Abstract
Plants are inescapably exposed to environmental stress because of their sessile lifestyle. Such stress induces the production of reactive oxygen species (ROS), which are in turn a source of genotoxic stress. ROS are also generated intrinsically during photosynthesis in the chloroplasts. Furthermore, plants are affected by the UV component of sunlight, which damages their genomes. To protect their genomic integrity from DNA damage, plants activate a DNA damage response (DDR) system that regulates cell cycle arrest, DNA repair, and programmed cell death. Although plants have orthologs of several of the DDR factors that are found in animals, certain critical animal DDR factors, notably the tumor suppressor p53 and the DDR kinases CHK1 and CHK2, have not been found in plants. In this mini-review, we summarize the functions and regulatory mechanism of Arabidopsis thaliana SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), a plant-specific transcription factor that plays a central role in the DDR. The characteristics of SOG1 are similar to those of animal p53, even though the proteins’ amino acid sequences are unrelated. We suggest that plants acquired the central transcriptional factor SOG1 as a functional homolog of p53 during the evolution of their DDR system.
Keywords: DNA damage response, Arabidopsis, SOG1, p53, genome stability, phosphorylation, ATM, ATR, Ionizing radiation
Introduction
Genomic DNA of living organism accumulates thousands of lesions every day.1 The majority of these lesions are caused by a variety of environmental and endogenous genotoxic insults, such as UV (UV) light, ionizing radiation (IR), and byproducts of normal cell metabolism, notably reactive oxygen species (ROS).2 DNA damage can have deleterious effects, as it prevents DNA replication and transcription and can finally result in mutations and chromosomal aberrations.3 Hence, protecting genomic DNA is indispensable not only to protect cells from DNA damage but also to ensure faithful transmission of genetic information from one generation to the next. To maintain the integrity of genomic DNA, therefore, eukaryotes have evolved a highly coordinated cellular system called the DNA damage response (DDR), which ultimately activates cell cycle checkpoints, DNA repair, and cell death.2 Defects in this system in animals contribute to various disorders, including cancer and developmental defects, which highlights the critical importance of an efficient DDR for the viability of both the cell and the organism.4
An appropriate and effective DDR system is also critical for plants. Unlike animals, plants are sessile and therefore under continuous exposure to environmental stresses that include drought, high and low temperatures, and high light intensity, all of which induce the production of ROS.5 ROS include singlet oxygen, superoxide anions and hydroxyl radicals, and these species damage DNA. Furthermore, although sunlight is required for plants to produce energy through photosynthesis, its UV component damages the genome; and photosynthesis itself generates ROS.6 Another source of environmental DNA damage is chemicals from soil. High levels of aluminum and boron in soil, for example, are known to negatively affect plant growth, and also to induce DNA damage.7,8 Plants are therefore, thought to be constantly exposed to DNA damage. Given their contrasting situations, plants and animals are likely to have evolved different strategies to minimize the deleterious effects of genotoxic environmental agents. The DDR pathway contains several key components: DNA damage sensors, signal transducers, mediators, and effectors.9 Most DNA damage sensors (e.g., MRE11/RAD50/NBS1 [MRN], RAD9/RAD1/HUS1 [9–1-1]) and several signal transducers (e.g., ataxia telangiectasia mutated [ATM], and ATM and Rad3-related [ATR]) are conserved between animals and plants.10,11,12,13,14 However, other signal transducers (CHK1 and CHK2) and also p53, an effector that acts as a transcription factor to regulate many genes, have not been identified in plants. These observations imply that the mechanisms for detection of DNA damage are well conserved between animals and plants, while the mechanisms for signal transduction and gene regulation have diverged.
This mini-review focuses on a plant-specific DDR factor, SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), which in Arabidopsis thaliana is a transcription factor playing an important role in the DDR. We outline recent progress in characterizing the function and regulatory mechanism of SOG1 in the DDR. From these collective results, we observed the striking functional similarities between plant SOG1 and animal p53. Because the amino acid sequences of SOG1 and p53 are unrelated, plants and animals subsequently and independently invented their own mechanisms for the transcriptional regulation of DDR. Therefore, the comparison between SOG1 and p53 would be a good model to study convergent evolution. To gain insight into when plants acquired the SOG1 system, we compared the amino acid sequences of SOG1 orthologs found in other plant species.
DNA Damage Responses in A. thaliana
Plants experiencing DNA damage activate various responses. Gamma irradiation of A. thaliana inhibits the expression of genes that promote M-phase progression, such as CDKB2;1 and KNOLLE.15 This observation suggests that DNA damage induces cell cycle arrest. Plants possess orthologs of most of the genes involved in the widely conserved DNA repair pathways, some of which (such as BRCA1, RAD51, PARP-1) are rapidly induced by treatment with agents that induce DNA double-strand breaks (DSBs), such as bleomycin and gamma irradiation, meaning that plants activate a S phase and G2 phase associated homologous recombination for repairing DSBs.15,16,17,18 Stem cell death is also observed at roots and shoots in A. thaliana after treatment with UV or DSB inducers,19,20 and the latter agents induce endoreduplication in roots and sepals, which enables meristematic cells to stop dividing and promotes cell expansion.21 The most remarkable response to gamma irradiation in A. thaliana is a rapid (within 3 h of irradiation) and robust transcriptional regulation of numerous genes.15,17,22 This rapid transcriptional response is almost completely dependent on ATM, a protein kinase in the DDR pathway.15,17 More than 100 genes are upregulated, and about 30 downregulated, compared with unirradiated plants (1.5 h after irradiation, fold change cutoff (≥ 4), q-value < 0.05).15 Some of upregulated genes are involved in genome maintenance and metabolism (e.g., ribonucleotide reductase, DNA polymerases (delta) and (epsilon), and RPA-like genes), chromatin structure and maintenance (SYN2, a cohesin family protein), and DNA repair (e.g., BRCA1, RAD51, RAD17, and PARP-1). Some of the downregulated genes are involved in cell cycle regulation at late-S/G2 (CYCB1;2 and CDKB1;2), G2/M (CDKB2;1) and cytokinesis (KNOLLE). The suppression of these cell cycle-related genes probably reflects cell cycle arrest to allow time for DNA repair. As seen from the above, wild-type plants activate a wide range of responses - cell cycle arrest, DNA repair, stem cell death, endoreduplication and transcriptional regulation - to maintain their genome integrity.
The Role of the Plant-Specific DDR Factor SOG1: A Key Transcription Factor in the DDR System
SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1) is a plant-specific regulator that functions in the DDR.22 A. thaliana sog1–1 was originally isolated as a suppressor mutation of xpf-2, which is defective in DNA repair endonuclease activity.23 Gamma-irradiated xpf-2 mutant seeds display delayed development as seedlings: they germinate but cannot form true leaves for several days. However, the sog1–1 mutation suppresses the radiation-induced developmental delay. By map-based cloning, SOG1 was identified and found to be one of the NAC (NAM, ATAF1/2, and CUC2) proteins, which constitute one of the largest families of plant-specific transcription factors.22 Chromatin immunoprecipitation analysis, after treatment of seedlings with the DSB-inducing drug zeocin, showed that SOG1 acts as a transcription factor the binds to the promoters of SIAMESE-RELATED 5 and 7, two plant-specific CDK inhibitors that inhibit cell division.24 Plants carrying the pSOG1::SOG1-GUS construct showed GUS staining in the shoot and root apical meristems, and in lateral root primordia, demonstrating that SOG1 functions mainly in tissues displaying cell division activity.25 Fluorescence from a functional SOG1-GFP fusion protein was observed in the nucleus but not the cytoplasm, and neither the intensity nor the localization of the SOG1-GFP signal were affected by DNA damage,25 indicating that SOG1 function is independent of protein accumulation and subcellular localization.
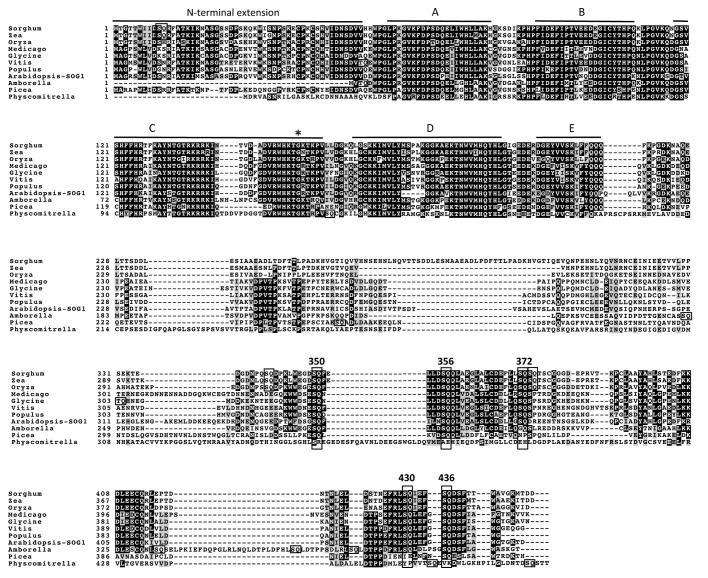
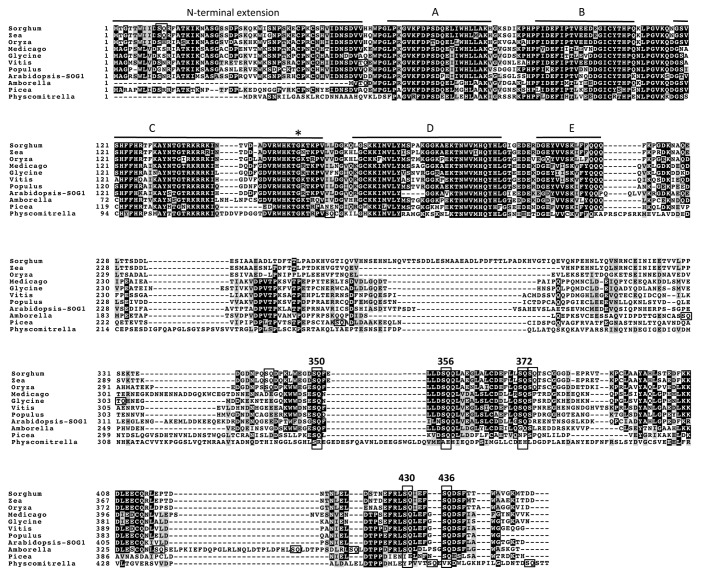
Many of the responses induced in wild-type plants by DNA damage were not observed in sog1–1 mutant plants. The sog1–1 mutation impairs transcriptional repression of the cell cycle-related genes CDKB2;1 and KNOLLE during the first 24 h after irradiation.22 Additionally, while root growth was inhibited after transfer of wild-type seedlings to zeocin-containing medium, sog1–1 seedlings kept growing, indicating that the sog1–1 mutant is more resistant to zeocin than wild-type, in terms of root growth (Fig. 1).21 These results suggest that SOG1 is involved in DNA damage-induced cell cycle arrest. The sog1–1 mutant does not show programmed cell death in response to DSB inducers or UV-B at the stem cells in the root apical meristem 24 h after treatment,20 indicating that SOG1 is involved in the stem cell death observed in wild-type. However, non-stem cell death is observed in the sog1–1 mutant 48 h after treatment.20 Because this non-stem cell death does not require SOG1 function, it may be a response to uncontrolled DNA damage. Furthermore, the sog1–1 mutant does not exhibit endoreduplication in roots 24 h after treatment with the DSB inducer zeocin, indicating that SOG1 is involved in this endoreduplication.21 Although numerous genes are up- and downregulated 1.5 h after irradiation in wild-type, this regulation is almost abolished in sog1–1 mutants,22 indicating that the great majority of the rapid transcriptional modulation response to DNA damage is regulated through SOG1. These results show that SOG1 is an important regulator of the DDR in plants. Indeed, the sog1–1 mutation exacerbated IR-induced loss of heterozygosity,22 indicating that the transcriptional response through SOG1 contributes to the maintenance of genetic integrity after DNA damage. Because SOG1 is required for the induction of endoreduplication of epidermal and cortex cells and cell death of stem cells in roots 24 h after DNA-damaging treatment, distinct sets of target genes may be regulated by SOG1 in different cell types. The sog1–1 mutation is a single base change (G to A) that creates a missense mutation (Gly (GGA) to Arg (AGA)),22 affecting a highly conserved amino acid in the NAC subdomain C that may be involved in DNA binding (Fig. 2).26 Therefore, the sog1–1 mutation may lead to deficiency of SOG1 binding to many target genes, and consequenctly, the misregulation of target genes may cause the pleiotropic phenotype observed in sog1–1 mutant.
Figure 1. Phosphorylation of SQ motifs is required for zeocin sensitivity of root growth. Four-day-old seedlings were transferred onto media lacking (A) or containing 5 µM (B) zeocin, and root growth was measured. Data represent mean values ± standard deviation from 3 independent experiments.
Figure 2. Sequence alignment of SOG1 with the six other NAC proteins in its subfamily. Numbers on the left indicate the adjacent residue position for each row. Black and gray boxes represent identical and similar residues, respectively. NAC subdomains A-E are shown by bars above the sequences. The alignment was performed with Clustal X and is displayed using BoxShade. The site of the A. thaliana sog1–1 mutation is marked with an asterisk, and the five SQ motifs found in SOG1 are boxed and numbered. SQ/TQ motifs found in the other aligned sequences are also boxed.
The Regulatory Mechanism of SOG1
SOG1-GFP expression and SOG1 immunoblotting experiments showed that the amount of SOG1 protein is not increased by DNA damage.25 Immunoblotting experiments also indicated that a small amount of SOG1 is phosphorylated independently of DNA stress by an unidentified kinase. In response to DSB-inducer treatment, part of the SOG1 population becomes hyperphosphorylated in an ATM-dependent and ATR-independent manner.25 These observations indicate that SOG1 is controlled by post-translational modification, not by transcriptional regulation. As ATR is known to be involved in the response to genotoxic stress that occurs during S phase,14,27 SOG1 phosphorylation status was examined after treatment with the replication inhibitors hydroxyurea and aphidicolin. Hyperphosphorylation was not observed after either treatment,25 suggesting that SOG1 is hyperphosphorylated in response to DSBs, but not to replication stress. Because no satisfactory antibody against A. thaliana ATM was available, ATM from human lymphoblastoid cells was immunoprecipitated with anti-ATM antibody and used for an in vitro kinase assay. SOG1 was phosphorylated by the human ATM immunoprecipitates in vitro.25 As the phosphatidylinositol 3-kinase domain of A. thaliana ATM shares 67% amino acid similarity with that of human ATM,28 A. thaliana ATM may directly hyperphosphorylate SOG1. The C-terminal region of SOG1 has five serine-glutamine (SQ) motifs,25 which are the preferred target for phosphorylation by human ATM and ATR.29,30 The DNA damage-dependent SOG1 hyperphosphorylation observed in wild-type plants disappeared in transgenic plants carrying mutant SOG1(5A), which encodes serine-to-alanine substitutions at all five SQ motifs,25 meaning that one or more of the SQ motifs are targets for the hyperphosphorylation. Although sog1–1 mutants carrying wild-type SOG1 (sog1–1/SOG1-Myc) can complement various sog1–1 phenotypes, namely the gamma-resistant leaf phenotype of xpf-2 sog1–1, the defective transcriptional response to irradiation, the impaired programmed cell death and the zeocin-resistant character of root growth, sog1–1 mutants carrying SOG1(5A) construct (sog1–1/SOG1(5A)-Myc) cannot complement them (Fig. 1).25 These results indicate that ATM-dependent hyperphosphorylation of the SQ motif(s) is essential for SOG1 functions. It will be necessary to clarify which SQ motif(s) are actually phosphorylated in response to DNA damage, and how SQ phosphorylation contributes to SOG1 function. SOG1 also seems to act in concert with ATR, because cell cycle arrest in xpf-2 seedlings grown from irradiated seeds requires ATR and SOG1 but not ATM.22 These results suggest that SOG1 participates in the pathways governed by both ATM and ATR DDR kinases.
Comparison Between Plant SOG1 and Animal p53
The animal tumor suppressor p53 is a critical transcription factor that governs the expression of many target genes involved in cell cycle control, DNA repair, apoptosis and senescence in response to DNA damage.31 The importance of the role of p53 in maintaining genome stability is exemplified by the finding that this molecule is mutated in more than 50% of human cancers.32 p53 is present at low levels under unperturbed conditions, but after exposure to DNA-damaging agents it is stabilized by post-translational modifications, and the stabilized p53 activates its target genes.33 Phosphorylation is the most important modification for p53 stabilization, and is performed by four DDR kinases: ATM, ATR, CHK1, and CHK2.31,34,35 Surveying the functions and regulatory mechanism of SOG1 demonstrates that its roles in the DDR system are comparable to those of p53 (Table 1). An obvious difference is that SOG1 function is not regulated by its own accumulation, as is the case for p53. These similarities between SOG1 and p53 lead us to propose that SOG1 is a functional homolog of p53. However, the amino acid sequences of the two proteins are unrelated to each other,25 suggesting that plants acquired plant-specific SOG1 during the evolution of the DDR.
Table 1. Comparison between plant SOG1 and animal p53.
| Animal p53 | Arabidopsis SOG1 |
| Functions | |
| Transcription factor | Transcription factor (NAC protein) |
| Activates transcriptional response | Activates transcriptional response |
| Involved in cell-cycle arrest | Involved in cell-cycle arrest |
| Stimulates apoptosis | Stimulates programmed cell death |
| Required for genome stability | Required for genome stability |
| Mechanisms of regulation (in response to DNA damage) | |
| Phosphorylated by ATM, ATR and CHK1/2 | Phosphorylated via ATM |
| p53 is stabilized | The amount of SOG1 does not change |
Alignment of SOG1 Orthologs Found in Other Plants
When did plants acquire the SOG1 during their evolution? Members of the NAC protein family have been identified in model plants like A. thaliana, Oryza sativa (rice), Pinus taeda (conifer), Selaginella moellendorffii (fern) and Physcomitrella patens (moss), but not in the unicellular green alga Chlamydomonas reinhardtii or in the colonial alga Volvox carteri.36 A. thaliana possesses 117 NAC genes, and phylogenetic analyses for NAC domains reveal that they comprise 21 subfamilies.36 Given that A. thaliana encodes so many NAC proteins, it is possible that some of them have similar roles to SOG1. The SOG1 (At1g25580, ANAC008) subfamily in A. thaliana includes seven NAC proteins (ANAC008 (SOG1), ANAC010, ANAC044, ANA073, ANAC075, ANAC085, and ANAC099).26 SOG1 has five SQ motifs in its transcription regulatory region at the C terminus, and the hyperphosphorylation of SQ motifs plays an important role in activating the DDR.37 To evaluate the possibility that other NAC proteins belonging to the SOG1 subfamily have similar roles to SOG1, we examined whether the other six members in same subfamily also have SQ motifs. Alignment of the amino acid sequences of all seven members of the SOG1 subfamily shows that the SQ motifs are not conserved (Fig. 2). Although some of the other subfamily members (ANAC010, ANAC073, ANAC075, ANAC085, ANAC099) have one or two C-terminal SQ/TQ motifs (both of these motifs are animal ATM/ATR substrates), their locations are unrelated to those of the SOG1 motifs, suggesting that they are not recent duplications and the DDR-related function of SOG1 is unique among A. thaliana NAC proteins. This conclusion is supported by the observation that the single mutation in sog1–1 plants causes various phenotypes, none of which can be complemented by the function of other NAC proteins. By identifying conserved SQ motifs, we may be able to find SOG1 orthologs in other plant species.
When the entire SOG1 amino acid sequence was used to search for SOG1-like proteins in other plants, and predicted SOG1 orthologs could be identified in most land plants, although we do not know whether the function of SOG1 is conserved in these species (Fig. 3).22 We tried to examine whether the SQ motifs found in SOG1 are conserved in these predicted SOG1 orthologs. Due to incomplete cDNA sequence information for S. moellendorffii (fern), we could not determine whether SOG1 orthologs exist in a fern. We used soybean (Glycine max) and barrel medic (Medicago truncatula) as eudicots, grape (Vitis vinifera) and black cottonwood (Populus trichocarpa) as eudicot trees, rice (O. sativa), maize (Zea mays) and sorghum (Sorghum bicolor) as monocots, Amborella (Amborella trichopoda) as an ancient flowering plant, conifer (P. glauca) as a gymnosperm, and moss (P. patens) as an embryophyte. All eudicot and monocot SOG1-like sequences have five C-terminal SQ motifs at conserved positions (Fig. 4). Furthermore, these predicted SOG1s have conserved N-terminal extensions of approximately 40 amino acid residues, which is another feature of SOG1 that distinguishes it from other NAC proteins.37 We also found two additional SQ motifs in the monocot sequences, which may be phosphorylated in response to DNA damage. Amborella, an ancestor of the angiosperms, possesses four of the five conserved C-terminal SOG1 SQ motifs. Although Amborella lacks the third motif (372SQ), it has four additional SQ motifs in its C-terminal region. Interestingly, P. glauca has a conserved N-terminal extension even though Amborella lacks the extension, and P. glauca has three SQ motifs (350SQ, 356SQ, and 436SQ) in its C-terminal region. Although two SQ motifs were found in the C-terminal region of the P. patens SOG1 ortholog, their positions differ from those of any of the angiosperm SOG1 SQ motifs. Furthermore, this SOG1 ortholog does not have N-terminal extension and it is not the only NAC protein that has SQ motifs in C-terminal region. It is thus unclear whether this ortholog is a SOG1-like protein, or whether other NAC protein(s) supply SOG1-like functions in mosses. Taken together, the results of these alignments lead us to propose that SOG1 had already been acquired in the gymnosperms.
Figure 3. Phylogenetic relationships of SOG-like proteins. The phylogenetic tree was constructed by the neighbor-joining method in MEGA5.2.2 based on the cDNA sequence of NAC domains. The NAC proteins in A. thaliana are labeled “ANAC.” The bracketed names are AGI_codes from TAIR (http://www.arabidopsis.org). ANAC005 and ANAC006 were used as outgroups. Bootstrap probabilities are given at each branching point. Values < 50 are not significant. The parenthetical numbers are DDBJ, NCBI, GenBank accession numbers (http://www.ddbj.nig.ac.jp, https://www.ncbi.nlm.nih.gov). The amino acid sequence of the starred (*) genes were used for alignment analysis in Figure 4.
Figure 4. Sequence alignment of A. thaliana SOG1 with predicted SOG1s in other species. For notation, see the legend to Figure 2. See the text for species names.
Summary
The transcription factor SOG1 is a plant-specific DDR factor, and many of its functions and regulatory mechanisms have been elucidated. SOG1 plays important roles in various aspects of the DDR, including cell cycle arrest, DNA repair, programmed cell death and endoreduplication. SOG1 is regulated by ATM-mediated hyperphosphorylation. The roles and regulatory mechanism of SOG1 are analogous with those of animal p53. Because the amino acid sequences of these two proteins are unrelated, it seems that plants have independently acquired SOG1 as a central transcriptional regulator of the DDR.
No NAC protein family members have been identified in unicellular plants suggesting that NAC may have arisen after the transition from water to land.36 Phylogenetic analyses of NAC proteins identified in land plants (eudicots, monocots, ferns, and mosses) showed that several of major subfamilies already existed in early-diverged land plants (mosses or ferns), whereas the others diverged within the angiosperms.36 Although we previously reported that predicted SOG1 orthologs were found in most land plants,22 the alignment analysis of SQ motifs showed that it is difficult to determine if mosses have a SOG1-like protein. Because SQ motifs found in SOG1 are conserved in eudicots, eudicot trees, monocots, an ancient flowering plant (Amborella) and gymnosperms, we propose that SOG1 had already been acquired in the gymnosperms. More sequence data of ferns is required to perform detailed analyses to determine when SOG1 genes arose in the evolution of plants.
As the DDR described here was usually triggered experimentally by two DSB inducers, gamma irradiation and zeocin, DSBs probably caused the activation of the DDR response. However, it is not known whether more generic environmental stresses can also induce DSBs. Although ROS production is likely to be a major source of DNA damage under stress conditions in plants,5,38 DSBs are not thought to be a frequent consequence of ROS induction. Furthermore, methyl viologen, which is often used to catalyze the formation of ROS, does not induce BRCA1 or RAD51 expression (Genevestigator: https://www.genevestigator.com). Therefore, ROS seems not to activate DDR. Because IR also produces other DNA lesions such as AP sites, DNA-DNA cross-links, and clustered damage sites that are complex and less readily repaired,39 these types of damage may be more important inducers of the DDR system in plants.
Plants transcriptionally induce numerous genes in response to DNA damage. For instance, some DNA repair genes (e.g., BRCA1, RAD17, and PARP-1) are strongly induced by gamma irradiation in plants.15 However, the transcripts induced by gamma irradiation of human cells bear little relation to those induced in plants. Although BRCA1 is induced by gamma irradiation in human cells, the induction is not as strong as that in Arabidopsis.40 RAD17 and PARP-1 are not induced in human cells.41 This observation implies that the human orthologs of these genes are regulated in a different fashion, such as post-translationally. Furthermore, although this robust transcriptional response in plants is regulated by a single protein, SOG1, the transcriptional response to DNA damage in animals is not exclusively regulated by p53. It remains unclear why plants have such a strong transcriptional responses regulated only by SOG1. However, plants may, under normal conditions, need to repress more genes than animals do in order to save energy, or they may need to change gene expression drastically in response to a stressful environment.
To deepen our understanding of plant-specific DDR, it is necessary to study SOG1 in greater detail. The identification of genes that are directly regulated by SOG1 is important to know how signal transduction occurs in response to DNA damage. Furthermore, we need to identify the factors that interact with SOG1 to understand how SOG1 activity is regulated. Further studies are required to determine which SQ motif(s) is/are phosphorylated and to determine the significance of each phosphorylation event in SOG1 activation. Another remaining question is whether plants acquired SOG1 before the divergence of gymnosperms. If so, it would be interesting to determine whether SOG1’s functions differ in different species. In Arabidopsis, SOG1’s functions clearly differ in different cell types. Because most of the work on the transcriptional aspects of DDR in plants was performed with whole seedlings, it remains to be determined whether SOG1 triggers different transcriptional response in different cell types. Addressing these open questions will have profound implications for our understanding of the evolution of DDR in plants, and how plants’ specific responses to DNA damage have helped them succeed in stressful environments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank H. Nakayama (Kyoto Sangyo Univ.) for helpful discussion and advice. This work was supported in part by grants from the Japan Society for the Promotion of Science to K. O. Y., and from MEXT (KAKENHI, Grant Number 22119009) and JST, CREST to M. U. Thanks go to K. Kaminoyama for analyzing the root sensitivity of sog1–1 mutant lines to a DNA damage agent. We thank I. Smith for refining the English.
References
- 1.Lindahl T, Barnes DE. . Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000; 65:127 - 33; http://dx.doi.org/ 10.1101/sqb.2000.65.127; PMID: 12760027 [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. . The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179 - 204; http://dx.doi.org/ 10.1016/j.molcel.2010.09.019; PMID: 20965415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis Second Edition. Amer Society for Microbiology, Washington, DC USA 2005. [Google Scholar]
- 4.Jackson SP, Bartek J. . The DNA-damage response in human biology and disease. Nature 2009; 461:1071 - 8; http://dx.doi.org/ 10.1038/nature08467; PMID: 19847258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter A, Mittler R, Suzuki N. . ROS as key players in plant stress signalling. J Exp Bot 2013; PMID: 24253197 [DOI] [PubMed] [Google Scholar]
- 6.Foyer CH, Shigeoka S. . Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol 2011; 155:93 - 100; http://dx.doi.org/ 10.1104/pp.110.166181; PMID: 21045124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nezames CD, Sjogren CA, Barajas JF, Larsen PB. . The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell 2012; 24:608 - 21; http://dx.doi.org/ 10.1105/tpc.112.095596; PMID: 22345493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T. . Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell 2011; 23:3533 - 46; http://dx.doi.org/ 10.1105/tpc.111.086314; PMID: 21917552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. . Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73:39 - 85; http://dx.doi.org/ 10.1146/annurev.biochem.73.011303.073723; PMID: 15189136 [DOI] [PubMed] [Google Scholar]
- 10.Gallego ME, White CI. . RAD50 function is essential for telomere maintenance in Arabidopsis. Proc Natl Acad Sci U S A 2001; 98:1711 - 6; http://dx.doi.org/ 10.1073/pnas.98.4.1711; PMID: 11172016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heitzeberg F, Chen IP, Hartung F, Orel N, Angelis KJ, Puchta H. . The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 2004; 38:954 - 68; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02097.x; PMID: 15165187 [DOI] [PubMed] [Google Scholar]
- 12.Bundock P, Hooykaas P. . Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 2002; 14:2451 - 62; http://dx.doi.org/ 10.1105/tpc.005959; PMID: 12368497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A. . AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003; 15:119 - 32; http://dx.doi.org/ 10.1105/tpc.006577; PMID: 12509526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culligan K, Tissier A, Britt A. . ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004; 16:1091 - 104; http://dx.doi.org/ 10.1105/tpc.018903; PMID: 15075397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. . ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 2006; 48:947 - 61; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02931.x; PMID: 17227549 [DOI] [PubMed] [Google Scholar]
- 16.Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. . The transcriptional response of Arabidopsis to genotoxic stress - a high-density colony array study (HDCA). Plant J 2003; 35:771 - 86; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01847.x; PMID: 12969430 [DOI] [PubMed] [Google Scholar]
- 17.Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH. . ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS One 2007; 2:e430; http://dx.doi.org/ 10.1371/journal.pone.0000430; PMID: 17487278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West CE, Waterworth WM, Sunderland PA, Bray CM. . Arabidopsis DNA double-strand break repair pathways. Biochem Soc Trans 2004; 32:964 - 6; http://dx.doi.org/ 10.1042/BST0320964; PMID: 15506937 [DOI] [PubMed] [Google Scholar]
- 19.Fulcher N, Sablowski R. . Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci U S A 2009; 106:20984 - 8; http://dx.doi.org/ 10.1073/pnas.0909218106; PMID: 19933334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BW, Aggoune D, Hays JB, Britt AB. . A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 2010; 9:940 - 8; http://dx.doi.org/ 10.1016/j.dnarep.2010.06.006; PMID: 20634150 [DOI] [PubMed] [Google Scholar]
- 21.Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. . Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci U S A 2011; 108:10004 - 9; http://dx.doi.org/ 10.1073/pnas.1103584108; PMID: 21613568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshiyama K, Conklin PA, Huefner ND, Britt AB. . Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A 2009; 106:12843 - 8; http://dx.doi.org/ 10.1073/pnas.0810304106; PMID: 19549833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preuss SB, Britt AB. . A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 2003; 164:323 - 34; PMID: 12750343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al. . The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 2014; 26:296 - 309; http://dx.doi.org/ 10.1105/tpc.113.118943; PMID: 24399300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M. . ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 2013; 14:817 - 22; http://dx.doi.org/ 10.1038/embor.2013.112; PMID: 23907539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. . Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 2003; 10:239 - 47; http://dx.doi.org/ 10.1093/dnares/10.6.239; PMID: 15029955 [DOI] [PubMed] [Google Scholar]
- 27.Cimprich KA, Cortez D. . ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616 - 27; http://dx.doi.org/ 10.1038/nrm2450; PMID: 18594563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia V, Salanoubat M, Choisne N, Tissier A. . An ATM homologue from Arabidopsis thaliana: complete genomic organisation and expression analysis. Nucleic Acids Res 2000; 28:1692 - 9; http://dx.doi.org/ 10.1093/nar/28.8.1692; PMID: 10734187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. . A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 1999; 13:152 - 7; http://dx.doi.org/ 10.1101/gad.13.2.152; PMID: 9925639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. . Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 1998; 281:1677 - 9; http://dx.doi.org/ 10.1126/science.281.5383.1677; PMID: 9733515 [DOI] [PubMed] [Google Scholar]
- 31.Rozan LM, El-Deiry WS. . p53 downstream target genes and tumor suppression: a classical view in evolution. Cell Death Differ 2007; 14:3 - 9; http://dx.doi.org/ 10.1038/sj.cdd.4402058; PMID: 17068503 [DOI] [PubMed] [Google Scholar]
- 32.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. . Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007; 28:622 - 9; http://dx.doi.org/ 10.1002/humu.20495; PMID: 17311302 [DOI] [PubMed] [Google Scholar]
- 33.Lavin MF, Gueven N. . The complexity of p53 stabilization and activation. Cell Death Differ 2006; 13:941 - 50; http://dx.doi.org/ 10.1038/sj.cdd.4401925; PMID: 16601750 [DOI] [PubMed] [Google Scholar]
- 34.Shieh SY, Ikeda M, Taya Y, Prives C. . DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91:325 - 34; http://dx.doi.org/ 10.1016/S0092-8674(00)80416-X; PMID: 9363941 [DOI] [PubMed] [Google Scholar]
- 35.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. . The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 2000; 14:289 - 300; PMID: 10673501 [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu T, Nevo E, Sun D, Peng J. . Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 2012; 66:1833 - 48; http://dx.doi.org/ 10.1111/j.1558-5646.2011.01553.x; PMID: 22671550 [DOI] [PubMed] [Google Scholar]
- 37.Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. . The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 2010; 426:183 - 96; http://dx.doi.org/ 10.1042/BJ20091234; PMID: 19995345 [DOI] [PubMed] [Google Scholar]
- 38.Gill SS, Tuteja N. . Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 2010; 48:909 - 30; http://dx.doi.org/ 10.1016/j.plaphy.2010.08.016; PMID: 20870416 [DOI] [PubMed] [Google Scholar]
- 39.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. . Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res 2002; 30:3464 - 72; http://dx.doi.org/ 10.1093/nar/gkf467; PMID: 12140332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding LH, Park S, Peyton M, Girard L, Xie Y, Minna JD, Story MD. . Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to γ-rays and different elemental particles of high Z and energy. BMC Genomics 2013; 14:372; http://dx.doi.org/ 10.1186/1471-2164-14-372; PMID: 23724988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieger KE, Chu G. . Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res 2004; 32:4786 - 803; http://dx.doi.org/ 10.1093/nar/gkh783; PMID: 15356296 [DOI] [PMC free article] [PubMed] [Google Scholar]