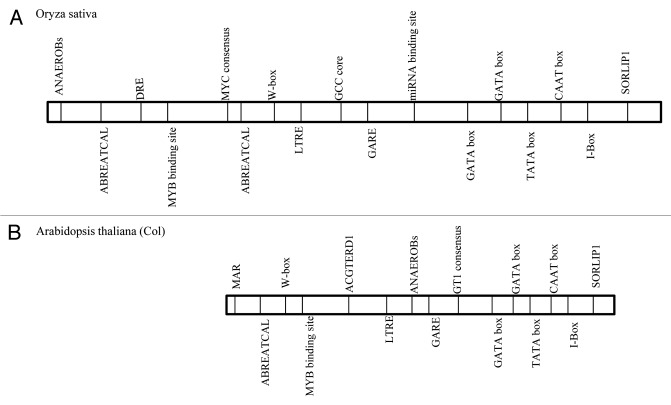
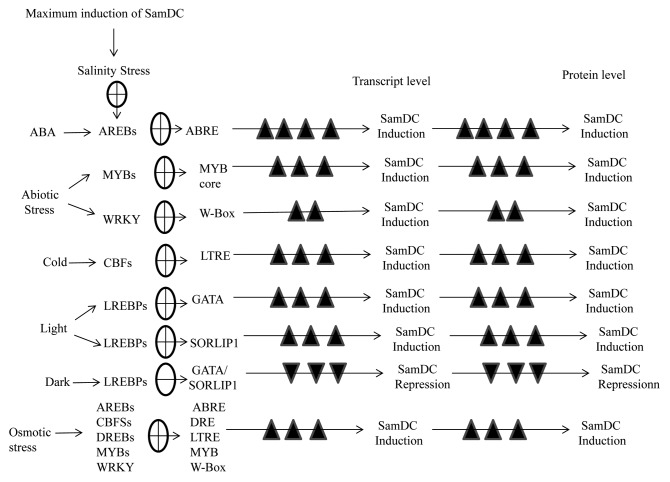
Polyamines are large nitrogenous compounds known to play a variety of roles in many physiological processes as well as response to environmental stresses. Previously we have shown that exogenous application of polyamines (Spd and Spm) helps in alleviating salinity stress in rice.5 To further study the polyamine biosynthesis pathway in detail we have done a comparative analysis of SamDC promoter from rice and Arabidopsis (using Plant Care and Place databases) which revealed the presence of several regulatory elements like ABRE, DRE, LTREs, LREs etc. (Fig. 1). Furthermore we have amplified and compared the upstream region of SamDC from IR-29 and Pokkali which revealed that there was no significant difference in the sequences (HM102353, HM102354). This further emphasizes the presence of several trans-acting factors that are induced upon exposure to abiotic stress that binds to the different cis-elements present in the upstream region and thereby regulate SamDC gene expression. Figure 2 shows the various possible signaling cascades controlling SamDC gene expression under different conditions.
Figure 1. Comparative analysis of the promoter region of SamDC gene from Arabdiopsis thaliana (Col) and rice (Oryza sativa) showing the presence of different regulatory elements. (A) Oryza sativa SamDC promoter (HM102354). (B) Arabidopsis thaliana SamDC promoter (AT3G02470). ABRE- Abscisic acid Acid Responsive Elements, ANAEROB- Anaerobic responsive element, DRE- Dehydration responsive elements, GARE- Gibberellic acid responsive element, LTRE- Low temperature responsive elements, MAR- Matrix attachment region.
Figure 2. Schematic representation of regulation of SamDC gene expression and modulation of polyamine biosynthesis in response to various environmental conditions.
Abscisic acid Acid Responsive Elements (ABRE elements)
ABA is an important phytohormone that plays a key role in the various physiological and developmental stages of rice. During vegetative stage ABA plays a key role in eliciting response to various environmental stresses like salinity, drought and cold. ABA mediated stress response involves stomata closure and ABA induced gene expression. The upstream analysis of ABA regulated genes has shown the presence of ABA responsive elements or ABREs sharing a (C/T) ACGTGGC consensus sequence. bZip type transcription factors which are inducible by osmotic stresses have been shown to bind ABREs in the promoter of rd29A and are categorized as AREB1, AREB2 and AREB3.6 The upstream analysis of SamDC promoter showed the presence of ABREATCAL elements. DNA protein interaction analysis showed the presence of two complexes which were inducible by salinity and drought stress treatments in the salt tolerant rice cultivar Nonabokra.1 Previously a bZip transcription factor designated OSBZ8 has been shown to bind to the promoter region of Rab16A gene.7 Southwestern Blot analysis performed with nuclear extract prepared from salt and PEG treated lamina of Nonabokra showed the presence of 38 and 28 kDa AREBs. The 38kDa AREB was highly induced upon PEG treatment thereby clearly suggesting a definitive role for this transcription factor in drought stress signaling of SamDC.1
WRKY and MYB Cis Regulatory Elements
WRY superfamily of transcription factors are one of the multigene families of transcriptional regulators in plants absent in prokaryotes, animals and fungi.8 The upstream of many defense related genes showed the presence of W-Box [(C/T)TGAC(T/C)] an element known to be the binding site of WRKY transcription factors. There are considerable reports on several WRKY transcription factors being induced by cold, salinity or wounding.9,10,11 Besides the WRKY another superfamily of transcription factors in plants are the MYBs. In plants the MYB transcription factors contains R2 and R3 domains and the binding sequence varies from four to 14 nucleotides.12 In Arabidopsis it has been shown that the β-glucoronidase gene driven by AtMYB2 promoter was inducible by osmotic stresses.13 The upstream analysis of SamDC promoter showed the presence of W-Box elements and MYB regulatory cis elements (TAACTG). Gel mobility shift assay performed with nuclear extract prepared from salt and PEG treated lamina of IR-29 (salt susceptible) and Nonabokra (salt tolerant) showed the presence of inducible complexes with varying intensity between the cultivars1. Southwestern blot analysis revealed the presence of 25kDa and 16kDa WRKY transcription factors and a 40kDa MYB.1
Dehydration Responsive Elements (DRE) and Low Temperature Responsive Elements (LTREs)
Rice plants exposed to low temperatures faces three major problems: disorientation of biological membranes, alteration in the physiological and biochemical reactions, changes in the water level. These responses are also symptomatic to drought stress as well thereby indicating that cold tolerance and drought stress have some mechanism in common. The promoter region of cold responsive genes contains the dehydration response element (DRE) with a core sequence of CCGAC.14 Previously it has been shown that the upstream of cor15A gene in Arabidopsis is weakly active under drought but becomes highly active under cold stress.14 The promoter regions of cold stress induced genes contain a core GCCGAC motif called Low Temperature Responsive Element (LTREs) that also forms the core of DRE sequence.15 Analysis of the promoter region of SamDC showed the presence of DRE as well as LTRE elements. Gel retardation assay with LTRE elements as probe revealed the presence of an inducible complex of varying intensity among susceptible and tolerant rice cultivars.1 Southwestern Blot analysis using nuclear extract prepared from cold stressed lamina of Nonabokra identified a 30kDa CBF.1
Light Responsive Elements (LREs)
Plants thrive on light as a source of energy for photosynthesis. Light plays a crucial role in the growth and developmental pathway of pathway of plants from carbon metabolism to photosynthesis. Previously it has been shown that SamDC is regulated by blue and red light indicating the involvement of blue light photoreceptor and phytochrome mediated regulation of SamDC at the transcriptional level.16 In rice the transcript level of SamDC was induced upon exposure to light and got repressed in the dark1 indicating a clear role of LREs.
Role of GATA Box and SORLIPs in the Light Mediated Regulation of SamDC
GATA box and the I-Box (GATAAG) are commonly found in the promoter region of light inducible genes. SORLIP1 (sequences over-represented in light-induced promoters) is another unique light responsive cis element controlling light induced gene expression. DNA-protein interaction studies performed with rice nuclear extracts after light treatments showed the presence of two GATA box binding trans-acting factors of 25 and 16kDa respectively and a 50kDa SORLIP binding transcription factor.1
Splicing and miRNA Mediated Regulation of SamDC
The upstream region of SamDC showed the presence of miRNA binding sites thereby clearly suggesting the existence of a probable posttranscriptional mode of regulation. Furthermore the presence of a splice junction sequence (CAAG) indicates the possibility of an alternative splicing mechanism giving rise to an isoform that can be active under different treatments.
Conclusion and Outlook
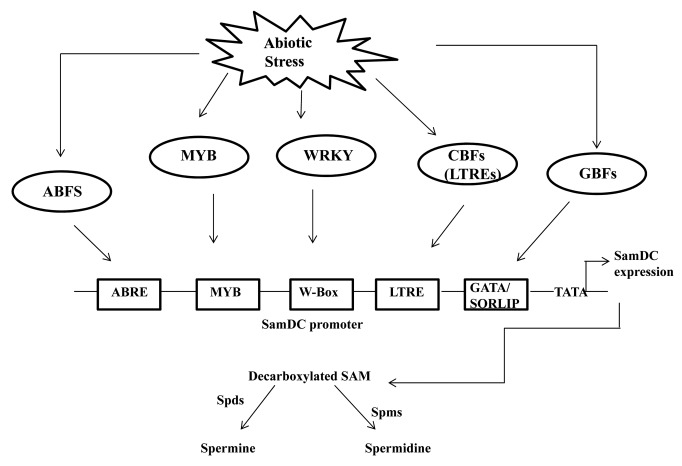
Rice is an important crop feeding almost one-third of the world. Consequently abiotic stress management is the area of major research in the recent years to develop high yielding crops resistant to the abiotic stresses. Hence in the context of present scenario it is very likely to study the underlying molecular mechanism governing abiotic stress induced gene expression. In this context our research on regulation of SamDC gene expression provides valuable insights into the polyamine biosynthesis pathway in rice. Finally our study will help in dissecting the polyamine biosynthesis pathway and hence their role in stress response (Fig. 3).
Figure 3. Hypothetical model of transcriptional regulation of Polyamine biosynthesis in response to abiotic stress in rice. ABF represents the ABRE binding transcription factors, CBFS, MYB, WRKY represents the LTRE binding TFs, MYB TFs and W-box binding trans-acting factors respectively. GBFs represent the transcription factors binding to the GATA and SORLIPs respectively. Spds and Spms are spermidine synthase and spermine synthase respectively.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
Financial assistance from Bose Institute, Kolkata and Department of Biotechnology, Government of India, New Delhi is gratefully acknowledged.
References
- 1.Basu S, Roychoudhury A, Sengupta DN. Identification of trans-acting factors regulating SamDC expression in Oryza sativa. Biochemical and Biophysical Research Communication. 2014 doi: 10.1016/j.bbrc.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–99. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 3.Hu WW, Gong H, Pua EC. The pivotal roles of the plant S-adenosylmethionine decarboxylase 5′ untranslated leader sequence in regulation of gene expression at the transcriptional and posttranscriptional levels. Plant Physiol. 2005;138:276–86. doi: 10.1104/pp.104.056770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z-Y, Chen S-Y. Isolation and characterization of a salt- and drought-inducible gene for S-adenosylmethionine decarboxylase from wheat (triticum aestivum L.) J Plant Physiol. 2000;156:386–93. [Google Scholar]
- 5.Roychoudhury A, Basu S, Sengupta DN. Amelioration of salinity stress by exogenously applied spermidine or spermine in three varieties of indica rice differing in their level of salt tolerance. J Plant Physiol. 2011;168:317–28. doi: 10.1016/j.jplph.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97:11632–7. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roychoudhury A, Gupta B, Sengupta DN. Trans-acting factor designated OSBZ8 interacts with both typical abscisic acid responsive elements as well as abscisic acid responsive element-like sequences in the vegetative tissues of indica rice cultivars. Plant Cell Rep. 2008;27:779–94. doi: 10.1007/s00299-007-0498-1. [DOI] [PubMed] [Google Scholar]
- 8.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 9.Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet. 2000;263:30–7. doi: 10.1007/PL00008673. [DOI] [PubMed] [Google Scholar]
- 10.Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem. 2004;279:11736–43. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- 11.Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002;31:319–30. doi: 10.1046/j.1365-313X.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Guan S, Zhu Z, Wang Y, Lu Y. A valid strategy for precise identifications of transcription factor binding sites in combinatorial regulation using bioinformatic and experimental approaches. Plant Methods. 2013;9:34. doi: 10.1186/1746-4811-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 14.Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–13. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 15.Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–99. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida I, Yamagata H, Hirasawa E. Light-regulated gene expression of S-adenosylmethionine decarboxylase in Pharbitis nil. J Exp Bot. 1998;49:617–20. [PubMed] [Google Scholar]