Abstract
The mitochondrial uniporter is a highly selective calcium channel in the organelle’s inner membrane. Its molecular components include the EF-hand containing proteins mitochondrial calcium uptake 1 (MICU1) and MICU2 and the pore forming subunit mitochondrial calcium uniporter (MCU). We sought to achieve a full molecular characterization of the uniporter holocomplex (uniplex). Quantitative mass spectrometry of affinity-purified uniplex recovered MICU1 and MICU2, MCU and its paralog MCUb, and essential MCU regulator (EMRE), a previously uncharacterized protein. EMRE is a 10 kD, metazoan specific protein with a single transmembrane domain. In its absence, uniporter channel activity was lost despite intact MCU expression and oligomerization. EMRE was required for the interaction of MCU with MICU1 and MICU2. Hence, EMRE is essential for in vivo uniporter current and additionally bridges the calcium-sensing role of MICU1 and MICU2 with the calcium conducting role of MCU.
The mitochondrial calcium uniporter is a highly selective channel that moves calcium ions across mitochondrial inner membrane (1). Although its physiology has been studied for decades, a complete description of its molecular composition has remained elusive. Recently, integrative genomics methods enabled the discovery of the uniporter pore, mitochondrial calcium uniporter (MCU), and its regulatory subunits, mitochondrial calcium uptake 1 and 2 (MICU1 and 2) (2–5). MCU is an integral membrane protein that is essential for the electrophysiologically defined uniporter current (6); it has two transmembrane domains, and orients both its N and C termini into the matrix (3, 7). MICU1 contains an EF-hand calcium binding domain and is found in the mitochondrial intermembrane space (IMS), where it serves as a calcium-sensing gatekeeper, keeping the channel closed when calcium levels are low and opening the channel in response to transient rises (2, 5, 8, 9). Its paralog and binding partner, MICU2, has not been extensively characterized (5). Other proteins, including leucine-zipper EF-hand containing transmembrane protein 1 (LETM1), mitochondrial calcium uniporter regulator 1 (MCUR1), mitochondrial sodium calcium exchanger (NCLX), transient receptor potential 3 (TRCP3), and uncoupling protein 2 and 3 (UPC2 and 3) are also crucial for mitochondrial calcium physiology, but their physical relation to the uniplex are unclear (10–14).
We took a biochemical approach to fully characterize composition the uniporter complex. We stably expressed MCU tagged with the FLAG epitope at its carboxy terminus (MCU-FLAG) in human embryonic kidney (HEK)-293T cells. MCU-FLAG restored mitochondrial calcium uptake in cells in which MCU was depleted with RNAi, and even caused a gain-of-function phenotype compared to that of cells that expressed a control protein (Fig. S1A). MCU exists in a large protein complex when isolated by digitonin permeabilization and native gel electrophoresis of mitochondria from HeLa cells or mouse liver (3). Similarly, in HEK-293T cells that stably expressed MCU-FLAG, MCU migrated at ~480 kD (Fig. 1A). Immunoprecipitation of MCU-FLAG, but not that of a control protein, yielded a protein complex of comparable size (Fig. 1A). Hence, MCU-FLAG associates with the apparent uniporter holocomplex, which we call the uniplex (uniporter complex).
Fig. 1. Affinity purification and proteomic analysis of the uniporter complex, uniplex.
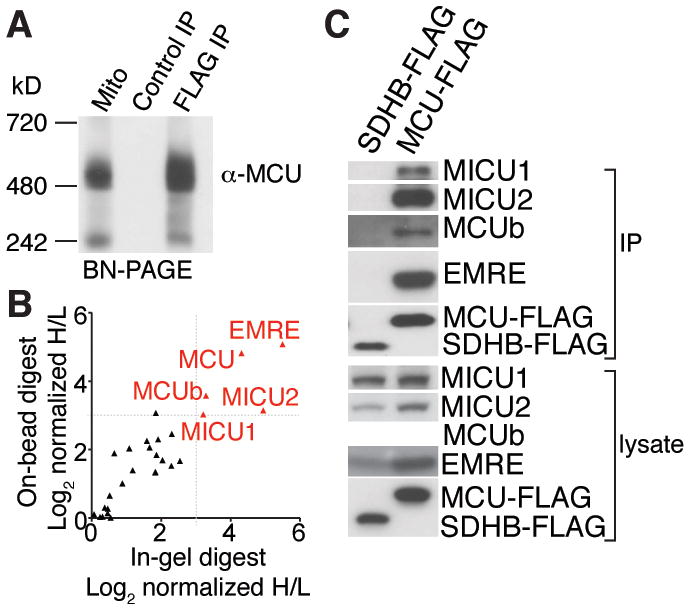
(A) MCU-FLAG or control SDHB-FLAG was stably expressed in HEK-293T cells. Proteins from digitonin-permeabilized mitochondria from MCU-FLAG expressing cells and FLAG immunoprecipitations from SDHB-FLAG and MCU-FLAG expressing cells were subjected to Blue Native-PAGE (BN-PAGE) and immunoblotted with MCU antibody. (B) Identification of proteins that interact with MCU-FLAG. MCU-FLAG expressing cells were grown in the presence of heavy amino acids, control HEK-293T cells were grown in the presence of light amino acids. FLAG immunoprecipitates from both samples were mixed and analyzed by mass spectrometry. The ratios of heavy and light proteins annotated with mitochondrial localization from two replicates are shown. Proteins that show significant enrichment in heavy samples are shown in red. (C) Interaction of MCUb, MICU1, MICU2 and EMRE with MCU. MCU-FLAG or control SDHB-FLAG was immunoprecipitated from HEK-293T cells. Immunoprecipitates and cell lysates were analyzed by immunoblotting for the indicated proteins.
To define the components of the uniplex, we used a quantitative mass spectrometry approach using stable isotope labelling by amino acids in cell culture (SILAC) (15). We grew MCU-FLAG expressing and wild-type HEK-293T cells in medium containing heavy or light amino acid isotopes, respectively, immunoprecipitated proteins with FLAG antibody-conjugated beads and mixed heavy and light samples before they were processed for mass spectrometry. Two independent quantitative mass spectrometry experiments, with two different digestion protocols, reproducibly identified only five known or predicted mitochondrial proteins (Fig. 1B, Table S1). MCU was enriched in MCU-FLAG samples, as well as its known regulators MICU1 and MICU2. The uniplex also contains MCUb, a paralog of MCU (16). The only other mitochondrial protein revealed through this analysis was C22orf32, a previously uncharacterized protein we call essential mcu regulator (EMRE). MICU1, MICU2, MCUb and EMRE were detected by protein immunoblotting after immunoprecipitation of MCU-FLAG from HEK-293T and HeLa cells, but not with succinate dehydrogenase iron-sulfur subunit SDHB-FLAG, a protein also localized to the inner membrane (Fig. 1C, S1B). Moreover, MCU overexpression resulted in increased abundance of MICU1, MICU2 and EMRE (Fig 1C), probably because it stabilized its binding partners.
EMRE is a ~10 kD protein with a predicted mitochondrial targeting sequence, a predicted transmembrane domain (17, 18), and a highly conserved C-terminus rich in aspartate residues (Fig. 2A). MCU and MICU1 are found in all major eukaryotic taxa, with lineage specific losses in several clades. We did not find EMRE homologs in plants or protozoa. Analysis of recently sequenced opisthokonts and basal fungi revealed that EMRE homologs are not found in any fungi, indicting that it most likely arose in the metazoan lineage (Fig. 2B). EMRE RNA was broadly expressed in all mouse tissues (Fig. S2). Similar to MCU and MICU1, EMRE was identified by proteomic analysis of mitochondria (19) with a pattern suggesting it is ubiquitously expressed in the mitochondria of all mammalian tissues. EMRE appears to be a bona fide transmembrane protein as it was resistant to carbonate extraction at high pH, as is MCU (Fig. 2C).
Fig. 2. Domain architecture, phylogeny and membrane association of EMRE.
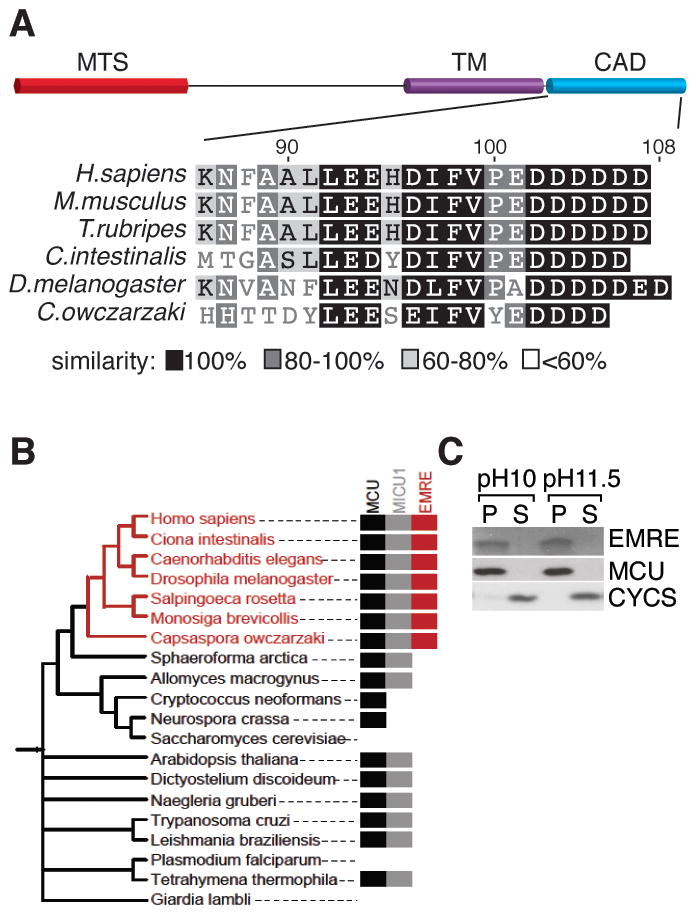
(A) Schematics showing predicted mitochondrial targeting sequence (MTS), transmembrane (TM) and conserved carboxy terminal acidic domain (CAD) of EMRE. The CAD of EMRE from six species was aligned using BLOSUM similarity matrix. (B) EMRE is metazoan-specific. Presence of homologs of MCU/MCUb, MICU1/MICU2 and EMRE across 20 selected species spanning the NCBI taxonomy tree, indicating that EMRE is a metazoan innovation. (C) EMRE is a membrane protein. HEK-293T cell mitochondria were isolated and proteins were extracted with 0.1M Na2CO3 at pH 10 and pH 11.5. EMRE is observed in the insoluble pellet (P) similar to MCU. Cytochrome c (CYCS) is loosely associated with the inner membrane and is detected in the soluble fraction (S).
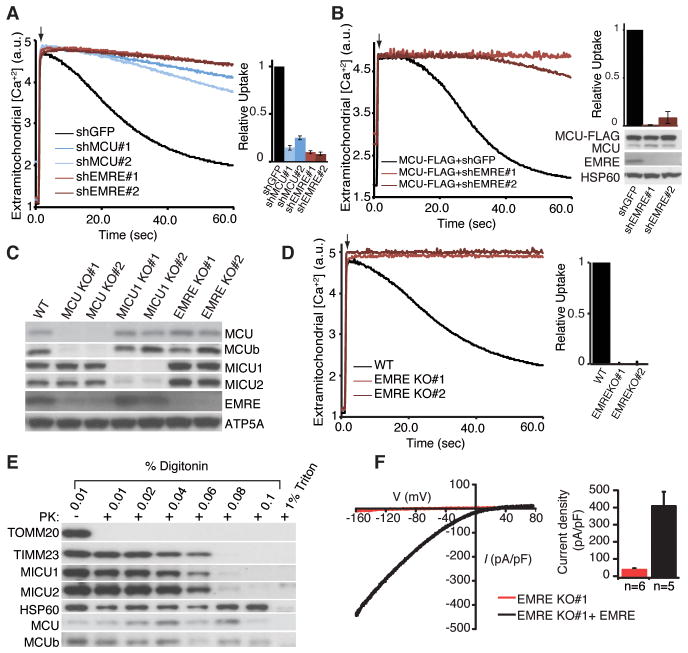
We tested the impact of loss of EMRE on uniplex function by RNAi mediated silencing of EMRE. Depletion of EMRE with each of two, sequence-independent hairpins led to loss of mitochondrial calcium uptake equivalent to MCU silencing in permeabilized HEK-293T and HeLa cells, as well as in intact HeLa cells after histamine stimulation (Fig. 3A, S3A, B). The appearance of the cells and their rates of proliferation were normal after EMRE silencing. The mitochondrial membrane potential was intact and could be depolarized in cells depleted of EMRE (Fig. S3C), indicating that loss of calcium uptake was not a trivial consequence of loss of the mitochondrial membrane potential. Overexpression of MCU in cells depleted of EMRE failed to restore mitochondrial calcium uptake (Fig. 3B), suggesting that MCU is not sufficient for in vivo uniporter current as previously proposed (4).
Fig. 3. Requirement of EMRE for uniporter activity.
(A) EMRE knock down impairs mitochondrial calcium uptake. Digitonin permeabilized HEK-293T cells were incubated with the calcium indicator Oregon Green Bapta 6F (OGB6F). After addition of 50 μM CaCl2 (arrow), the depletion of extracellular calcium due to mitochondrial uptake was monitored by OGB6F fluorescence. Representative traces of mitochondrial calcium uptake from HEK-293T cells with GFP (control), MCU and EMRE knock down are shown. The bar graph shows the rate of calcium uptake relative to shGFP cells (mean ± s.d., n=4). (B) MCU overexpression cannot rescue mitochondrial calcium uptake in EMRE knock down cells. EMRE was knocked down in HEK-293T cells that overexpress MCU-FLAG. The samples were treated as in (A) (n=4). Lysates were analyzed by immunoblotting for indicated proteins. (C) Loss of MCU decreases EMRE and MCUb abundance. MCU, MICU1 and EMRE knockout (KO) HEK-293T cells were generated using TALEN technology, cell lysates from wild type (WT) and two independent knockout clones were prepared and analyzed by immunoblotting for control ATP5A and uniplex proteins (D) Mitochondrial calcium uptake is severely impaired in cells that lack EMRE. Mitochondrial calcium uptake was measured in WT and two independent EMRE KO cell lines as in (A). (E) Loss of EMRE does not change MCU, MCUb, MICU1 and MICU2 submitochondrial localization. Mitochondria were isolated from cells lacking EMRE, incubated with increasing concentrations of digitonin in the presence of proteinase K (PK), samples were analyzed by immunoblotting for outer membrane protein TOMM20, inner membrane protein TIMM23, matrix protein HSP60, as well as uniplex proteins (F) Exemplar trace (red) of a mitoplast derived from EMRE knockout cells demonstrates absent calcium current (IMiCa), whereas typical IMiCa is seen after EMRE is knocked back in (black). Voltage ramps were delivered from −160mV to +80mV for 750ms, using a holding potential of 0mV. (Right) Summary data. Error bars report SEM.
To characterize complete loss-of-function phenotypes of uniplex components, we generated HEK-293T cells lacking MCU, MICU1 or EMRE with TALEN technology (Fig. 3C). Surprisingly, in cells lacking MCU, abundance of EMRE was decreased compared to that of wild type cells (Fig. 3C). However, in these cells, abundance of EMRE mRNA was similar to that in wild type cells (Fig. S3D), suggesting that loss of EMRE occurred post-transcriptionally. Thus, EMRE may be destabilized when its binding partner MCU is lost, analogous to the dependence of MICU2 protein expression on MICU1 (Fig. 3C and (5)). Cells lacking EMRE, similar to cells lacking MCU, exhibited severe defects in mitochondrial calcium uptake (Fig. 3D, S3E). In cells lacking MICU1, depletion of EMRE also leads to loss of mitochondrial calcium uptake (Fig. S3F). Digitonin permeabilization of isolated mitochondria from control cell or cells lacking EMRE followed by proteolysis and western blotting showed that abundance of other uniplex proteins and their mitochondrial localization did not change after loss of EMRE (Fig. 3C, 3E, S3G). These experiments confirmed the topology of MCU (3, 7) and MICU1 (9) and showed that the topology of MCUb is similar to the topology of MCU and MICU2 localizes to the intermembrane space like MICU1 (Fig. 3E, SG). To show that loss of EMRE specifically affects the uniporter conductance and not other mitochondrial parameters that will impact mitochondrial calcium uptake (e.g., proton motive force, pH, buffering capacity), we voltage-clamped mitoplasts isolated from cells lacking EMRE or these same cells after stable expression of EMRE, and measured mitochondrial calcium current (IMiCa). Mitoplasts from cells lacking EMRE has significantly reduced IMiCa whereas typical IMiCa was seen after EMRE re-expression (Fig. 3F) (6).
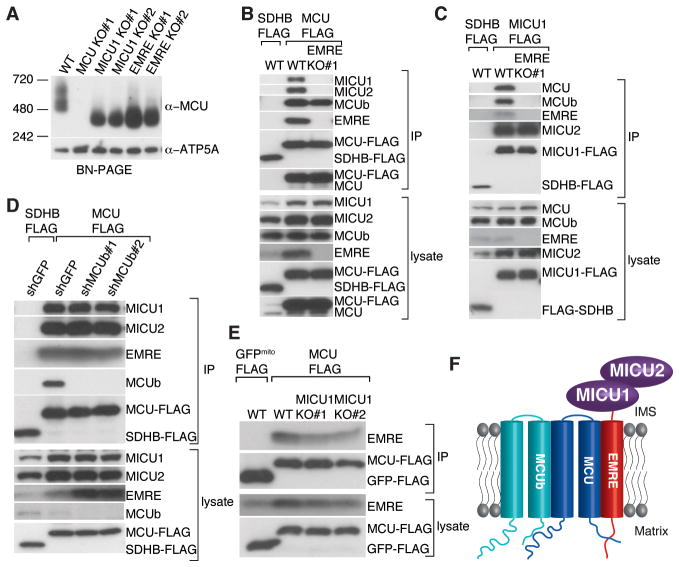
Consistent with EMRE being a core component of the uniplex, when EMRE was lost, the size of the complex on a native gel reduced to ~300 kDa, similar to that in cells lacking MICU1 (Fig. 4A). The observation that loss of either EMRE or MICU1 reduced the size of the native complex to a similar extent raised the hypothesis that EMRE may mediate interaction between MCU and MICU1 and MICU2. To test this, we stably expressed MCU-FLAG or a control protein in HEK-293T or HeLa wild-type cells or cells lacking EMRE and performed FLAG immunoprecipitations. In wild-type cells, MICU1 and MICU2 immunoprecipitated with MCU-FLAG, but not with control SDHB-FLAG (Fig. 4B, S4). However, in cells lacking EMRE, the interaction between MCU-FLAG and MICU1 and MICU2 was completely lost (Fig. 4B, S4). Similarly, MCU was not associated with immunoprecipitated MICU1-FLAG in cells lacking EMRE (Fig. 4C). In the absence of EMRE, MCU still oligomerized (Fig. 4A, B) and interacted with MCUb. Moreover, in the absence of EMRE, the interaction between MICU1 and MICU2 was intact (Fig. 4C). These data indicate that loss of EMRE specifically interrupted the association of MCU with MICU1 and MICU2. Furthermore, MCUb, MICU1 and MICU2 appear to be dispensable for MCU-EMRE interaction because in cells lacking MCUb or MICU1, EMRE was still associated with immunoprecipitated MCU-FLAG (Fig. 4D, E).
Fig. 4. Requirement of EMRE for the interaction of MCU with MICU1 and MICU2.
(A) Loss of EMRE or MICU1 reduces the size of the uniplex to ~300 kD. Mitochondria from indicated cell lines were isolated, digitonin permeabilized and immunoblotted for MCU after BN-PAGE. EMRE mediates MCU-MICU1 and MICU2 interaction. Stably expressed SDHB-FLAG (control) or MCU-FLAG (B) or MICU1-FLAG (C) was immunoprecipitated from WT or EMRE knockout cells, lysates and immunoprecipitates were analyzed by immunoblotting for indicated proteins. (D) Loss of MCUb does not alter MCU-EMRE, MICU1 and MICU2 interaction. Stably expressed SDHB-FLAG or MCU-FLAG was immunoprecipitated from cells after GFP (control) or MCUb knock down with two different hairpins. Samples were treated and analyzed as in (B). (E) Loss of MICU1 does not alter MCU-EMRE interaction. Plasmid for EMRE expression was co-transfected with plasmids for control mitochondrial GFP-FLAG or MCU-FLAG expression, in WT and MICU1 knockout cells. Samples were treated and analyzed as in (B). (F) Model showing submitochondrial localization and organization of uniplex proteins. MCU is the pore-forming component of the uniplex; MICU1 and MICU2 reside in the IMS where they sense outside calcium and regulate MCU through EMRE. EMRE is a single-pass transmembrane protein. It bridges MCU and MICU1 and MICU2 and is essential for the activity of the uniporter. MCU-MCUb interaction is not mediated by EMRE.
We propose a model where EMRE interacts with MICU1 and MICU2 in the IMS and with MCU oligomers in the inner membrane, thus linking the calcium sensing activity of MICU1 and MICU2 to the channel activity of MCU (Fig. 4F). The fact that our immunoprecipitated complex migrates at the same apparent molecular size as the uniporter from purified mitochondria (Fig. 1A), and recovers the founding members of the complex, suggests that we immunoprecipiated all of its components. It is notable that although other proteins have been reported to participate in mitochondrial calcium handling, such as LETM1, NCLX, UCP2 and 3, MCUR1 and TRCP3, they were not recovered in our proteomic assay, suggesting that they play key roles outside of the uniplex. A key insight from the current work is that EMRE is essential for in vivo uniporter current (IMiCa) and that MCU oligomers alone are not sufficient for in vivo uniporter activity, in contrast to what has been suggested using in vitro bilayer studies (4). Why MCU needs association with EMRE for in vivo calcium conductance requires further investigation. Phylogenetic analysis indicate that the uniporter must have been a feature of the earliest mitochondria, since MCU and MICU1 are found within all major eukaryotic taxa, with lineage specific losses (20). EMRE is unique amongst human uniplex components in that it appears to have emerged more recently and represents a metazoan innovation. Its identification should help us understand how the activity and regulation of this ancient channel evolved.
Supplementary Material
Acknowledgments
We thank Z. Grabarek and D.M. Shechner for helpful discussions. Y.S. received support from Helen Hay Whitney Foundation. D.C. received support from NIH F32HL107021. D.E.C. is an Investigator of the Howard Hughes Medical Institute. This work was supported by grants to V.K.M from NIH DK080261 and a gift from W. Dan and Pat Wright.
References and Notes
- 1.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004 Jan 22;427:360. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 2.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010 Sep 16;467:291. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011 Aug 18;476:341. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011 Aug 18;476:336. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plovanich M, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri D, Sancak Y, Mootha VK, Clapham DE. MCU encodes the pore conducting mitochondrial calcium currents. Elife. 2013;2:e00704. doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012 Nov;30:1143. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallilankaraman K, et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012 Oct 26;151:630. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csordas G, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca(2+) Uniporter. Cell Metab. 2013 Jun 4;17:976. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palty R, et al. Lithium-calcium exchange is mediated by a distinct potassium-independent sodium-calcium exchanger. J Biol Chem. 2004 Jun 11;279:25234. doi: 10.1074/jbc.M401229200. [DOI] [PubMed] [Google Scholar]
- 11.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009 Oct 2;326:144. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallilankaraman K, et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012 Dec;14:1336. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng S, et al. Canonical transient receptor potential 3 channels regulate mitochondrial calcium uptake. Proc Natl Acad Sci U S A. 2013 Jul 2;110:11011. doi: 10.1073/pnas.1309531110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trenker M, Malli R, Fertschai I, Levak-Frank S, Graier WF. Uncoupling proteins 2 and 3 are fundamental for mitochondrial Ca2+ uniport. Nat Cell Biol. 2007 Apr;9:445. doi: 10.1038/ncb1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002 May;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 16.Raffaello A, et al. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. Embo J. 2013 Jul 30; doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001 Jan 19;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 18.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996 Nov 1;241:779. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 19.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008 Jul 11;134:112. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012 May 18;336:886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.