Abstract
The REIL1 and REIL2 proteins of Arabidopsis thaliana are evolutionarily conserved homologs of the cytosolic 60S ribosomal maturation factors Rei1 and its paralog Reh1 of Saccharomyces cerevisiae. We previously demonstrated that the REIL proteins like the yeast homologs are required for the growth of both organisms at suboptimal temperatures. In addition, the cold sensitivity of the yeast Δrei1 mutant was almost fully rescued by heterologous expression of the REIL1 protein. These phenomena and conservation of co-expressed genes linked the function of REIL proteins to the maturation of the eukaryotic ribosome in A. thaliana. Here we demonstrate that REIL proteins interact in yeast-2-hybrid assays with A. thaliana homologs of the yeast proteins, Rlp24, Rpl24A, Rlp24B, Arx1, and Jjj1. These proteins take part in the cytosolic 60S ribosomal maturation process within yeast and physically interact with Rei1. Our study does not provide proof but is consistent with a conserved role of the A. thaliana REIL proteins in ribosomal maturation and demonstrates the potential of future investigations that aim to unravel the protein interactions of REIL proteins in planta.
Keywords: Cold sensitivity, REIL proteins, cytosolic maturation of the eukaryotic ribosome, leaf growth, protein–protein interaction, yeast-2-hybrid assay
The REIL1 and REIL2 proteins are encoded by the genes At4g31420 and At2g24500, respectively. REIL homologs occur in eukaryotes and have 4 conserved zinc finger domains. REIL proteins of A. thaliana are homologs of Rei1, a cytosolic 60S ribosomal maturation factor from yeast, and its paralog Reh1. We recently demonstrated that T-DNA insertion mutations located in the exons of REIL1 and REIL2 are silent at optimal growth temperature but are required for leaf growth at 10 °C.1 A reil1–1 reil2–1 double mutant is arrested at 10 °C prior to the emergence of the first rosette leaf, 2 allelic reil2 mutants, reil2–1 and reil2–2, are dwarfed in the cold compared with the A. thaliana Col-0 wild type and form small spoon-shaped leaves during the vegetative growth phase. In contrast, the reil1–1 mutant grows normally at 10 °C except for slightly delayed germination.1 The reil mutants of A. thaliana are equivalent in these aspects to the homologous yeast deletion mutants. The yeast Δreh1 mutant has no effect on growth in the cold, the Δrei1 mutant is cold sensitive even at moderately suboptimal temperatures, whereas the Δrei1Δreh1 double mutant is more cold sensitive.2-4 Furthermore, the REIL2 gene of A. thaliana and the Rei1 gene of yeast are both co-expressed with genes that are phylogenetically conserved in both species.1 The co-expressed genes are mostly part of the complex maturation machinery of eukaryotic ribosomes. The conserved co-expression pattern in A. thaliana points to a conserved function of REIL proteins. We supported this conclusion by heterologous complementation of the cold sensitivity of the yeast Δrei1 deletion mutant. Specifically, the heterologous expression of REIL1 complemented the growth deficit of Δrei1 mutant at suboptimal temperatures almost completely. In this study we explore whether the physical protein–protein interactions of the yeast Rei1 protein have an informative value in regard to its function and whether the REIL proteins of A. thaliana may have conserved interacting proteins.
Protein interactions play a crucial role for ribosomal maturation in yeast. The current model of the cytosolic maturation steps that are necessary after nuclear export to render the pre-60S subunit of the eukaryotic ribosome translational active contains 20 proteins (Fig. 1). These proteins are recruited from the cytosol to the 60S subunit or are released from the pre-60S subunit after nuclear export.5 A part of the proteins is only transiently associated with the 60S subunit during the process. The maturation follows a main path that first releases the nuclear export proteins, Rlp24 and Nog1, and then removes factors that inhibit further maturation and translational activity, namely the Arx1/Alb1 complex, Tif6, and Nmd3. A side branch removes Mrt4 and thereby enables the assembly of the ribosome stalk (Fig. 1).
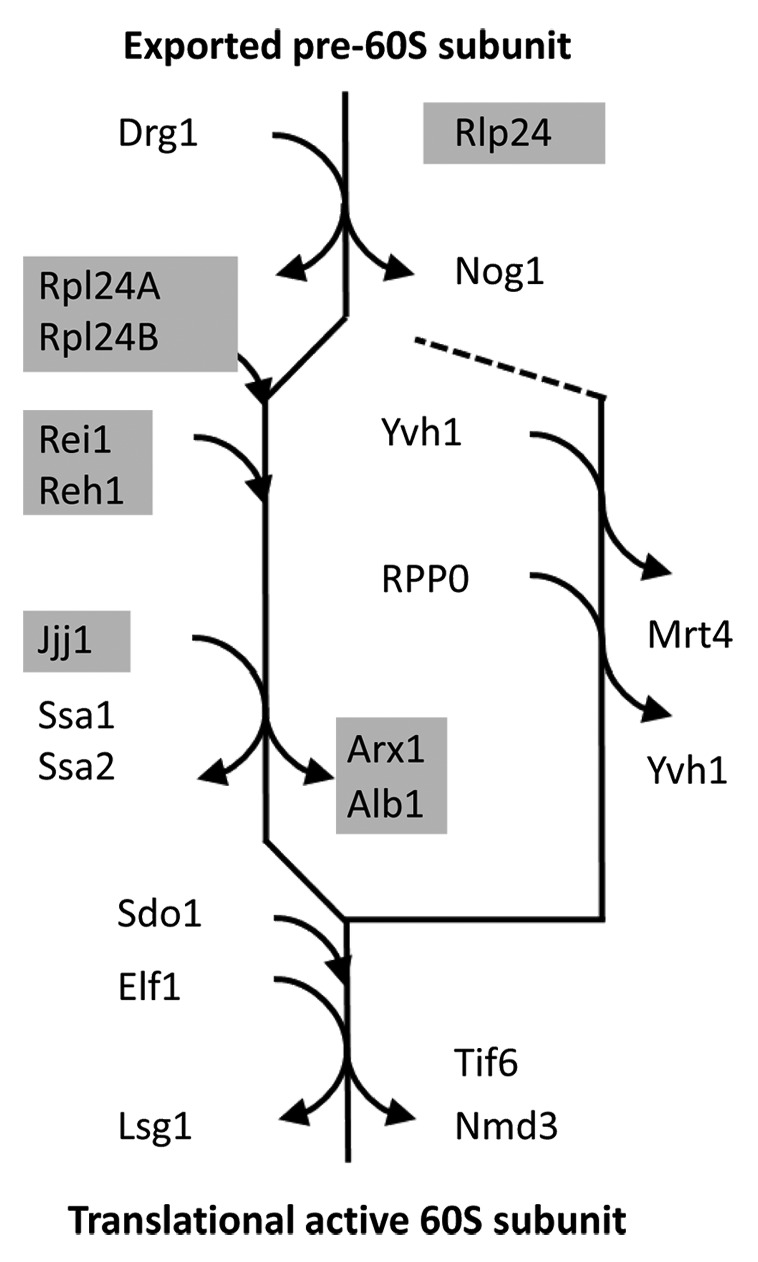
Figure 1. The cytosolic maturation machinery of the 60S large ribosomal subunit from Saccharomyces cerevisiae. The flow scheme represents the current model of the cytosolic maturation machinery5 starting with the pre-60S subunit after export from the nucleus (top) and ending with the translational active 60S ribosomal subunit (bottom). The direct mechanistic interaction partners of the yeast Rei1 protein within the model are indicated by light gray underlay. The cytosolic maturation releases inhibitory factors, such as the Arx1/Alb1 complex, Tif6, and Nmd3, (main path on the left) and enables assembly of the ribosome stalk (side branch on the right).
The Rei1 protein has the role of a transiently associated release factor. Rei1 acts together with Jjj1 in the Ssa1/Ssa2-dependent release of the Arx1/Alb1 complex from the pre-60S subunit.6 Deletion of Rei1 or Jjj1 in yeast blocks the release of Arx1/Alb1 and thereby arrests maturation. The Rei1 protein is recruited from the cytosol to pre-60S subunits after release of Rlp247 and replacement of Rlp24 by the closely related paralogs, Rpl24A or Rpl24B. Y2H-interactions of Rei1 with Rpl24B explain the recruitment of Rei1 to the pre-60S subunit of the eukaryotic ribosome, but the binding of Rei1 seems to involve additional factors.3,5 The role of Reh1 is partially redundant to Rei1. However, the details of functional diversification between the Rei1 and Reh1 paralogs in yeast still need to be investigated.4
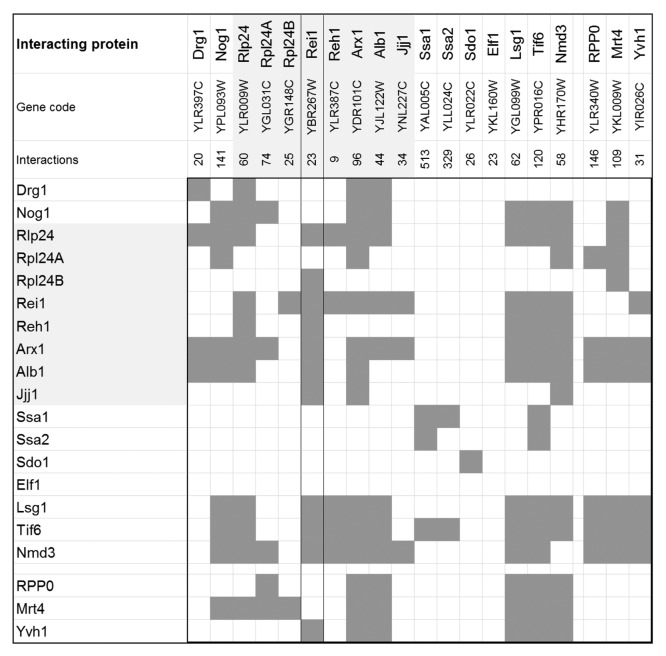
Physical protein–protein interactions play a crucial role not only for Rei1 function but for many cytosolic maturation steps of the 60S ribosomal subunit. This fact is reflected by the currently known physical protein–protein interactions among the 20 members of the cytosolic maturation machinery (Fig. 2). The underlying protein interaction data of the interaction matrix compiled in this figure were retrieved from the BioGRID database version 3.2.106.8 The members of the yeast maturation machinery have, on average, 97 physical interaction partners (Fig. 2). The Hsp70 family ATPases, Ssa1 and Ssa2, are the most highly interconnected components of the machinery with as many as 329 and 513 interacting proteins, respectively. In contrast, Rei1 and Reh1, have comparatively few interactions. Many of these interactions are with other members of the cytosolic maturation machinery. In detail, 11 of 23 physical interaction partners of Rei1 belong to the cytosolic maturation machinery. Reh1 has 5 out of 9 respective interactions (Fig. 2). Rei1 shows self-interaction and interacts with Reh1 as well as with most direct functional neighbors in the maturation pathway, namely with Rlp24, Rpl24B, Arx1, Alb1, and Jjj1. The downstream release factors Ssa1, Ssa2, Sdo1, and Elf1, neither interact with Rei1 nor with any other member of the maturation factors. Rei1 interacts also with Yvh1 of the stalk formation branch and furthermore, with Lsg1, Tif6, and Nmd3. These proteins represent the last maturation steps that render the 60S subunit active.
Figure 2. Protein–protein interaction matrix of the cytosolic maturation machinery of the 60S large ribosomal subunit from Saccharomyces cerevisiae. Each dark gray box within the matrix indicates a physical interaction between the cytosolic maturation factors. Interactions were retrieved and consolidated from BioGRID (Version 3.2.106). The protein names, corresponding gene codes, and the total number of physically interacting proteins are listed in the first 3 rows. The rows and columns are symmetrically ordered according to the sequence of the proteins acting within the main path of the model followed by the proteins relevant for stalk assembly, namely RPP0, Mrt4, and Yvh1. The interactions of the Rei1 protein are highlighted by a framed column. The light gray underlay accentuates the direct mechanistic interaction partners of Rei1, also refer to (Fig. 1).
Our in silico analysis demonstrates that the Rei1 and Reh1 proteins interact with few yeast proteins and that many of the few interactions reflect their specific function. We can therefore expect that the protein–protein interactions of the REIL proteins in A. thaliana will reflect their conserved function.
Information on the protein interactions of REIL proteins is scarce. The REIL proteins like the yeast homologs appear to interact with only few proteins. The current version of the BioGRID database lists no interaction for REIL1 and only 3 interactions for REIL2. The 3 known interacting proteins, namely At2g37150, a RING/U-box superfamily protein, At5g51910, a TCP family transcription factor, and At5g61010, a member of the exocyst subunit EXO70 gene family, so far do not provide clear information on the function of REIL proteins.9 The Y2H technology allows rapid testing of protein interactions by heterologous expression of interaction partners in yeast. Y2H can be applied before molecular tools, e.g., tagged proteins or specific antibodies, are ready for in planta analysis of protein–protein interactions. Y2H technology appeared to be feasible for the targeted analysis of REIL interactions, because this technology had previously been used to study interactions that involve the yeast Rei1 protein.2,3 To investigate the potential of future, more sophisticated protein interaction studies of REIL proteins in A. thaliana we analyzed the Y2H interactions of REIL1 and REIL2 with the plant homologs of the direct mechanistic interaction partners of Rei1 (Figs. 1 and 2), namely the gene products of At2g44860 (homolog of Rlp24), At2g36620, At3g53020 (homologs of Rpl24A and Rpl24B), At3g51800 (homolog of Arx1), and At1g74250 (homolog of Jjj1). A homolog of Alb1 had not been found by sequence homology searches.1
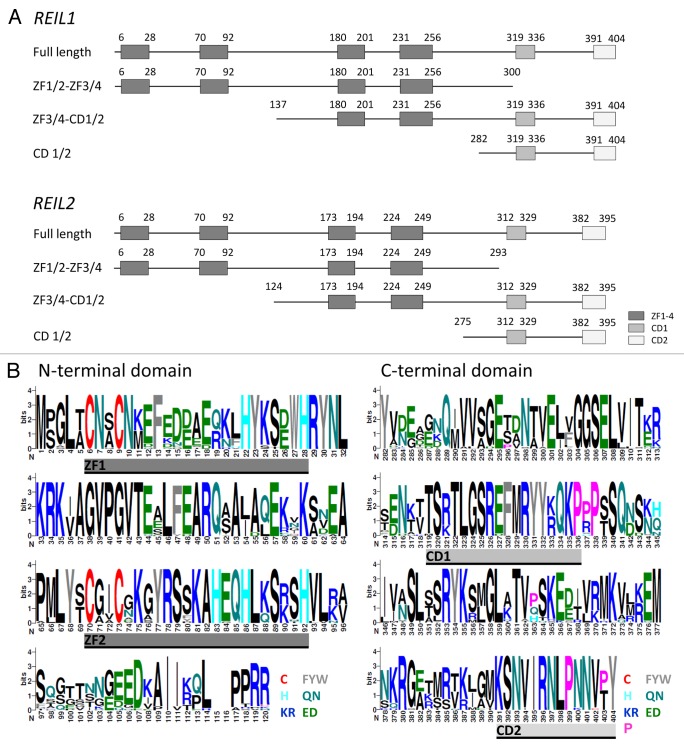
REIL proteins are composed of 4 conserved zinc finger domains and of 2 more specific domains conserved among the Brassicales, which we provisionally named CD1 and CD2 (Fig. 3A). The domains are evenly spread across the length of the REIL proteins and notably the N- and the C-terminus are highly conserved (Fig. 3B). Following the general organization of REIL protein topology (Fig. 3) we used 4 constructs in our Y2H study: 1) The full-length sequence of each REIL paralog, 2) a C-terminal truncation containing only the 4 zinc finger domains, i.e., ZF1/2-ZF3/4, with amino acids 1 to 300 of REIL1 or 1 to 293 of REIL2, 3) an N-terminal truncation, i.e., ZF3/4-CD1/2, comprising amino acids 137 to 404 of REIL1 or 124 to 395 of REIL2, and 4) an extended N-terminal truncation with only the 2 specific CD domains remaining, i.e., CD1/2, which contains amino acids 282 to 404 of REIL1 or 275 to 395 of REIL2 (Fig. 3A).
Figure 3. Topology of the REIL (Rei1-like) proteins from A. thaliana and of the partial proteins which were analyzed in this study by yeast-2-hybrid assays (Table 1).(A) The 404 amino acid REIL1 and the 395 amino acid REIL2 protein each contain 4 zinc finger domains and 2 additional conserved domains, provisionally named CD1 and CD2. The truncated proteins, ZF1/2-ZF3/4, contain the 4 zinc finger domains. The CD 1/2 truncations contain only the C-terminal parts with the conserved domains CD1 and CD2. The ZF3/4-CD1/2 partial proteins lack the N-terminal pair of zinc finger domains. (B) N-terminal and C-terminal alignment of REIL1 and REIL2 homologs of Brassicales species. Zinc finger domains, ZF1 and ZF2, and conserved domains, CD1 and CD2, are indicated. The color coding of the sequence logos highlights cysteine and histidine residues, aromatic amino acids, amide, acidic, basic residues, and the conserved proline residues of the C-terminus. Color coding is given with the insert. Sequence logos were generated at http://weblogo.berkeley.edu/logo.cgi after alignment of the REIL proteins from Arabidopsis thaliana, Arabidopsis lyrata subsp. lyrata, Eutrema parvulum, Eutrema salsugineum, Brassica rapa subsp. pekinensis, Capsella rubella, Camelina sativa, Leavenworthia alabamica, Sisymbrium irio, and Aethionema arabicum.
The full-length and all partial proteins do not auto-activate the Y2H system. Most of the tested interactions generate viable 2-hybrid positive yeast lines, even when using the most stringent selection medium. To differentiate the strength of interaction we quantified the colorimetric response of the X-Gal reporter activity relative to a positive control (Table 1). In agreement with the known interactions of yeast Rei1 (Fig. 2), both REIL1 and REIL2 exhibit self-interactions. In addition, the REIL paralogs interact with each other. The self-interaction of REIL1 is weaker than the self-interaction of REIL2. A possible function of the zinc finger domains for homo- and hetero-dimer formation of the REIL paralogs is indicated by enhanced self-interactions and respectively enhanced between-paralog interactions of the ZF1/2-ZF3/4 truncations with the respective full-length REIL proteins. Both REIL proteins also interact with most of the tested proteins. The interactions of the full-length REIL1 with the Arabidopsis homologs of Arx1p and Jjj1p are stronger than the respective interactions with full-length REIL2. The truncations do not reveal further information on domain functions. The observed interactions are in most cases gradually weakened rather than fully abolished. Exceptions are the interactions of REIL1 and REIL2 with the Rlp24 homolog. These interactions are absent or weak in the case of full-length REIL constructs but can apparently be enhanced by different truncations of the REIL proteins.
Table 1. Protein–protein interaction matrix of REIL1, REIL2 proteins with A. thaliana homologs of the yeast proteins, Rlp24p, Rpl24Ap, Rpl24B, Arx1p, and Jjj1p.
| Protein Name | REIL1 | REIL2 | |||||
|---|---|---|---|---|---|---|---|
| Yeast homologs | Rei1p | Reh1p | Rpl24Ap | Rpl24Bp | Rlp24p | Arx1p | Jjj1p |
| Gene Code | At4g31420 | At2g24500 | At2g36620 | At3g53020 | At2g44860 | At3g51800 | At1g74250 |
| Full Length | |||||||
| REIL1 | 7.9 | 11.9 | 5.1 | 5.1 | - a | 10.3 | 14.1 |
| REIL2 | 2.0 | 16.1 | - | 6.6 | 3.3 | 4.5 | 2.2 |
| Partial REIL1 | |||||||
| ZF1/2-ZF3/4b | 14.6 | 35.0 | 5.6 | 3.1 | 18.5 | 5.0 | 9.5 |
| ZF3/4-CD1/2 | 6.6 | 2.5 | 4.0 | 3.9 | 5.9 | 4.5 | 6.5 |
| CD1/2 | 5.4 | 8.9 | 3.1 | 5.4 | 5.5 | 3.9 | 9.5 |
| Partial REIL2 | |||||||
| ZF1/2-ZF3/4 | 6.2 | 32.8 | 5.9 | 7.7 | 2.9 | 7.4 | - |
| ZF3/4-CD1/2 | 8.9 | - | 4.9 | 7.4 | 13.3 | 4.6 | - |
| CD1/2 | 3.0 | 9.1 | 5.2 | 6.3 | 3.2 | 2.3 | - |
Full-length REIL bait proteins and C- or N-terminal truncations, rows also see (Fig. 3), were tested against full-length prey proteins, columns, using the yeast 2-hybrid assay. Values represent α-galactosidase activity of strains containing the respectively tested pair of heterologously expressed proteins. The activity is expressed as percent relative to a positive control strain carrying the plasmids pGBKT7–53 and pGADT7-T (Table S1)a Few interactions tested negative and did not yield viable yeast strains (-). b The full and partial proteins were cloned in to the bait vector using specific primers (Table S2). The names of the truncated proteins indicate the retained zinc finger domains (ZF) and REIL-specific conserved domains (CD). The α-galactosidase activity was assayed spectrophotometrically on microtiter plates using 4–8 replications per combination and controls. The technical standard deviation of this assay was 3.5%.
The tested Y2H interactions of the heterologous expressed REIL proteins are difficult to interpret. Interpretation is in this case complicated by the inherent problems of analyzing conserved protein interactions. We demonstrated previously that REIL1 can almost fully rescue the cold sensitivity of the yeast Δrei1 mutant.1 For this reason, we must assume that the REIL proteins are fully in the case of REIL1 or possibly in part in the case of REIL2 competent to physically interact with the endogenous components of the yeast 60S maturation machinery. As a consequence the interpretation of observed Y2H interactions needs to consider first that the interactions may be suppressed or weakened by interference between the heterologous expressed plant proteins and the endogenous yeast homologs. Second, we also must take into regard that endogenous yeast proteins or likely the 60S ribosomal subunit may act as scaffolds which can generate false positive Y2H results via indirect protein interactions.
In conclusion, the Y2H data of this study are consistent with a conserved role of the A. thaliana REIL proteins for ribosomal maturation. The Y2H results, however, do not provide proof. But the data encourage future investigations of the proteins that interact with the REIL proteins in planta.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We kindly acknowledge funding by the Max-Planck Society and the longstanding support of Prof L Willmitzer.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/28224
Glossary
Abbreviations:
- Alb1
Arx1 little brother1
- Arx1
Associated with ribosomal export complex1
- Jjj1
Type III j-protein1
- Nmd3
Nonsense-mediated mRNA decay3
- Nog1
Nucleolar G-protein1
- Mrt4
mRNA turnover4
- Reh1
Rei1 homologue1
- Rei1
Required for isotropic bud growth1
- REIL
REI1-LIKE
- Rlp24
Ribosomal-like protein24
- Rpl24A/B
Ribosomal protein of the large subunit24A/B
- Ssa1/2
Stress-Seventy subfamily A1/2
- Tif6
Translation initiation factor6
- Y2H
yeast-2-hybrid
References
- 1.Schmidt S, Dethloff F, Beine-Golovchuk O, Kopka J. The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold. Plant Physiol. 2013;163:1623–39. doi: 10.1104/pp.113.223925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwase M, Toh-e A. Ybr267w is a new cytoplasmic protein belonging to the mitotic signaling network of Saccharomyces cerevisiae. Cell Struct Funct. 2004;29:1–15. doi: 10.1247/csf.29.1. [DOI] [PubMed] [Google Scholar]
- 3.Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol. 2006;173:349–60. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parnell KM, Bass BL. Functional redundancy of yeast proteins Reh1 and Rei1 in cytoplasmic 60S subunit maturation. Mol Cell Biol. 2009;29:4014–23. doi: 10.1128/MCB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer AE, Hoover LA, Craig EA. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J Biol Chem. 2010;285:961–8. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol. 2007;27:6581–92. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–7. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.