Abstract
Mitogen activated protein kinase kinase (MAPKK) is the central module of MAPK cascade and also point of signal integration and divergence. To investigate the regulatory role of OsMKK6, the regulon of genes controlled by OsMKK6 was constructed by microarray analysis between constitutively activated overexpressing transgenic lines and the wild type rice. Regulated genes were identified in overexpressed constitutively activated OsMKK6 and they were further subdivided on the basis of functional categories, viz. transcription, metabolism, signaling, defense and unknown function. These findings suggest the possible physiological role of OsMKK6 in modulating gene expression and signaling pathways during different stresses.
Keywords: Oryza sativa, Rice, Microarray, Mitogen activated protein kinase kinase, MAPKK, MAPKK6
Plants have developed sophisticated signaling machineries to protect them and to adapt their cellular metabolism in response to various forms of environmental biotic and abiotic stresses. The mitogen activated protein kinase (MAPK) transduction cascades connect the perception of these external environmental stimuli to physiological and cellular responses and have important mechanisms for stress adaptation by control of gene expression. The well conserved MAPK pathway consist of a protein kinase module that phosphorylate in an ordered cascade advanced from a MAPK kinase kinase (MAPK3K, MAPKKK, MKKK, MEKK) to a MAPK kinase (MAP2K, MAPKK, MKK, MEK), and then to a MAP kinase (MAPK).1,2 MAPK are activated by phosphorylation on their threonine and tyrosine (TXY) residues located in the activation loop (T-loop) by MAP2K. MAP2K are dual specificity kinases and gets activated by phosphorylating two amino acids in the S/T-X3–5-S/T motif of the activation loop.3
Arabidopsis genome analysis revealed the presence of 20 members of MAPKs, 10 MAP2Ks and around 80 MAP3Ks.1,4,5 The 10 MAP2K genes of Arabidopsis are classified into four groups (A-D) based on their protein sequence alignments. Rice (Oryza sativa L.), a monocot cereal research model, is one of the most important food crops worldwide.6,7 The completion of sequencing project revealed the presence of 15 MAPKs, 8 MAP2Ks, and 75 MAP3Ks in rice genome.5,8 Out of eight MAP2Ks reported from rice, a partial functional characterization of only OsMKK6 has been performed so far. OsMKK6 (earlier named as OsMEK1) specifically and physically interacted with OsMPK3 (earlier named as OsMAP1) in moderate low temperature stress signaling as shown by yeast two hybrid assay.9 Transcript studies have revealed that OsMKK4 and OsMKK6 are strongly regulated by salt and cold stress.10 OsMKK4-OsMPK3 module was shown to be involved in arsenite stress signal transduction.11 A MAP2K (OsMEK1), identified as OsWNK1 (With No Lysine kinase) in rice and its probable role in circadian rhythm and abiotic stress has been reported.12 Dominant mutants of constitutively active form of protein kinases are shown to be useful for determining various protein functions.13,14 The functional role of OsMKK6 in salinity stress using transgenic approach was showed in our report recently.15 A constitutively active form of OsMKK6 was generated by substituting putative phosphorylation sites from S and T to E residues (S221E and T227E by site directed mutagenesis). Transgenic rice overexpressing constitutive active OsMKK6EE exhibited higher expression of phytoalexin biosynthesis pathway genes upon UV stress and infection with Magnaporthe oryzae suggesting their role in biotic stress.16 In the present study, global gene expression analysis was performed for the overexpressed constitutive active OsMKK6 lines.
To define the total regulon of genes controlled by OsMKK6, DNA microarray analysis was performed in three constitutively active OsMKK6EE overexpressing lines and three wild type Pusa Basmati (PB1) plants. Two week old overexpressed lines and wild types rice shoots were subjected for total RNA extraction using the Qiagen RNeasy Mini kit (Qiagen, USA) according to manufacture’s protocols. Quality and integrity of RNA was checked by measuring A260/280 and RNA integrity number by Agilent BioAnalyzer (Agilent Technologies, USA). Samples were labelled using the Agilent low input RNA amplification kit and QC was performed using ND-1000 spectrophotometer measurements (NanoDrop Technologies, USA), and profile of amplified RNA was checked using Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Three independent transgenic lines of OsMKK6EE and three independent wild type rice shoots were used for the experiment. The transcriptome analysis was performed using the Agilent’s rice custom 44k array AMADID (Agilent Technologies, USA). Microarray experiments including the data analysis were performed according to manufacturer’s instruction (Agilent technologies, USA). Feature extraction and image analysis software (v7.3; Agilent Technologies, USA) was used to locate and delineate every spot in the array and to integrate each spots intensity, filtering and normalization by the Lowess method (P value cut off as 0.01). The statistical significance of gene expression was tested using a one way analysis of variance test combined with Benjamini and Hochberg false discovery rate multiple correction algorithm (Genespring 7.3) with a corrected P < 0.05 set as cutoff. The normalization was done using Genespring GX using the recommended per chip and per gene data. Changes in signal intensity between transgenic and wild type experiments exceeding 3-fold or higher difference in repeated experiments were considered significant. The genes with a signal ratio of OsMKK6EE/WT ≥ 2 (upregulated) or ≤ 0.5 (downregulated) were considered differentially expressed. Genes were considered differentially expressed for P ≤ 0.05 in the t test.
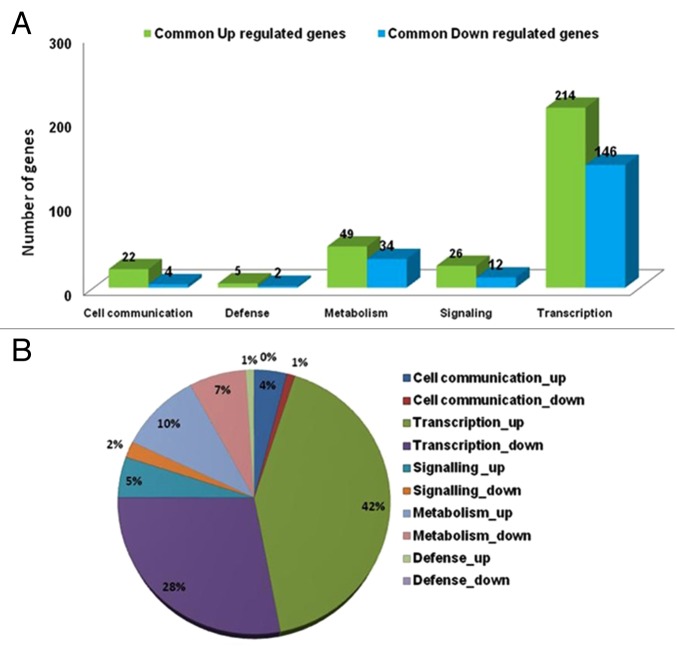
Gene expression ratios were calculated and expression levels of transgenic and wild type were compared. A final set of 316 common genes in all transgenic lines showed upregulation more than 2-folds, while 198 common genes in all transgenic lines showed downregulation and these genes were further subdivided into five functional categories (Fig. 1A). 214 genes constituting 42% of upregulated genes and 146 genes constituting 28% of downregulated genes were involved in transcription. 10% of upregulated genes and 7% of downregulated genes were involved in metabolism. 5% of upregulated and 2% of downregulated genes were involved in signaling, 4% of upregulated and 1% of downregulated genes were involved in cell communication and 1% of upregulated genes were involved in defense responses (Fig. 1A, 1B). Nevertheless, about one third of upregulated and downregulated genes were annotated with unknown function. Some of the common upregulated and downregulated genes with their putative function are listed (Table S1) and plotted as heat map in supporting information (Fig. S1).
Figure 1. Functional classification of upregulated and downregulated genes in the OsMKK6EE overexpressed transgenic plant. (A) Bar diagram representing number of genes upregulated and dow-regulated in relation to function in overexpressed lines compared with wild types. (B) Pie-diagram showing functional classification of common upregulated and downregulated genes in the overexpressed transgenic plant.
Validation of microarray data was performed by real-time quantitative RT–PCR (qRT–PCR) analysis of 2 week old rice shoots using StepOne real-time PCR systems (Applied Biosystems, USA). The specific primers used in this qRT-PCR study are listed in supporting information (Table S2). Pathogenesis related-1 protein gene (PR1), Lipoxygenase 9 (Lip9), WRKY transcription factor 50 (WRKY50), MAPK1, Salt responsive gene (SalT), abscisic acid responsive gene (ABA), cytosolic ascorbate peroxidase (APX) and dehydration responsive element binding protein 1 (DREB1) were upregulated in overexpressed plants compared with wild type plants (Fig. 2). SalT is involved in salt stress17 and Lip9 has role in low temperature signal transduction pathway.18 ABA, APX, and the transcription factor OsDREB1 are known to be upregulated in abiotic stresses.19,20 PR1 is involved in pathogen response and Ribulose biphosphate carboxylase (RbCs) transcript remain unaltered in overexpressed lines compared with wild type control. In microarray analysis, WRKY 50, Lip9, MAPK1, SalT, DREB1, and ABA showed upregulation more than 3-folds. RbCs showed no changes in transgenic plant compared with wild type. The transcript abundance from qRT-PCR and microarray of 10 selected genes were positively correlated, which suggests that transcripts quantified by qRT-PCR showed similar regulation pattern as in DNA chip assay.
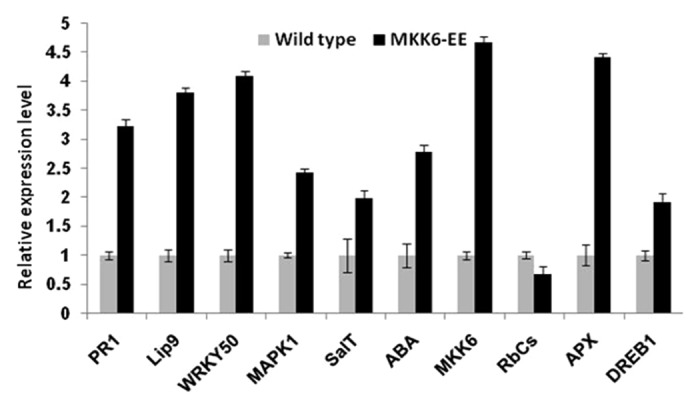
Figure 2. Real-time PCR quantification of relative expression levels of important genes regulated under different abiotic stress. Black bar denotes control wild type plant and gray bar denotes OsMKK6EE overexpressed transgenic plants. Error bars indicate standard error of three independent biological samples.
A final set of 316 genes showed upregulation and 198 genes showed downregulation in the constitutively active OsMKK6EE transgenic plants as compared with the wild types. Comprehensive expression analysis by gene chip microarray revealed that a substantial proportion of transcription factors were expressed differentially in OsMKK6EE transgenics. In agreement with the previous transcriptome analysis toward plant stress, multiple types of transcription factors constitute the largest group of upregulated genes.21,22As 1% of upregulated genes were also involved in defense, it suggest putative involvement of OsMKK6 in plant pathogen defense along with abiotic stress tolerance.16 OsMAPK1 also showed enhanced expression level in microarray analysis as well as in qRT-PCR analysis which has already been reported as OsMKK6 interacting partner.10 The genes quantified by qRT-PCR analysis showed similar expression pattern as in DNA chip analysis. OsMKK6EE plants showed enhanced expression of PR1, Lip9, APX, DREB1, SalT, and WRKY50 suggesting that these plants might be more tolerant to abiotic and biotic stresses. The constitutive expression of the OsMKK6 gene suggests that a pathways involving OsMKK6 might regulate the activity of downstream MAPKs and the transcription factors, which may ultimately regulate various abiotic and biotic stress responsive genes.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Department of Biotechnology, Government of India. KK acknowledge University Grant Commission, India for providing senior research fellowship during the course of study.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/28502
References
- 1.Jonak C, Okrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5:415–24. doi: 10.1016/S1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–49. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 3.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 4.MAPK Group Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–8. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 5.Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 2011;6:196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 8.Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–8. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Wen JQ, Oono K, Imai R. Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 2002;129:1880–91. doi: 10.1104/pp.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar K, Rao KP, Sharma P, Sinha AK. Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiol Biochem. 2008;46:891–7. doi: 10.1016/j.plaphy.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Rao KP, Vani G, Kumar K, Wankhede DP, Misra M, Gupta M, Sinha AK. Arsenic stress activates MAP kinase in rice roots and leaves. Arch Biochem Biophys. 2011;506:73–82. doi: 10.1016/j.abb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Kumar K, Rao KP, Biswas DK, Sinha AK. Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signal Behav. 2011;6:316–20. doi: 10.4161/psb.6.3.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 2002;29:637–47. doi: 10.1046/j.0960-7412.2001.01246.x. [DOI] [PubMed] [Google Scholar]
- 14.Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15:141–52. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Kumar K, Sinha AK. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice (N Y) 2013;6:25. doi: 10.1186/1939-8433-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wankhede DP, Kumar K, Singh P, Sinha AK. Involvement of mitogen activated protein kinase kinase 6 in UV induced transcripts accumulation of genes in phytoalexin biosynthesis in rice. Rice (N Y) 2013;6:35. doi: 10.1186/1939-8433-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia AB, Engler JdeA, Claes B, Villarroel R, Van Montagu M, Gerats T, Caplan A. The expression of the salt-responsive gene salT from rice is regulated by hormonal and developmental cues. Planta. 1998;207:172–80. doi: 10.1007/s004250050470. [DOI] [PubMed] [Google Scholar]
- 18.Lee SC, Huh KW, An K, An G, Kim SR. Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.) Mol Cells. 2004;18:107–14. [PubMed] [Google Scholar]
- 19.Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–53. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y, Masuta Y, Saito K, Murayama S, Ozawa K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 2011;30:399–406. doi: 10.1007/s00299-010-0985-7. [DOI] [PubMed] [Google Scholar]
- 21.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–90. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.