Abstract
In this study we analyzed the relationship between malate valve capacities, N-assimilation, and energy metabolism. We used transgenic plants either lacking the chloroplast NADP-dependent malate dehydrogenase or mutants with a decreased transcript level of the plastid-localized NAD-dependent malate dehydrogenase. Plants were grown on nitrate or ammonium, respectively, as the sole N-source and transcripts were analyzed by qRT-PCR. We could show that the lack of malate valve capacities enhances N-assimilation and plastidial glycolysis by increasing transcript levels of Fd-GOGATs or NADH-GOGAT and plastidic NAD-GAPDHs (GapCps), respectively. Based on our results, we conclude that the lack of malate valve capacities is balanced by an increase of the activity of plastid-localized glycolysis in order to cover the high demand for plastidial ATP, stressing the importance of the plastids for energy metabolism in plant cells.
Keywords: malate valve, plastidial glycolysis, ammonium assimilation, nitrate assimilation, energy supply, redox-balance
Malate valves are an essential feature for balancing metabolic fluxes due to their function in indirect transport of reducing equivalents. Therefore, malate dehydrogenases (MDHs) play a key role in the metabolism of plants. They belong to the group of the oxidoreductases and catalyze the interconversion of oxaloacetate (OAA) and malate in a reversible reaction, using NADH or NADPH depending on the enzyme’s coenzyme specificity.1
Plant cells possess several MDH isoforms. NAD-dependent isoforms are located in chloroplasts, mitochondria, microbodies, and cytosol.2,3 In addition, chloroplasts contain an NADP-dependent MDH.4 This redox-modulated isoprotein forms the malate valve in illuminated chloroplasts and is responsible for balancing the ATP/NADPH ratio and for maintaining redox homeostasis. During illumination, ATP and NADPH are generated by the photosynthetic electron transport (PET). The removal of excessive NADPH through the malate valve allows for continued ATP production via PET, since NADP-MDH regenerates NADP+ as an electron acceptor. Based on the fact that the NADP-MDH is light-activated and that its activation is inhibited by NADP+, the enzyme is only active when NADPH increases.5
The NAD-MDH (plNAD-MDH) plays an important role in the metabolism of dark chloroplasts. In the dark, glycolysis, respiration and the oxidative pentose-phosphate pathway (OPP) generate ATP and NADPH independently. ATP can be imported by ATP/ADP transporters (NTT1 and NTT2)6 or is produced through the activity of plastidial glycolytic enzymes. Triose-phosphate oxidation and concomitant ATP generation in the plastids requires NAD+. NADH production during glycolysis is catalyzed by the bispecific NAD(P)-GAPDH (GapA/B), which is active with NAD+ even in dark chloroplasts,7 or by the plastidic NAD-GAPDH (GapCp) in non-green tissues.8-10 Removal of NADH and regeneration of NAD+ is possible by the activity of plNAD-MDH.
Recently, we could demonstrate that a homozygous knockout of the plNAD-MDH is embryo lethal and leads to an impaired pollen-tube growth in vitro, which can be overcome by adding the substrates of NADH-GOGAT.11 The NADH-GOGAT is a key enzyme of ammonium assimilation and is mainly expressed in plastids of heterotrophic tissues comparable to plNAD-MDH. This enzyme represents a good candidate for an alternative system for the removal of NADH generated during plastidial glycolysis by GapCp. In addition, growth experiments on different nitrogen sources revealed that heterozygous knockout mutants with a decreased transcript level of plNAD-MDH (line 159) show an improved growth on ammonium as the sole N-source compared with the wild type.11 Such experiments were also performed using nadp-mdh knockout plants.12 In contrast to line 159, knockout mutants lacking NADP-MDH had an increased biomass on nitrate as the sole N-source. These results support the idea that plastidic malate valves could be involved in N-assimilation.
In this study, we analyzed various transcript levels of genes related to plastidial glycolysis, malate valves, and N-assimilation in nadp-mdh knockout mutants grown on nitrate and heterozygous plNAD-MDH knockout plants grown on ammonium. These investigations should lead to an explanation for the advantages of plants grown on nitrate or ammonium when lacking plastidic malate valve capacities.
In order to obtain more insight into a possible relationship between N-assimilation, energy metabolism, and malate valve capacities, heterozygous knockout seedlings with a decreased transcript level of plNAD-MDH (line 159) grown on 5 mM N solely as ammonia were analyzed by qRT-PCR for transcript abundance of plNAD-MDH, NADH-GOGAT (GLT1), NADP-MDH, Fd-GOGAT 1 (GLU1) and Fd-GOGAT 2 (GLU2), and GAPCP 1 and GAPCP 2 (Fig. 1). Under these conditions, the transcript levels of GLT1 and GAPCP1 were dramatically increased in line 159 compared with the wild type (Fig. 1). This might indicate that NADH-GOGAT can act as an alternative system for the removal of NADH from an even increased glycolytic flux through GapCp when plNAD-MDH capacity is reduced. This correlation is only based on GapCp1. In contrast, the GapCp2 transcript level does not significantly change in line 159 indicating an isoform-specific response.
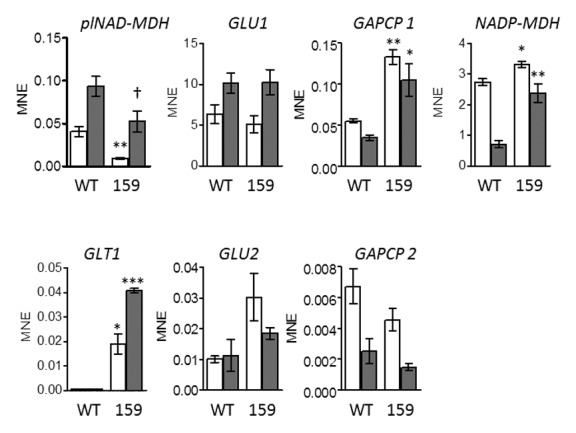
Figure 1. Expression of plastidic MDHs, GOGATs, and GapCps in leaves of heterozygous plNAD-MDH knockout mutants (line 159) and wild types (WT; ecotype Landsberg) grown on 2.5 mM (NH4)2SO4. Plants were cultivated under sterile conditions with a light period of 7.5 h (short day), a light intensity of 150 µE m−2 s−1 and 20 °C for 2 wk. Leaf material was harvested in the middle of the light period (white bars) and after 4 h into the dark period (gray bars). Total RNA was isolated from 100 mg frozen leaf material using TRI-Reagent. Afterwards, DNase digest and cDNA synthesis were performed. Samples were analyzed for contaminations with genomic DNA by PCR. Primers for the detection of NADP-MDH (for: 5′-GCTCCCAACA TTCCTGCAAA-3′, rev: 5′-CACCTGAGTC GTGGAGTGAT-3′), GLU1 (for: 5′-AGAGGCAAAG CTGGAGAGAG-3′, rev: 5′-GCAACGTTTC TTCCCACCTT-3′) and GLU2 (for: 5′-TCTGGGTGAG GGCATTTTCT-3′, rev: 5′-GTCTCACTCT TTTCGCGGAC-3′) were tested for their efficiency before they were used for qRT-PCR. All other primers were already tested.10 Each reaction of qRT-PCR was performed in triplicates. The average CT values for all genes and the reference gene RAN3 were used to calculate the mean normalized expression (MNE).13 Asterisks (* P < 0.05; ** P < 0.01; *** P < 0.001) and crosses († P < 0.1) indicate that the differences between WT and line 159 are statistically significant as determined by t-test.
Besides the NADH-GOGAT, 2 isoforms of Fd-GOGAT are localized in chloroplasts of higher plants. These isoproteins represent the predominant form in leaves.14 The analysis of transcript levels of Fd-GOGAT1 and Fd-GOGAT2 was included in our investigations because of their involvement in ammonium assimilation, but no differences between heterozygous plNAD-MDH knockout plants and wild types could be observed (Fig. 1). While the NADH-GOGAT is crucial for ammonium assimilation in roots,15 Fd-GOGATs contribute to the assimilation of ammonium derived from photorespiration which doesn’t seem to be influenced in line 159.
Furthermore, the transcript level of NADP-MDH was analyzed in heterozygous plNAD-MDH knockout mutants, since this enzyme represents the malate valve in the light. Interestingly, the expression of NADP-MDH is significantly increased compared with wild type (Fig. 1). Especially in the dark, transcription of NADP-MDH is very high. This result could represent a precaution for the light phase so that a faster adjustment for the demand of reducing equivalents in other cell compartments is possible when plNAD-MDH capacity is low.
Transcript analysis was also performed with nadp-mdh knockout plants grown on 5 mM N provided exclusively as nitrate, since these mutants show an increased biomass on nitrate.12 This result indicated that there could be a relationship between the malate valve capacity in the light and N-assimilation as shown for the plNAD-MDH as part of the dark malate valve. Therefore, transcript abundance of the same set of genes as chosen for line 159 was analyzed in nadp-mdh knockout mutants grown on nitrate.
In our previous study using Western-blot analysis, the absence of the NADP-MDH protein was confirmed in the nadp-mdh knockout plants. Northern-blot analysis and RT-PCR had revealed the lack of NADP-MDH transcript.12 In this study, we verified the transcript abundance of NADP-MDH in the knockout mutants quantitatively by qRT-PCR (Fig. 2). Again, no transcript for this gene could be detected in the mutants so that these plants exhibit a total knockout of NADP-MDH.
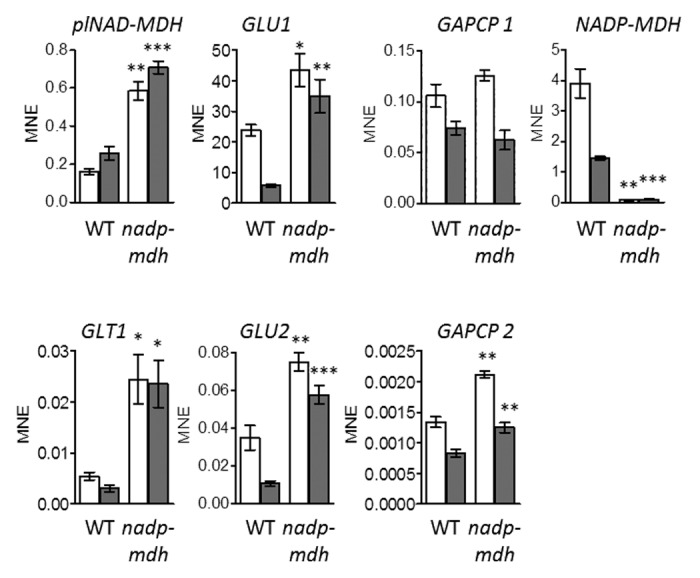
Figure 2. Expression of plastid-localized MDHs, GOGATs, and GapCps in leaves of wild types (WT; ecotype Columbia) and nadp-mdh knockout plants grown on 5 mM KNO3. Experimental procedure was the same as described in Figure 1. Asterisks (* P < 0.05; ** P < 0.01; *** P < 0.001) indicate that the differences between WT and nadp-mdh knockout mutants are statistically significant as determined by t-test.
Under low-oxygen conditions (non-photorespiratory conditions), isolated protoplasts of nadp-mdh knockout plants showed a decrease in photosynthesis compared with wild type. Furthermore, a high expression of glycine decarboxylase (GDC) and a shift in the glycine-to-serine ratio could be observed. These results indicate altered patterns in photorespiration in mutants lacking NADP-MDH.12 Since Fd-GOGATs play an important role in photorespiratory ammonium assimilation,15 the expression pattern of both Fd-GOGAT isoforms was analyzed in nadp-mdh knockout plants (Fig. 2). Compared with the wild type, transcript levels of GLU1 and GLU2 are significantly increased in plants lacking NADP-MDH. This result indicates that the reduced form of ferredoxin (Fdred) can provide electrons for the reduction of nitrite to ammonium as well as for the reductive transfer of the amido group of glutamine (Gln) to 2-oxoglutarate (2-OG) catalyzed by Fd-GOGAT. Therefore, Fd-GOGATs could represent an alternative way to remove excess electrons generated during photosynthesis when the NADP-MDH is lacking.
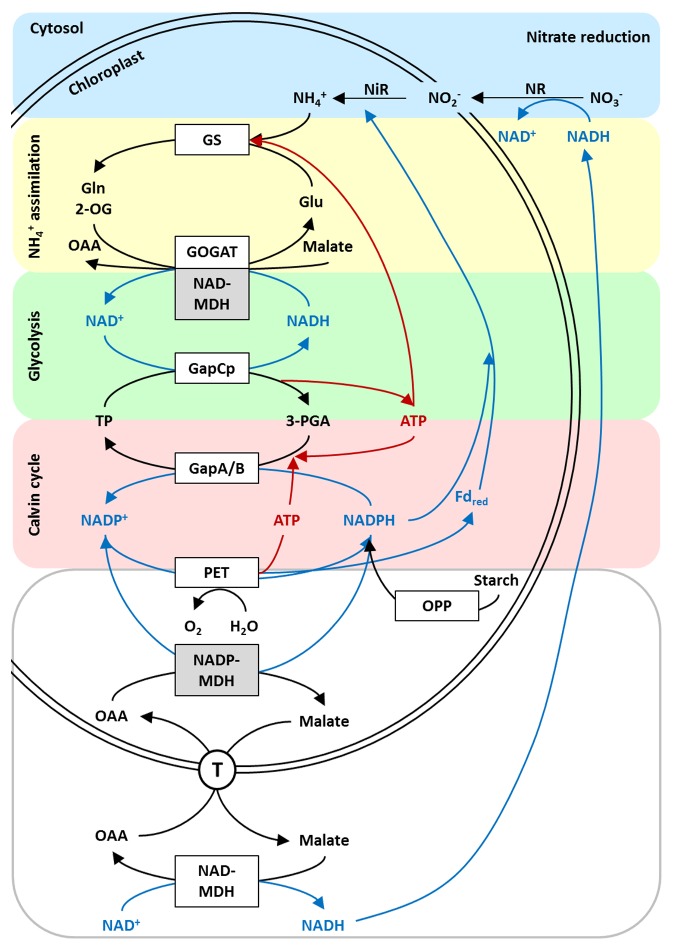
Since the expression of NADP-MDH is increased in heterozygous knockout plants with a reduced transcript level of plNAD-MDH, we were interested to investigate the transcription pattern vice versa. In nadp-mdh knockout mutants, transcript levels of plNAD-MDH, and GLT1, respectively, are significantly increased (Fig. 2), demonstrating that the plNAD-MDH could be responsible for supplying the other cell compartments with reducing equivalents when the NADP-MDH is missing. In addition, there seems to be a high demand for the regeneration of NAD+. In this context, transcript levels of GAPCP1 and GAPCP2 were also analyzed based on the fact that GapCps produce NADH during glycolysis, which is a coenzyme of plNAD-MDH and NADH-GOGAT, respectively. Again the transcription of GAPCP1 and GAPCP2 is isoform-specific, which is also the case in line 159 (Fig. 1). The level of GAPCP1 remains the same as in the wild type, but in contrast, GAPCP2 expression is significantly increased in nadp-mdh knockout plants (Fig. 2). This result provides an explanation for the higher transcript levels of plNAD-MDH and GLT1, since the production of NADH by the glycolytic GapCp is increased and NAD+ regeneration will therefore not become limiting. ATP from the concomitantly increased plastidial glycolysis is then available in addition to the ATP production by photosynthesis. ATP can be used in the Calvin cycle and for ammonium assimilation (Fig. 3). Nitrate reduction in the cytosol can be driven by NADH delivered via NAD-driven malate valve. Nitrite reduction leads to even increased electron flow through PET, resulting in more ATP as well.
Figure 3. Relationship between plastidic malate valves, glycolysis, and C- and N-assimilation.
Our recently published data11 and the results of this study both concerning the plNAD-MDH emphasize the relationship between the dark malate valve, ammonium assimilation and ATP generation via glycolysis (Fig. 3). Transgenic plants with a reduced transcript level of plNAD-MDH exhibit an increased level of NADH-GOGAT. This enzyme could serve as an alternative system for the removal of NADH, thereby regenerating NAD+ as a substrate for GapCp even allowing an increased rate of N-assimilation. The results concerning the GapCp isoforms in this study do not confirm the recently published data about GAPCP1 and GAPCP2 transcript levels in line 159 grown on half-strength MS medium.11 Nevertheless, in these plants the expression of NADH-GOGAT was significantly increased compared with wild type as well, but less pronounced compared with plants of line 159 grown on ammonium as the only N-source.11 An explanation for this discrepancy could be that plants prefer to assimilate nitrate rather than ammonium when both substances are available in the growth medium which is the case for MS medium. Under these conditions, plants of line 159 have no advantage compared with the wild type, which could already be observed during growth experiments on nitrate.11
Due to the fact that plNAD-MDH as well as NADH-GOGAT are both mainly expressed in plastids of non-green tissues,11,14 it was very interesting that recently a tissue-specific expression pattern during the life cycle of Arabidopsis for NADH-GOGAT was demonstrated15 which is very similar to our findings concerning the plNAD-MDH expression during plant growth.11 Both analyses were performed using gene promoter-GUS fusions and show a high β-glucuronidase activity in the tip of leaves and the root tip.11,15 Furthermore, in both studies the promoter activity in pollen was shown to be very high. These results again strengthen our hypothesis that the NADH-GOGAT can act as an alternative system for the regeneration of NAD+ to keep ATP production via glycolysis running (Fig. 3) which was already shown by transcript analysis and the compensatory effect on impeded in vitro pollen tube growth in line 159 by feeding the NADH-GOGAT substrates 2-OG and Gln.11 In this context, it is also very interesting that both promoter studies show a high β-glucuronidase activity in the stigma of the flowers indicating that pollen tube growth of line 159 is not affected in vivo. Therefore, pollen tube growth must be supported with the required substrates by the maternal tissue (stigma of heterozygous parent plant) so that tube growth of knockout pollen is not abolished in vivo.
A very high expression of NADH-GOGAT was also found in endosperm as well as developing and mature embryos, respectively,15 emphasizing our postulation that the lack of chlorophyll in homozygous plNAD-MDH knockout embryos might be due to the competition for Glu that appears to be consumed for the operation of the “glutamate valve” when plNAD-MDH is lacking and NADH-GOGAT expression is increased.11
Based on the fact that nadp-mdh knockout mutants show an improved growth on nitrate as the sole N-source, we also analyzed transcript levels of various genes related to N-assimilation, energy metabolism and malate valve capacity in these mutants when grown on nitrate (Fig. 2). The NADP-MDH is responsible for the removal of excess electrons (e.g under high light conditions) and therefore facilitates the regeneration of NADP+ as the electron acceptor for PET in chloroplasts. In nadp-mdh knockout mutants, this regenerating step is lacking. Surprisingly, the nadp-mdh knockout plants show no different phenotype under high light treatment compared with wild type. Nevertheless, we could uncover a combination of different mechanisms to cope with excess reducing equivalents by increased fluxes through NTRC/2-Cys peroxiredoxin system, proline biosynthesis, and photorespiration, when the NADP-MDH is lacking.12 Increased growth of the nadp-mdh knockout line on nitrate as compared with the wild type was observed. With reference to the altered photorespiration in nadp-mdh knockout plants it was very interesting to observe a higher expression of both Fd-GOGAT isoforms in these mutants when grown on nitrate as the sole N-source (Fig. 2). These enzymes play an important role during photorespiratory ammonium assimilation16 and at the same time represent an alternative mechanism to cope with excess electrons. A lack of NADP-MDH appears to be compensated for by nitrite reduction and transamination of Gln and 2-OG catalyzed by Fd-GOGAT when reduced ferredoxin directly provides electrons for these pathways and therefore nitrate assimilation is enhanced (Fig. 3).
The interconversion of Glu and glutamyl phosphate represents the first step during proline biosynthesis. The phosphorylation of Glu is catalyzed by the glutamate kinase under ATP consumption. In addition, ATP is needed for the phosphorylation of glycerate to 3-PGA during photorespiration. Furthermore, N-assimilation is also dependent on ATP. During the transfer of NH4+ to Glu catalyzed by GS, ATP is needed. This means that there might be a very high demand for ATP production in nadp-mdh knockout plants to keep all alternative systems running for the removal of excess electrons and reducing equivalents. In this context, it was very interesting that we could observe a very high expression of plNAD-MDH, NADH-GOGAT, and GAPCP2, respectively (Fig. 2). The plNAD-MDH as well as the NADH-GOGAT are responsible for the removal of NADH produced by GapCp to keep ATP production via plastidial glycolysis running (Fig. 3). In addition, the plNAD-MDH could serve as an alternative system to provide reducing equivalents for other cell compartments when NADP-MDH is lacking. Although these pathways are thought to be needed only in the dark, it is conceivable that glycolysis is also active during the day on a basal level to increase ATP availability for various metabolic pathways. At first sight, this hypothesis seems to be inconsistent because the simultaneous activities of the glycolytic enzyme GapCp and the Calvin-cycle enzyme GapA/B would result in a futile cycle (Fig. 3).
Apparently, the nadp-mdh knock out per se has resulted in the change at the transcriptional level, as shown in this study, and transcript was already present under growth-light conditions without any high-light treatment. Nevertheless, the amount of protein and its post-translational modifications will also pose a high impact on the activation state of enzymes. For example, under oxidizing conditions GapA/B is present as an inactive hexadecamer. Only after incubation with a reductant and 1,3-bisphosphoglycerate (1,3bisPGA), which causes the dissociation of the oligomer into a heterotetramer, the enzyme is fully activated.17,18 For the phosphorylation of 3-PGA to form 1,3bisPGA ATP is needed. Therefore, additional ATP serves to drive a feed-forward loop for increased flow through the reductive step in carbon assimilation.
Due to the fact that all of the alternative pathways which cope with excess reducing equivalents in plants lacking the NADP-MDH depend on ATP, it is conceivable that glycolysis runs at a basal level during the light phase to keep ATP production and therefore also the Calvin cycle running. In this context, it is also very interesting to note that for example in fd2 knockout plants, a decreased activation state of NADP-MDH and an activation of the OPP enzyme glucose-6-phosphate dehydrogenase were shown to occur even in the light.19 This finding strengthens our hypothesis that enzymes which are normally only active in the dark can also be active during the day to adapt and balance the ATP/NADPH ratio according to the requirements of the cell. In conclusion, all our results indicate an interplay between malate valves, N-assimilation, and energy metabolism which enables the plant to adapt to short-term and long-term metabolic and environmental changes by optimally balancing the ATP/NADPH ratio in each case.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We greatly acknowledge support from the Deutsche Forschungsgemeinschaft (SFB 944, project P9, R.S.), the government of Lower Saxonia (Lichtenberg fellowship to J.S.), and the Frauenfoerderpool of the University of Osnabrueck (fellowship to J.S.).
Glossary
Abbreviations:
- 1,3bisPGA
1,3-bisphosphoglycerate
- 2-OG
2-oxoglutarate
- 3-PGA
3-phosphoglycerate
- Fd
ferredoxin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GDC
glycine decarboxylase
- Gln
glutamine
- GLT1
NADH-GOGAT
- Glu
glutamate
- GLU1/2
Fd-GOGAT ½
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- MDH
malate dehydrogenase
- MNE
mean normalized expression
- NiR
nitrite reductase
- NR
nitrate reductase
- OAA
oxaloacetate
- OPP
oxidative pentose-phosphate pathway
- PET
photosynthetic electron transport
- TP
triose phosphate
References
- 1.Ocheretina O, Scheibe R. Cloning and sequence analysis of cDNAs encoding plant cytosolic malate dehydrogenase. Gene. 1997;199:145–8. doi: 10.1016/S0378-1119(97)00361-2. [DOI] [PubMed] [Google Scholar]
- 2.Gietl C. Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim Biophys Acta. 1992;1100:217–34. doi: 10.1016/0167-4838(92)90476-T. [DOI] [PubMed] [Google Scholar]
- 3.Berkemeyer M, Scheibe R, Ocheretina O. A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J Biol Chem. 1998;273:27927–33. doi: 10.1074/jbc.273.43.27927. [DOI] [PubMed] [Google Scholar]
- 4.Scheibe R. NADP+-malate dehydrogenase in C3-plants: Regulation and role of a light-activated enzyme. Physiol Plant. 1987;71:393–400. doi: 10.1111/j.1399-3054.1987.tb04362.x. [DOI] [Google Scholar]
- 5.Scheibe R. Malate valves to balance cellular energy supply. Physiol Plant. 2004;120:21–6. doi: 10.1111/j.0031-9317.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- 6.Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE. Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 2004;136:3524–36. doi: 10.1104/pp.104.049502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baalmann E, Backhausen JE, Rak C, Vetter S, Scheibe R. Reductive modification and nonreductive activation of purified spinach chloroplast NADP-dependent glyceraldehyde-3-phosphate dehydrogenase. Arch Biochem Biophys. 1995;324:201–8. doi: 10.1006/abbi.1995.0031. [DOI] [PubMed] [Google Scholar]
- 8.Backhausen JE, Vetter S, Baalmann E, Kitzmann C, Scheibe R. NAD-dependent malate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta. 1998;205:359–66. doi: 10.1007/s004250050331. [DOI] [Google Scholar]
- 9.Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009;151:541–58. doi: 10.1104/pp.109.143701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 2010;152:1830–41. doi: 10.1104/pp.109.150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selinski J, König N, Wellmeyer B, Hanke GT, Linke V, Neuhaus HE, Scheibe R. The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol Plant. 2014;7:170–86. doi: 10.1093/mp/sst151. [DOI] [PubMed] [Google Scholar]
- 12.Hebbelmann I, Selinski J, Wehmeyer C, Goss T, Voss I, Mulo P, Kangasjärvi S, Aro EM, Oelze ML, Dietz KJ, et al. Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J Exp Bot. 2012;63:1445–59. doi: 10.1093/jxb/err386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–40. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 14.Lancien M, Martin M, Hsieh M-H, Leustek T, Goodman H, Coruzzi GM. Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 2002;29:347–58. doi: 10.1046/j.1365-313X.2002.01218.x. [DOI] [PubMed] [Google Scholar]
- 15.Konishi N, Ishiyama K, Matsuoka K, Maru I, Hayakawa T, Yamaya T, Kojima S. NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol Plant. 2014;••• doi: 10.1111/ppl.12177. [DOI] [PubMed] [Google Scholar]
- 16.Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;10:741–52. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baalmann E, Backhausen JE, Rak C, Vetter S, Scheibe R. Reductive modification and nonreductive activation of purified spinach chloroplast NADP-dependent glyceraldehyde-3-phosphate dehydrogenase. Arch Biochem Biophys. 1995;324:201–8. doi: 10.1006/abbi.1995.0031. [DOI] [PubMed] [Google Scholar]
- 18.Baalmann E, Backhausen JE, Kitzmann C, Scheibe R. Regulation of NADP-dependent glyceraldehyde 3-phosphate dehydrogenase activity in spinach chloroplasts. Bot Acta. 1994;107:313–20. doi: 10.1111/j.1438-8677.1994.tb00801.x. [DOI] [Google Scholar]
- 19.Voss I, Koelmann M, Wojtera J, Holtgrefe S, Kitzmann C, Backhausen JE, Scheibe R. Knockout of major leaf ferredoxin reveals new redox-regulatory adaptations in Arabidopsis thaliana. Physiol Plant. 2008;133:584–98. doi: 10.1111/j.1399-3054.2008.01112.x. [DOI] [PubMed] [Google Scholar]