Abstract
A strict control of abundance and localization of plasma membrane proteins is essential for plants to be able to respond quickly and accurately to a changing environment. The proteins responsible for the initial recognition and concentration of ubiquitinated plasma membrane proteins destined for degradation, are well characterized in mammals and yeast,1 yet no clear orthologs were found in plants.2 Recently, we have identified a family of proteins in higher plants, which function in vacuolar targeting and subsequent degradation of ubiquitinated plasma membrane proteins3,4 termed TOM1-like (TOL) proteins.
Keywords: Protein degradation, ESCRT-0, TOL, VHS- and GAT-domain, Arabidopsis
We show here an expression analysis of the 9 different TOLs by RT-PCR. For further more detailed expression analysis, we generated and examined reporter constructs for 2 family members. Overall, visualization of TOL expression points toward overlapping but also distinct individual function of the TOLs.
Due to their sessile lifestyle higher plants have evolved a plethora of mechanisms to be able to respond quickly and accurately to a varying, often stressful environment. Plasma membrane proteins, involved in perception of external stimuli as well as in transport processes, are of particular importance as they act at the interface between cellular compartment and the outside, and are therefore subjected to a tight regulation of localization and activity. A key function in sorting processes for the homeostatic regulation of plasma membrane proteins has been attributed to their ubiquitination.5 Cargo ubiquitination functions as regulator of endocytosis and vesicular trafficking toward the vacuole for degradation. Proteins are targeted to and sorted at vesicles via interactions of their ubiquitin moieties and different ubiquitin binding proteins.5-9 This process is controlled by an evolutionary conserved, multi-subunit complexes termed the Endosomal Sorting Complex Required for Transport (ESCRT).9,10
In plants, the ESCRT machinery is generally well conserved, with the exception of the ESCRT-0, constituted of 2 subunits, responsible for the initial targeting and concentration of the ubiquitinated cargo and the recruitment of the downstream ESCRT machinery.2,11 We have recently identified a family of 9 proteins termed TOLs, with a similar domain structure to the ESCRT-0, as they contain an N-terminal VHS (Vps27, Hrs, and STAM) domain followed by a GAT (GGAs and TOM) domain, and demonstrated their crucial function in vacuolar targeting and subsequent degradation of ubiquitinated plasma membrane proteins.3 According to our findings, members of the TOL protein family can be considered as a plant-specific functional substitute for the ESCRT-03 in the initial targeting of ubiquitinated plasma membrane proteins destined for degradation.
Phylogenetic analyses revealed that plant TOLs diverged from their animal and fungal counterparts, before the latter kingdoms evolved ESCRT-0 subunits and further proteins with a similar VHS-GAT domain structure, like the TOM1 (target of myb) adaptor proteins and the GGAs (Golgi-localized, γ-ear-containing Arf binding proteins).12,13 Unlike metazoa and fungi, plant genomes encode for a disproportionally large number of these VHS-GAT domain-containing proteins, exemplified by 9 TOL family members in Arabidopsis thaliana.12 Redundant but also divergent functions of these different TOL family members are quite likely, as several double and higher order mutants of tol T-DNA deletion lines have no apparent phenotype, while combinations of other tol loss-of-function alleles are quite detrimental to the plant.3 In addition, expression profiles from publicly available gene and protein expression data sets show overlapping but also distinct expression patterns for different members of the TOL family.14
To characterize expression of all TOL genes in planta, we investigated their transcription by generating cDNAs from different plant tissues followed by gene-specific RT-PCR after normalization to tubulin.15 TOL-specific transcripts were detectable in most organs tested, demonstrating that TOL family members are expressed ubiquitously in most adult organs, including roots, stem, leaves (rosette and cauline), flowers, and siliques as well as in seedlings 5 days after germination (Fig. 1). The observed expression pattern is in agreement with the proposed high degree of functional redundancies within this gene family.3 An interesting exception was TOL8, with an almost exclusive expression in siliques and flowers of adult plants. Homozygous single T-DNA insertion mutants for all loci are viable and show no obvious phenotype.3 Yet, 2 double-mutant combinations, which both include the mutant tol8 locus (tol5–1/tol8–1 and tol7–1/tol8–1), could not be obtained as homozygotes and showed aborted seed development.3 Both, TOL5 and TOL7, also have a more pronounced expression in either of these 2 organs (Fig. 1). This might reflect a distinct function of TOL8 in flowers and siliques, which, in combination with TOL5 and/or TOL7, might specify early events in plant development.
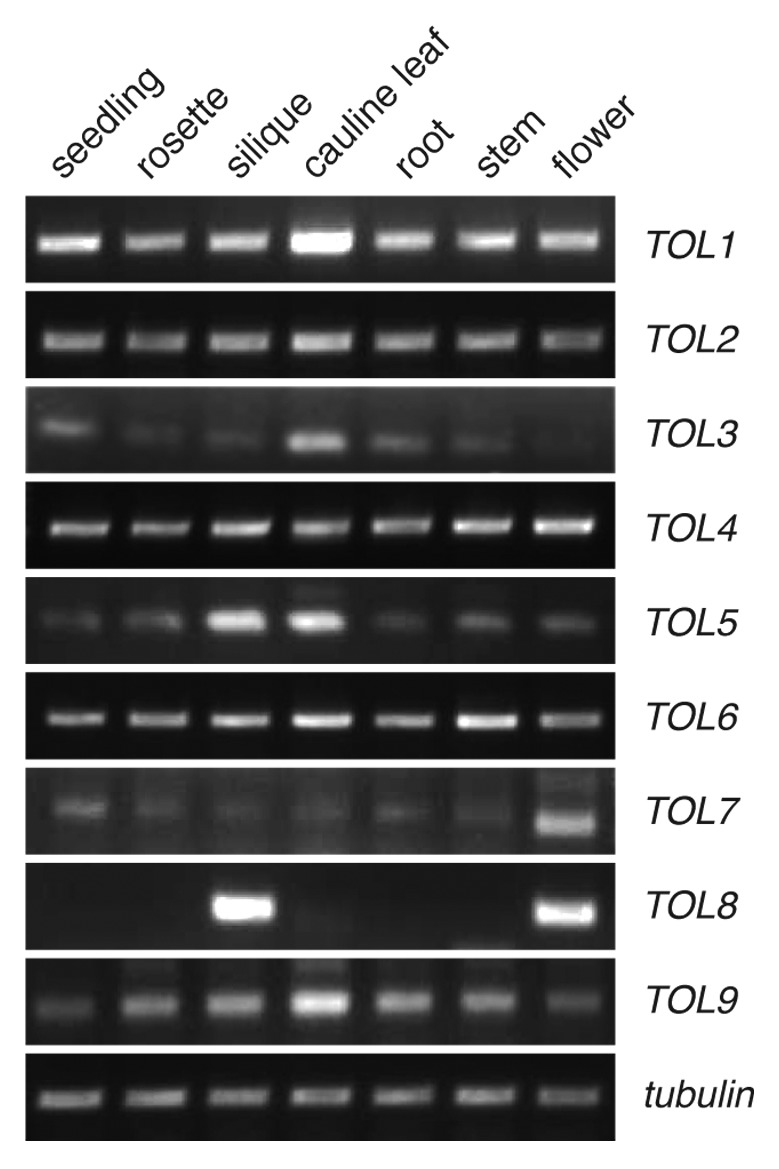
Figure 1. Expression analysis of TOLs. Expression of 9 TOLs in different tissues. RT-PCR performed on cDNA, derived from adult plants for the siliques, flowers, cauline leaves, and stems, from plants 13 days after germination (DAG) for the rosette leaves and roots or 5 DAG total seedlings. A TOL-specific fragment is detectable in most tissues tested. Tubulin was used as an internal standard.
For further analysis of TOL expression in a developmental and tissue-specific context, a set of promoter reporter constructs, with a fragment containing 2 kb upstream region of the TOL1 or TOL5 coding region fused to glucuronidase (GUS) was generated (TOL1p::GUS; TOL5p::GUS). These constructs were transformed into wild type Arabidopsis as described in ref 16. The analysis of the resulting transgenic lines showed that TOL1 and TOL5 are strongly expressed in leaves and in stamen (Fig. 2A and D, data not shown for TOL5p::GUS). Additionally, GUS assays revealed locally restricted expression in flower abscission zones after organ shed (Fig. 2B), which persisted late into silique maturation. Furthermore, GUS-staining revealed pronounced signals in roots of TOL1p::GUS and TOL5p::GUS plantlets, while comparably weaker staining was observed in the shoot portions (Fig. 2C and E for TOL1p::GUS;3 for TOL5p::GUS), with GUS activity strongest in the meristematic zones of the root apex (Fig. 2C).3 The expression of GUS-reporters, driven by either the TOL1 or TOL5 promoter did not show large discrepancies, once more indicative of their potential functional redundancies. One exception was a pronounced staining in the shoot apical meristem of transgenic TOL1p::GUS, not detectable in TOL5p::GUS plantlets, possibly indicative of a functional diversification.
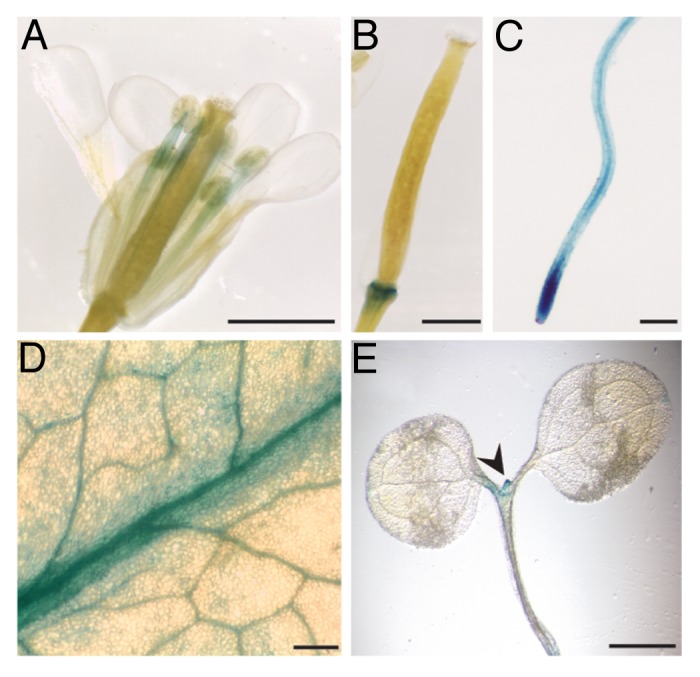
Figure 2. Expression and Localization of prTOL-GUS. (A-E) GUS activity in TOL1p::GUS plants. For this construct, approximately 2 kb upstream of the TOL1 ORF was amplified with primers prTOL1f/prTOL1r, ligated via SacI/XmaI into pPZP-GUS17 and confirmed by sequencing. (A) Stamen of the inflorescences (adult plant) (B) Floral abscission zone (C) Primary root meristem (5 DAG) (D) Rosette leave (E) Shoot apical meristem (5DAG). Scale bar: (A, B, E) = 2 mm (C, D) = 200µm
Here, we performed a crude expression analysis of members of the Arabidopsis TOL gene family, which function as potential substitutes for the elusive ESCRT-0 in plants.3 Taking into account the considerable expansion of the TOL gene families in plants, compared with other eukaryotes,11,13 it would not be surprising to find several redundancies, as well as unique functions of the TOLs. Indeed, analysis of the expression pattern of the TOLs revealed that some TOLs, like TOL8, are highly specific to certain organs, where they might perform a plant specific function, while others are expressed ubiquitously. This could reflect the involvement of different TOLs in different steps of protein sorting, which would explain the absence of other VHS-GAT domain subfamilies in plant genomes. While some TOL proteins seemingly mimic ESCRT-0 functions required for cargo degradation in conjunction with the ESCRT complex,3 others might be required for further cargo sorting steps at endosomes, the Golgi, the TGN or the PM or potentially might have acquired even additional, plant-specific functions. The data presented here should thus serve as a first impulse to allow us to speculate about the function of TOLs in different pathways of the distinctive endosomal system of plants. (Table 1)
Table 1. Oligonucleotides used in this study.
| Name | Sequence 5′-3′ | Purpose |
|---|---|---|
| TOL1RT- f1 | CCAGTGAACT TCGCTACCTA CC | RT-PCR TOL1 |
| TOL1-d | GGGTTTGTTC ATCTCCTCAT AC | RT-PCR TOL1 |
| TOM-L 10 RT-u | GCTGAAGACT GGTGGAGC | RT-PCR TOL2 |
| TOL2-d | TGGTGGAAAA CAGGAAGATA AG | RT-PCR TOL2 |
| TOL3-u | GCTCAGGCAA CTGCATCAG | RT-PCR TOL3 |
| TOL3-d | CGGTATTGGA GTGGGAGCTG | RT-PCR TOL3 |
| TOL4RT-f1 | GCATGTGCAG AAAGGGCTAC | RT-PCR TOL4 |
| TOL4RT-r1 | CAACGAAGCC TGAATAGCAG C | RT-PCR TOL4 |
| TOL5-u | CCTACGCTTG TGAAGATAG | RT-PCR TOL5 |
| TOM-L 31-RT-d | GCCTCGATGT CATGACGAG | RT-PCR TOL5 |
| TOL6-u | GTGGATATTT TCCCTCTGGA CC | RT-PCR TOL6 |
| TOL6-d | GCGACGGTGG CTGTTGATAA AG | RT-PCR TOL6 |
| TOL7RT-f1 | CTCTCAATTC TAATCGCTG | RT-PCR TOL7 |
| TOL7RT-r1 | GCTCTTGGTA TGCCCAGTTG | RT-PCR TOL7 |
| TOMl40-u | GGCTTACTAG TAGAACTTC | RT-PCR TOL8 |
| TOMl40–63RT-d | GGATACCTTG TCGAAGGACC | RT-PCR TOL8 |
| TOL9RT-f1 | CCTCAGTGGC GATGATCTTG | RT-PCR TOL9 |
| TOM- L-760-RT1-d | GGTTTCAGGC CAAGTGACCT TG | RT-PCR TOL9 |
| prTOL1f | GAGCTCGGTG ATATGGGTAG GCAG | Cloning |
| prTOL1r | CCCGGGGCTG ATACTCAAAA ACCTG | Cloning |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to C Luschnig for critically reading the manuscript. This work was supported by a Hertha Firnberg fellowship from the Austrian Science Fund (FWF T477).
References
- 1.Hurley JH. . The ESCRT complexes. Crit Rev Biochem Mol Biol 2010; 45:463 - 87; http://dx.doi.org/ 10.3109/10409238.2010.502516; PMID: 20653365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes FC, Buono R, Otegui MS. . Plant endosomal trafficking pathways. Curr Opin Plant Biol 2011; 14:666 - 73; http://dx.doi.org/ 10.1016/j.pbi.2011.07.009; PMID: 21821464 [DOI] [PubMed] [Google Scholar]
- 3.Korbei B, Moulinier-Anzola J, De-Araujo L, Lucyshyn D, Retzer K, Khan MA, Luschnig C. . Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr Biol 2013; 23:2500 - 5; http://dx.doi.org/ 10.1016/j.cub.2013.10.036; PMID: 24316203 [DOI] [PubMed] [Google Scholar]
- 4.Sauer M, Friml J. . Plant biology: gatekeepers of the road to protein perdition. Curr Biol 2014; 24:R27 - 9; http://dx.doi.org/ 10.1016/j.cub.2013.11.019; PMID: 24405674 [DOI] [PubMed] [Google Scholar]
- 5.Hicke L, Dunn R. . Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 2003; 19:141 - 72; http://dx.doi.org/ 10.1146/annurev.cellbio.19.110701.154617; PMID: 14570567 [DOI] [PubMed] [Google Scholar]
- 6.Katzmann DJ, Babst M, Emr SD. . Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001; 106:145 - 55; http://dx.doi.org/ 10.1016/S0092-8674(01)00434-2; PMID: 11511343 [DOI] [PubMed] [Google Scholar]
- 7.Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, Bonifacino JS, Hurley JH. . The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev Cell 2007; 12:973 - 86; http://dx.doi.org/ 10.1016/j.devcel.2007.04.013; PMID: 17543868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raiborg C, Rusten TE, Stenmark H. . Protein sorting into multivesicular endosomes. Curr Opin Cell Biol 2003; 15:446 - 55; http://dx.doi.org/ 10.1016/S0955-0674(03)00080-2; PMID: 12892785 [DOI] [PubMed] [Google Scholar]
- 9.Raiborg C, Stenmark H. . The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009; 458:445 - 52; http://dx.doi.org/ 10.1038/nature07961; PMID: 19325624 [DOI] [PubMed] [Google Scholar]
- 10.Williams RL, Urbé S. . The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 2007; 8:355 - 68; http://dx.doi.org/ 10.1038/nrm2162; PMID: 17450176 [DOI] [PubMed] [Google Scholar]
- 11.Winter V, Hauser MT. . Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci 2006; 11:115 - 23; http://dx.doi.org/ 10.1016/j.tplants.2006.01.008; PMID: 16488176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Craene JO, Ripp R, Lecompte O, Thompson JD, Poch O, Friant S. . Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics 2012; 13:297; http://dx.doi.org/ 10.1186/1471-2164-13-297; PMID: 22748146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman EK, Walker G, van der Giezen M, Dacks JB. . Multivesicular bodies in the enigmatic amoeboflagellate Breviata anathema and the evolution of ESCRT 0. J Cell Sci 2011; 124:613 - 21; http://dx.doi.org/ 10.1242/jcs.078436; PMID: 21266469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson LG, Mullen RT. . Meta-analysis of the expression profiles of the Arabidopsis ESCRT machinery. Plant Signal Behav 2011; 6:1897 - 903; http://dx.doi.org/ 10.4161/psb.6.12.18023; PMID: 22105035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, Müllner AE, Luschnig C. . Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc Natl Acad Sci U S A 2010; 107:10308 - 13; http://dx.doi.org/ 10.1073/pnas.0913918107; PMID: 20479223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clough SJ, Bent AF. . Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735 - 43; http://dx.doi.org/ 10.1046/j.1365-313x.1998.00343.x; PMID: 10069079 [DOI] [PubMed] [Google Scholar]
- 17.Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. . Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 2000; 12:853 - 70; PMID: 10852933 [DOI] [PMC free article] [PubMed] [Google Scholar]


