Abstract
The NKG2 family of NK receptors includes activating and inhibitory members. With the exception of the homodimer-forming NKG2D, NKG2 receptors recognize the nonclassical MHC class I molecule HLA-E, and can be subdivided into two groups: those that associate with and signal through DAP12 to activate cells and those that contain an ITIM motif to promote inhibition. The function of NKG2 family member NKG2E is unclear in humans and its surface expression has never been conclusively established, largely because there is no antibody that binds specifically to NKG2E. Seeking to determine a role for this molecule, we chose to investigate its expression and ability to form complexes with intracellular signaling molecules. We found that NKG2E was capable of associating with CD94 and DAP12 but that the complex was retained intracellularly at the ER instead of being expressed on cell surfaces, and that this localization was dependent on a sequence of hydrophobic amino acids in the extracellular domain of NKG2E. As this particular sequence has emerged and been conserved selectively among higher order primates evolutionarily, this observation raises the intriguing possibility that NKG2E may function as an intracellular protein.
Keywords: NKG2E, NKG2C, CD94, DAP12, HLA-E, natural killer cells, ER associated retention
Introduction
Natural killer (NK) receptors have a wide range of effects and are expressed by both natural killer cells(1, 2) and cytotoxic CD8+ T cells(3, 4). They are involved in protection against invading pathogens(5, 6) and dysregulation of certain NK receptors has been implicated in autoimmunity(3, 7). Consequently, a delicate balance has to be maintained between the activating and inhibitory receptors present.
One such group is the NKG2 family, which is comprised of seven receptors, known as NKG2A, -B, -C, -D, -E, -F, and –H(8). With the exception of the more distantly related and homodimer-forming NKG2D(9) and the orphan receptor NKG2F(10), NKG2 family proteins assemble into heterodimers with CD94, a C-type lectin-like molecule that allows for interaction of the complex with HLA-E(11, 12). CD94/NKG2 heterodimers are expressed on most NK cells, and the inhibitory family members are expressed by many subsets of CD8+ T cells. While CD94 is associated with all NKG2 signaling complexes save NKG2D and F, activating NKG2 receptors also require the binding of the adaptor molecule DAP12, also known as KARAP(13), for cell surface expression and signaling(14). DAP12 contains an ITAM motif, allowing for activation of NK cells by the NKG2C/CD94/DAP12 complex through the Syk and ZAP70 kinases(14). In contrast, NKG2A contains an ITIM motif in its intracellular portion, allowing it to directly activate the SHP1/2 phosphatases(15) and obviating the need for an additional adaptor molecule(16). Meanwhile, NKG2F associates with DAP12 but not CD94, and thus fails to be expressed on cell surfaces(10).
While NKG2A and NKG2C have been linked to autoimmune diseases such as celiac disease(17) and rheumatoid arthritis(18), as well as chronic viral infections(19), there is a very limited amount of data available for NKG2E. Crucially, NKG2E and NKG2C have identical transmembrane sequences (see fig. 1C), suggesting that DAP12 will associate with both with equal efficiency(20). Furthermore, NKG2E was reported to successfully bind to HLA-E-peptide complexes (21). Finally, NKG2E has been shown to have activating properties in response to Qa-1(22) and to be important for defense against viral infections in mice(23). Consequently, NKG2E is often cited in conjunction with NKG2C as being a de facto activating receptor.
Figure 1.
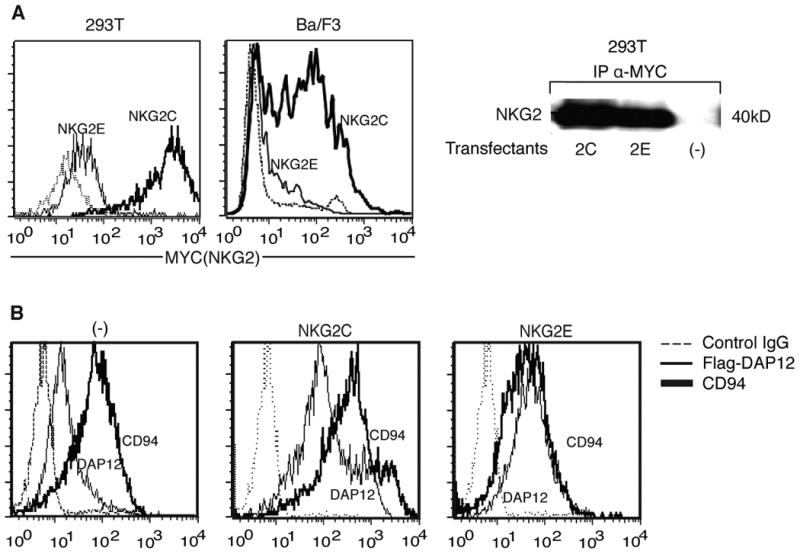
NKG2E is translated and associates with CD94, but fails to be expressed on cell surfaces. (A) NKG2C but not NKG2E is expressed on cell surfaces. 293T (left) and Ba/F3 (middle) cells were transfected with MYC tagged NKG2C or NKG2E in conjunction with DAP12 and CD94. Cells were surface stained using anti-MYC mAb and analyzed by flow cytometry. Protein expression was verified by western blot (right). Data are representative of at least three independent experiments. (B) CD94 and DAP12 are expressed on cell surfaces when transfected with NKG2C but not NKG2E. Ba/F3 cells were transfected with CD94 and FLAG-tagged DAP12 alone (left) or additionally with NKG2C (middle) or NKG2E (right) and analyzed for CD94 and DAP12 surface expression by flow cytometry using anti-CD94 or anti-FLAG mAb. Data are representative of at least three independent experiments.
However, there is no data that directly verifies NKG2E surface expression in humans, and the association of NKG2E with DAP12 has never been demonstrated. In addition, mouse NKG2E is unrelated to the human gene and it lacks a large portion of the extracellular domain that we dubbed the ECII region. Unfortunately, investigations into hNKG2E's function are hampered by the lack of a specific antibody, as those currently available also bind to hNKG2C and hNKG2A with similar affinity (Fig. S1A). Despite our best efforts, we were also unable to generate an antibody of adequate specificity. Consequently, it has yet to be determined whether NKG2E can be expressed at a protein level in human NK or T cells and, if so, whether or not it is able to function as a receptor. It has been previously noted that NKG2E transcripts are increased in NKG2A-negative CTLs that have undergone NK reprogramming in patients with celiac disease, but whether and how this may contribute to pathogenesis is unknown(17, 24).
We sought to address this gap in the literature by investigating NKG2E at a protein level, using FLAG and MYC tagging in absence of a specific antibody, as previously described(14). Intriguingly, we found that although NKG2E is translated, unlike NKG2C it fails to be expressed on cell surfaces efficiently and is instead retained intracellularly. Furthermore, we observed that this retention is dependent on the ECII portion of the extracellular domain. Finally, we demonstrate that NKG2E is assembled into a multi-protein complex with CD94 and DAP12.
Materials and Methods
Cell culture
The 293T cell line is a human renal epithelial cell line and was cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS) and antibiotics. The Ba/F3 cell line is a murine B cell line and was cultured in DMEM with 10% FBS, 1% L-glutamine, and 1% streptomycin/penicillin.
Transfection
Human CD94, NKG2C, NKG2E, and DAP12 were cloned for mammalian expression into pcDNA4/TO (Invitrogen), papuroFLAG (containing N-terminal FLAG tag), pcDNAmyc (Invitrogen, containing C-terminal MYC tag), pIRESEGFP (Clontech) and SX vectors by standard cloning techniques. 293T cells were transfected with plasmids using Lipofectamine 2000 Reagent (Invitrogen) after reaching approximately 80% confluency. The standard transfection protocol provided by the manufacturer was used. After transfection, cells were cultured for 24 to 30 hours before staining for flow cytometry. Ba/F3 cells were transfected with the Amaxa nucleofector using nucleofection solution V and program T-20. Cells were cultured for 24 to 30 hours post-transfection before staining for flow cytometry. Ba/F3 cells expressing CD94/DAP12 and CD94/NKG2C-IRES-EGFP/DAP12 were previously described(14). For the generation of stable transfectants of CD94/NKG2E-IRES-EGFP/DAP12, Ba/F3 cells were transfected with Amaxa and sorted for expression of EGFP, then cultured in medium with selective antibiotics.
Reagents for flow cytometry
PE and APC-conjugated anti-CD94 were purchased from BD Bioscience. Unconjugated anti-MYC antibody was purchased from Covance. Unconjugated anti-NKG2E antibody was purchased from Abnova. Fluorochrome-conjugated anti-FLAG antibody was purchased from Sigma. Anti-DAP12(14) and HLA-E tetramers loaded with HLA-G leader peptide(11) were kind gifts. Unconjugated F(ab')2 goat anti-mouse IgG2a isotype control was purchased from Southern Biotech.
Reagents for confocal microscopy
Anti-hCD94 mouse IgG2a (Beckman Coulter), anti-PDI rabbit (Abcam), and anti-pan-cadherin mouse IgG1 (Abcam) were used as the primary antibodies for confocal microscopy. The secondary antibodies used were goat anti-mouse IgG2a FITC, donkey anti-rabbit Cy5, and goat anti-mouse Igg1 Cy5 and were from Jackson ImmunoResearch.
Reagents for immunoprecipitation and western blot
Anti-hCD94 (Beckman Coulter) and anti-hNKG2-N-18 (Santa Cruz) were used for immunoprecipitation. Antibodies used for blotting were anti-hNKG2-N-18 and mouse anti-hDAP12. The associated secondary antibodies were horseradish peroxidase-conjugated donkey anti-goat and goat anti-mouse antibodies (from Jackson ImmunoResearch).
Flow cytometry analysis
For surface staining, cells were incubated with fluorochrome-conjugated antibodies according to standard protocols. Unconjugated antibodies were revealed with appropriate fluorochrome-conjugated F(ab) goat anti-mouse IgG isotype. For intracellular staining, cells were fixed and permeabilized using CytoFix/CytoPerm solution (BD Bioscience). Fluorescence was analyzed on a four-color FACSCanto or FACSCalibur with quadrants set to score as negative >99% of control Ig stained cells.
Confocal microscopy
293T cells were grown on coverslips overnight, followed by transfection with CD94, DAP12, and NKG2C or E using Lipofectamine-2000 (Invitrogen). Cells were fixed with 3.7% paraformeldahyde and stained with antibodies for receptor complex and subcellular localization markers for 1 hour at room temperature. Antibodies were chosen to have different species of origin or IgG isotype specificity if of mouse origin. Coverslips were washed in phosphate-buffered saline, mounted with ProLong antishade reagent (Invitrogen) and analyzed by confocal microscopy using a TCS SP2 AOBS confocal microscope and workstation (Leica). Samples were obtained with sequential acquisition of different wavelengths and 63°X NA 1.4 oil objective lens.
Western blot and immunoprecipitation
293T cells were transfected with human CD94, DAP12, and NKG2C or E. Ba/F3 cell lines were transfected with nucleofector solution V (Amaxa), sorted for high EGFP expression and selected by antibiotics. Cells were cultured, harvested and lysed. In brief, cells were lysed in buffer containing freshly added protease and phosphatase inhibitors (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10 mM iodoacetamide, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and protease inhibitor cocktail tablets). For immunoprecipitation, lysates in 0.5% Triton X-100 were incubated with anti-hCD94 or anti-hNKG2-N-18, specific for NKG2C and NKG2E but not NKG2A. Equal amounts of protein were subjected to SDS-PAGE under reducing conditions in 12% gels, transferred to nitrocellulose membranes (Bio-Rad Laboratories), and probed with goat anti-hNKG2 and mouse anti-hDAP12, followed by horseradish peroxidase-conjugated donkey anti-goat and goat anti-mouse antibodies. To control for loading differences, blots were probed for total DAP12 and NKG2. Binding of secondary antibodies was visualized using the enhanced chemiluminescence ECL kit (GE Healthcare).
Generation of reciprocal chimeras and mutants
NKG2C and NKG2E chimeras were created by recursive PCR as previously described (Current Protocols, John Wiley & Sons). In brief, primers spanning different regions of NKG2E and NKG2C were used to create DNA fragments, which were reciprocally hybridized and the final products amplified. In the NKG2E/C chimera the last seventeen amino acids of NKG2E were replaced with the last nine amino acids of NKG2C, while in the NKG2C/E chimera the reverse was performed. NKG2E mutants were created by standard PCR mutagenesis, using custom primers. Chimeras and mutants were cloned into pcDNAmyc and pIRESEGFP. Final constructs were confirmed by DNA sequencing.
Phylogenetic analysis of the NKG2C-E family members
The alignment of NKG2C and NKG2E among primates and two rodents (mouse and rat). was done using the software Muscle(25, 26) and visualized using ClustalX(27). The Phylogenetic tree based on protein sequence alignment was inferred by RaxML 7.2.8 under an LG+F+Gamma4 model of sequence evolution (empirical amino acid frequencies and four discrete gamma categories). Bootstrap values were based on 100 replicates. dN/dS was calculated using the previously described method of Li et al. (28)
Results
NKG2E is translated and associates with CD94 and DAP12, but does not translocate to the cell surface
To first assess whether or not NKG2E could form a complex with CD94 and DAP12, we transfected the human renal epithelial 293T cell line and the murine Ba/F3 B cell line previously used to study CD94 with CD94, DAP12, and either NKG2E or NKG2C expressing plasmids, with both NKG2 members tagged with C-terminal MYC(14). We then stained for surface MYC, and found that while NKG2C was present on the cell surface as expected, NKG2E-transfected cells expressed minimal NKG2E extracellularly (Fig. 1A, left panels). The presence of both proteins after transfection and lack of NKG2 expression in untransfected controls was confirmed by immunoblotting (Fig. 1A, right panel). To verify that the lack of NKG2E surface expression was not caused by an unanticipated side effect of tagging with MYC, we transfected Ba/F3 cells with N-terminal FLAG tagged DAP12, CD94, and NKG2E or C cloned into bicistronic IRES-EGFP. EGFP positive cells were then surface stained using anti-CD94 and anti-FLAG antibodies. We found that NKG2C transfection led to major upregulation of CD94 and DAP12 on the surface compared to cells transfected with CD94 and DAP12 alone, whereas the presence of NKG2E failed to induce surface expression (Fig. 1B). Overall, these data demonstrate that NKG2E transcripts are translated within cells, but NKG2E proteins fail to be expressed on cell surfaces in the presence of CD94 and DAP12.
NKG2E forms an intracellular complex with CD94 and DAP12
Given the finding that NKG2E is not present on cell surfaces, we were intrigued by the possibility that NKG2E might form intracellular complexes with CD94 and DAP12. We first sought to determine whether NKG2E is capable of retaining CD94 within the intracellular compartment. To study a potential effect of NKG2E on surface CD94 expression, we transfected 293T cells with CD94 alone, CD94 with DAP12, or CD94, DAP12, and NKG2E or C. Cells were then stained in two steps-first with anti-CD94 conjugated to APC for surface detection, then with anti-CD94-PE for intracellular detection after fixation and permeabilization. We found that there was a basal level of surface CD94 expression that was unaffected by the presence of DAP12, and that co-expression of NKG2C caused a significant increase in CD94 extracellularly (Fig. 2A). In contrast, transfection with NKG2E resulted in substantial downregulation of surface CD94 compared to cells transfected with CD94 alone or CD94 in conjunction with DAP12, suggesting that NKG2E not only fails to be expressed on cell surfaces, but that it additionally is capable of preventing surface CD94 expression.
Figure 2.
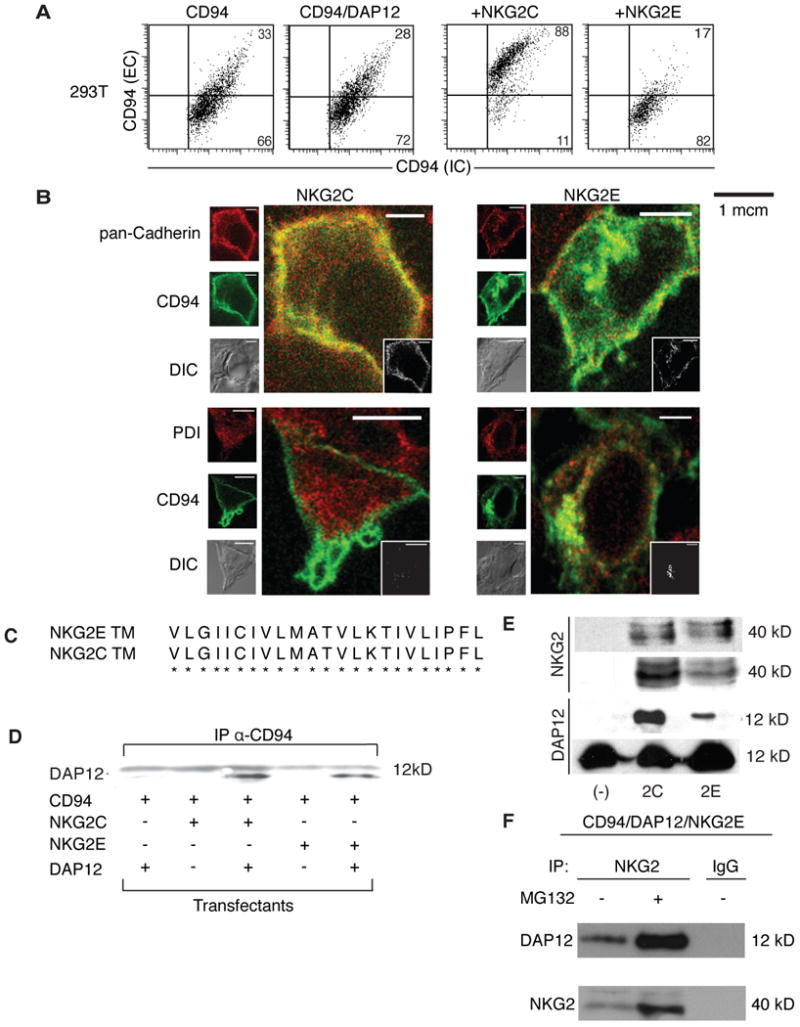
NKG2E forms an intracellular complex with DAP12 and CD94, and is trafficked to the ER but not the plasma membrane. (A) 293T cells were transfected with CD94 alone (first panel), CD94 and DAP12 (second panel), CD94/DAP12/NKG2C (third panel), or CD94/DAP12/NKG2E (fourth panel). Cells were stained for surface expression of CD94 and were permeabilized and stained for intracellular CD94. Data are representative of at least three independent experiments. (B) 293T cells were transfected with CD94, DAP12, and either NKG2C (left panels) or NKG2E (right panels) and assessed by confocal microscopy. CD94 is shown in green while differential interference contrast (DIC) is shown in gray for all samples. Pan-cadherin is shown in red in the top two panels, while protein disulfide-isomerase (PDI) is shown in red on the bottom two panels. Images are representative of at least three independent experiments. (C) NKG2C and NKG2E have identical transmembrane domains, suggesting that NKG2E can associate with CD94. (D) DAP12 associates with CD94 in the presence of NKG2C or NKG2E in 293T cells. 293T cells were transfected as in (B). Co-immunoprecipitation was performed on lysates, using an anti-CD94 mAb to pull down the NKG2/CD94/DAP12 complex, which was then probed using an anti-DAP12 mAb. Data are representative of at least three independent experiments. (E) NKG2E forms a complex with DAP12 and CD94 in Ba/F3 cells. Ba/F3 cells were transfected as in (B). Cell lysates were immunopreciptated with anti-CD94 and probed with anti-NKG2 (top row). Lysates were also immunoprecipitated with anti-NKG2 and probed with anti-DAP12 (third row). Total lysates were blotted with anti-NKG2 (second row) and anti-DAP12 (bottom row) as loading controls. Data are representative of at least three independent experiments. (F) Proteasomal inhibitor MG132 increases expression of CD94/NKG2E/DAP12 complexes. Ba/F3 cells were transfected with CD94, DAP12, and NKG2E, and were then treated with MG132 or with a vehicle control. Lysates were subsequently immunoprecipitated using an anti-NKG2 mAb or an IgG control. The resulting complex was probed using an anti-DAP12 mAb (top row) or an anti-NKG2 mAb (bottom row). Data are representative of at least three independent experiments.
To further elucidate the subcellular compartmentalization of CD94 in the presence of NKG2E, we performed confocal microscopy on 293T cells transfected with CD94, DAP12, and NKG2E or NKG2C. Cells were stained for CD94 (green) and PDI or pan-cadherin (both shown in red). PDI exclusively stains the ER, while both the ER and plasma membrane stain positively by pan-cadherin. We found that when CD94 was transfected with DAP12 and NKG2C, it colocalized with cadherin but not PDI, suggesting an association with the plasma membrane (Fig. 2B). In contrast, CD94 colocalized with both cadherin and PDI when transfected with NKG2E, indicating an association with the ER rather than the plasma membrane (Fig. 2B).
DAP12 is required for the surface expression of NKG2C(2) forming associationsvia interactionsbetween transmembrane residues(20). Because the transmembrane domain of NKG2E is identical to that of NKG2C (Fig. 2C), we wondered whether NKG2E was capable of forming a complex with DAP12 and CD94 despite its inability to be expressed on cell surfaces. To that end, we transfected 293T cells with various combinations of CD94, DAP12, and NKG2E or C. We then performed co-immunoprecipitation experiments, using an anti-CD94 antibody to pull down the protein complex and subsequently probing using anti-DAP12. As expected, we were able to confirm that CD94 formed a complex with DAP12 when cotransfected with NKG2C. Intriguingly, we also noted that NKG2E had a similar capacity to form complexes with DAP12 and CD94 despite its inability to localize to cell surfaces (Fig. 2D).
To verify our findings, we also tested the association between NKG2, CD94, and DAP12 in Ba/F3 cells. After transfection with CD94, DAP12, and/or NKG2E or C, we again used co-immunoprecipitation to analyze protein associations. When an anti-CD94 Ab was used to pull down the complex, we found that roughly the same amount of NKG2E bound CD94 as did NKG2C (Fig. 2E, top row), despite lower levels of expression overall (Fig. 2E, second row). Additionally, we used an anti-NKG2 antibody to pull down and probed using an anti-DAP12 antibody, and found that NKG2E was able to associate with DAP12 (Fig. 2E, third row). Total DAP12 levels are shown as a loading control (Fig. 2E, bottom row). Finally, NKG2E levels were increased upon incubation with the proteasomal inhibitor MG132, and we noted a corresponding rise in DAP12 in complex with NKG2E (Fig. 2F).
Overall, these data demonstrate that NKG2E forms an intracellular complex with CD94 and DAP12, even though it fails to reach the cell surface.
Hydrophobic residues in the extracellular portion of NKG2E are responsible for its intracellular retention
We next sought to determine which portion of NKG2E was responsible for its failure to be expressed on cell surfaces. Protein trafficking to membranes is dictated by the presence of specific recognition sequences within the transmembrane (TM), intracellular (IC), or extracellular (EC) segments. When comparing the sequences of NKG2E and NKG2C, we found that the majority of the non-homologous amino acid differences were localized to the end of the extracellular C-terminus in a region we dubbed the ECII segment (Fig. 3A). Furthermore, we noted that the ECII region of NKG2E contained a great deal of hydrophobic residues (Fig. 3A), which have previously been implicated in the retention of proteins in the endoplasmic reticulum(29).
Figure 3.
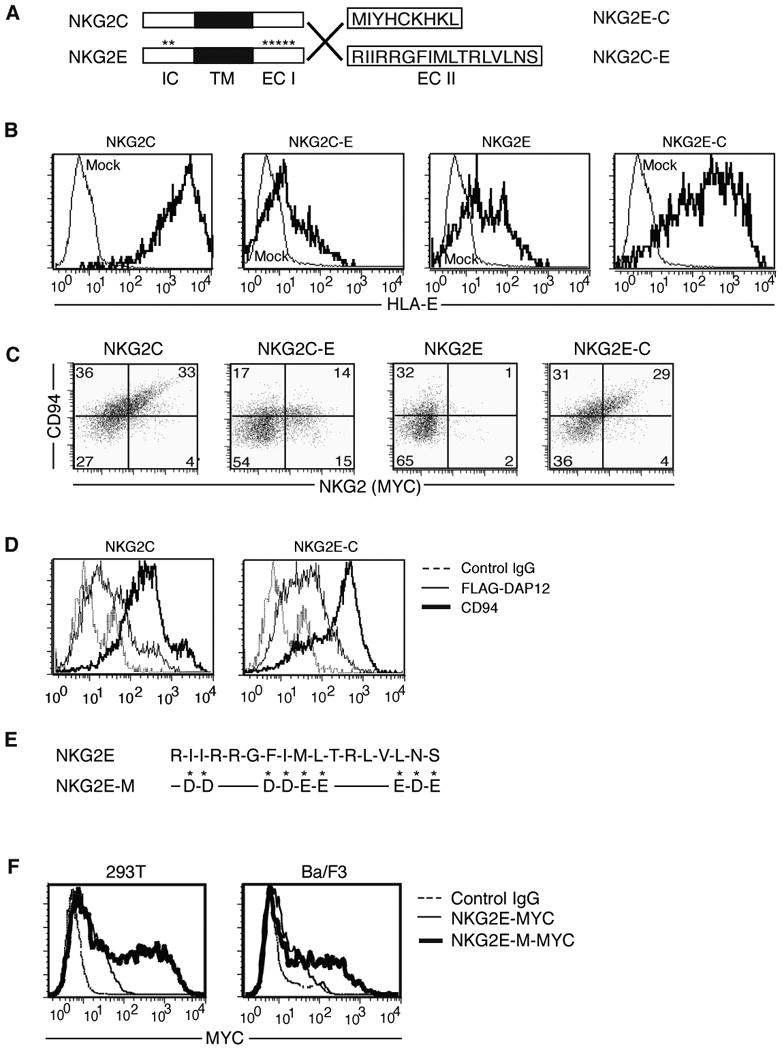
Hydrophobic residues in the extracellular portion of NKG2E cause it to be retained intracellularly. (A) Generation of chimeras is shown schematically. The final 17 amino acids of NKG2E were used to replace the final 9 amino acids of NKG2C to create the NKG2C-E chimera, while the reverse was performed to create the NKG2E-C fusion protein. (B) Evaluation of chimera function by flow cytometry. 293T cells were transfected with the indicated MYC-tagged fusion or control NKG2 in addition to CD94 and DAP12, and were evaluated for their ability to recognize HLA-E tetramers loaded with HLA-G leader peptide. Data are representative of at least three separate experiments. (C) Evaluation of chimera surface expression by flow cytometry. Ba/F3 cells were transfected as in (B), then stained with anti-CD94 and anti-MYC mAbs. Data are representative of at least three independent experiments. (D) Surface expression of DAP12 and CD94 was assessed to verify complex formation and translocation. Ba/F3 cells were transfected with untagged NKG2C or NKG2E-C, FLAG-tagged DAP12, and CD94, then analyzed by flow cytometry using anti-FLAG and anti-CD94 mAb. Data are representative of at least three independent experiments. (E) Schematic of the mutations generated in the ECII domain of NKG2E. Hydrophobic residues were replaced with either aspartic or glutamic acid according to whichever more closely approximated the size of the replaced residue. (F) Replacement of all three stretches of hydrophobic residues resulted in surface expression of NKG2E. MYC-tagged mutant or control NKG2E was transfected into 293T (left) and Ba/F3 (right) cells in conjunction with DAP12 and CD94, and surface expression was evaluated by flow cytometry using an anti-MYC antibody. Data are representative of at least three experiments.
To test whether the ECII region of NKG2E was responsible for its lack of expression on cell surfaces, we generated three pairs of chimeric molecules by exchanging the IC, ECI, or ECII segments of NKG2E and NKG2C (Fig. 3A and data not shown). We noted no change in receptor expression patterns between chimeras generated by switching either the IC or ECI sequences upon transfection into 293T cells along with CD94 and DAP12 (data not shown). However, when the ECII domain of NKG2E was replaced with that of NKG2C (NKG2E-C), we saw robust upregulation of surface expression compared to native NKG2E, as detected by tetramers of HLA-E loaded with HLA-G leader peptide (Fig. 3B) and by an anti-NKG2E antibody (Fig. S1B). Conversely, cells expressing NKG2C containing the ECII domain of NKG2E (NKG2C-E) were only weakly stained by the tetramer (Fig. 3B). Lysates were probed by western blot to verify chimera protein expression (data not shown).
We then sought to verify this expression pattern by using Ba/F3 cells, which better approximate physiological systems. Cells were transfected with CD94, DAP12, and MYC-tagged NKG2E, C, or one of our chimeras and then stained for CD94 and MYC expression. The inability of NKG2E to be expressed on cell surfaces was even more pronounced than in 293T cell line (Fig. 3C). We again noted that the surface association of CD94 and NKG2C was strongly reduced upon replacement of its ECII domain, while NKG2E gained the ability to coexpress with CD94 at cell surfaces upon receipt of the ECII domain of NKG2C (Fig. 3C). To further confirm that the ECII domain was critical for cellular localization and that our data was not an artifact from MYC-tagging, we transfected Ba/F3 cells with CD94, NKG2E-C or NKG2C, and N-terminal FLAG-tagged DAP12. Staining for CD94 revealed that its expression was highly upregulated in conjunction withNKG2E-C (Fig. 3D). Furthermore, DAP12 surface levels were increased when transfected with NKG2E possessing the NKG2C ECII domain (Fig. 3D).
Upon further examination of the ECII region of NKG2E, we found a large number of hydrophobic residues (9 of 17) assembled into three sequences (Fig. 3E). Suspecting that one or more of these sequences might be responsible for the intracellular retention of NKG2E, we generated substitutions of each hydrophobic sequence for negatively charged aspartic or glutamic acids, choosing either D or E to best approximate the size of the native residue replaced. In total, we created three partial mutants and one full mutant: NKG2E partial mutant 1 (NKG2E-PM1): I225I226 to D225D226; NKG2E-PM2: F230I231M232L233 to D230D231E232E233; NKG2E-PM3: L236N237S238 to E236D237E238; and NKG2E-M, which has all the hydrophobic residues replaced (Fig. S2A). We noted that while NKG2E-PM1 and NKG2E-PM3 were not efficiently expressed on cell surfaces after transfection with DAP12 and CD94, NKG2E-PM2 expression was increased roughly 10 fold, with surface expression of the NKG2E-M mutant lacking all hydrophobic sequences increased a further 10 fold (Fig. S2B). We transfected NKG2E-M into both Ba/F3 and 293T cells, noting that compared to native NKG2E, NKG2E-M was expressed at significantly higher levels on the surface when transfected with DAP12 and CD94 (Fig. 3F).
Taken together these data indicate that the hydrophobic residues of the ECII region are responsible for its intracellular retention.
NKG2E is present only in higher order primates and is strongly conserved
In the mouse, NKG2C and NKG2E are very closely related proteins with an overall homology of 91% in their amino acid sequences(22) and no differences in their ECII extracellular domains. This observation is in accordance with previous studies showing that, in mice, these two molecules serve identical functions(22). In contrast, the ECII extracellular domains of the human NKG2C and NKG2E are markedly divergent and, as our study shows, such divergence leads to major functional differences between these two molecules. Interestingly, phylogenetic analysis revealed that only Pan troglodytes (chimpanzees) and Pongo Abelii (orangutans) have a clearly homologous version of human NKG2E, as defined by the presence of the unique ECII extracellular domain (Fig. 4A). This places the appearance of the human-like NKG2E at only around 16 million years ago (Fig. 4B). Since its appearance; however, the distinctive amino-acid sequence of NKG2E and particularly the ECII extracellular domain appear to be strongly conserved. To formally test if NKG2E is evolving under strong selective constraints, we calculated dN/dS ratios using both the entire NKG2E sequences as well as restricting the analyses to the ECII region. We found that dN/dS ratios for the entire NKG2E were significantly lower than 1 in all pairwise comparisons (Human vs Chimpanzee: NKG2E dN/dS = 0.15; P = 4.98×10-6; Human vs Pongo: NKG2E dN/dS = 0.44; P = 0.03; Chimpanzee vs Pongo: NKG2E dN/dS = 0.45; P = 9.12×10-3). When focusing uniquely on the ECII region a similar trend was observed. Between Humans and Pongo we found dN/dS = 0.27 (lower than that observed for the entire sequence). This value was non-significant (P>0.05) which could be expected given the short length of the sequence (only 51bp). Between Humans and Chimpanzees, the nucleotide sequence coding the ECII region is 100% identical between the two species, which is again compatible with the notion that ECII region is probably evolving under strong selective constraints after the divergence between Human and Chimpanzees. Finally, we also noted that all the ECII hydrophobic sites that we showed to be important for the regulation of surface expression of NKG2E (Supplemental Figure 2) are 100% conserved across the three species. Collectively, these analyses provide statistical support that NKG2E has been evolutionary conserved since its recent emergence in the Hominidae family, which indicates that NKG2E plays an important and non-redundant role in host immunity.
Figure 4.
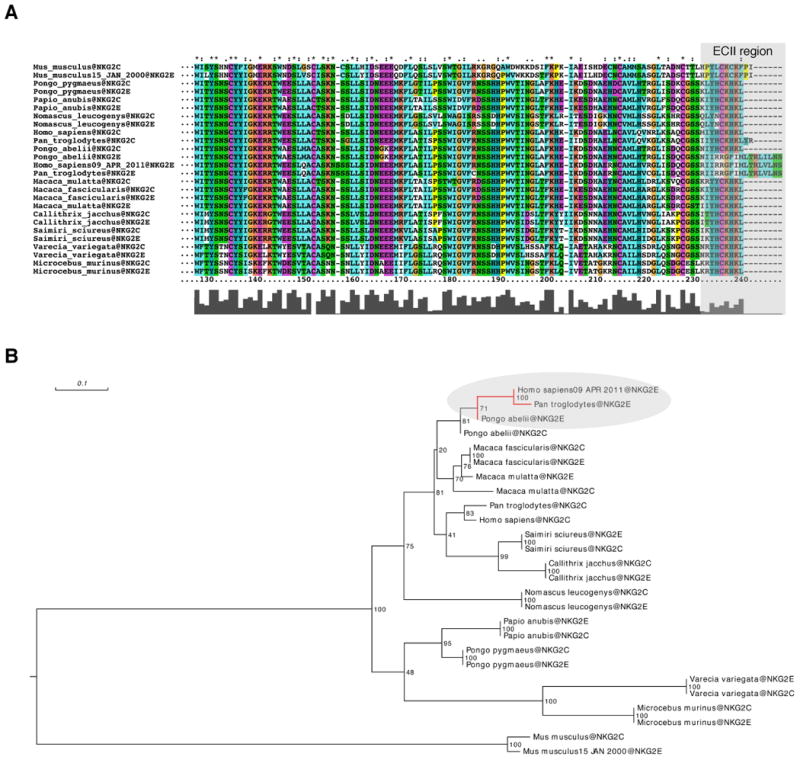
Phylogenetic analysis of the NKG2C-E family members. (A) Multiple sequence alignment of the protein sequence of NKG2C and NKG2E among primates and two rodents (mouse and rat). The alignment was done using the software Muscle and the visualization using ClustalX. (B) Phylogenetic tree based on protein sequence alignment. The tree was inferred by RaxML 7.2.8 under an LG+F+Gamma4 model of sequence evolution (empirical amino acid frequencies and four discrete gamma categories). The values indicated correspond to bootstrap support values based on 100 replicates.
Discussion
The exact role of NKG2E has remained poorly defined, largely due to technical limitations imposed by the lack of a specific antibody. Using tagged NKG2E and DAP12 as previously described, we found that NKG2E is translated and forms an intracellular complex with CD94 and DAP12.
DAP12 is a critical adaptor molecule required for surface expression and signaling through not only NKG2C and the activating members of the NKG2 family, but also through other activating NK receptors such as NKp44(30) and the non-ITIM bearing killer-cell inhibitory receptors (KIRs)(31). Consequently, it plays a key role in the activation of inflammatory and killing pathways by NK cells(1, 2, 6) and pathological cytotoxic T cells that have acquired an NK-like phenotype in the context of chronic viral infections(19) and autoimmune disorders such as celiac disease(17). Our experiments suggest that NKG2E associates with DAP12, suggesting two possible functions for NKG2E: it may play a role in the recognition of intracellular molecules, or it may act in a regulatory capacity by binding to adaptor molecules and preventing their association with other activating receptors on the cell surface. NKG2E was shown to have a higher affinity for HLA-E than the corresponding NKG2C complex (21). Given that NK cells themselves express HLA-E, it is possible that the role of the CD94/NKG2E/DAP12 receptor in NK cells is to recognize intracellular HLA-E complexed to peptides that direct trafficking to the ER rather than the cell surface.. Alternatively, soluble HLA-E could be taken up by NK cells and subsequently recognized by intracellular CD94/NKG2E/DAP12 complexes. Such a mechanism has been implicated in NK cell activation by HLA-G recognized by intracellular KIRD2L4 (32). However, the mechanism by which these complexes would be targeted to the endoplasmic reticulum remains unknown..
Another intriguing possibility is that NKG2E couldpotentially function as a regulatory molecule that limits destructive inflammatory responses by reducing free levels of DAP12 that could bind other inflammatory receptors. We did note that the proteasome inhibitor increased expression of DAP12 and NKG2E. We have also data suggesting that association with NKG2E results in increased ubiquitination of DAP12, but these studies were limited by the fact that we had no recourse but to transfect nonlimiting amounts of NKG2C and NKG2E, since there is no antibody that can differentiate between the two (Figure S1B). Furthermore, we were unable to show that NKG2E impacted in CD94/NKG2C-mediated activation of NK cells (data not shown).
Even though the specific role of NKG2E remains yet to be determined, our study demonstrates that NKG2E is functional in that it can retain CD94 in the intracellular compartment and form a complex with DAP12. Furthermore, our study reveals that NKG2E acts as an intracellular protein and that this characteristic is linked to the hydrophobic residues present in its ECII domain. Importantly, phylogenetic analysis suggests that NKG2E has evolved relative recently, is present only in Pan troglodytes (chimpanzees) and Pongo abelii (orangutans), and that the ECII region that is responsible for its intracellular retention is highly conserved, suggesting that the ECII region may play an important role in higher primates. Further studies may uncover whether NKG2E signaling represents a key intracellular activation pathway, or alternatively the molecule may play a novel regulatory role in restricting the availability of binding partners for other activating receptors.
Supplementary Material
Acknowledgments
LLL is an American Cancer Society Professor. This work was supported by the Digestive Disease Research Core Center at the University of Chicago.
Abbreviations
- NK cell
natural killer cell
- EC
extracellular
- TM
transmembrane
- IC
intracellular
- ITAM
immunoreceptor tyrosine-based activation motif
- ITIM
immunoreceptor tyrosine-based inhibition motif
Footnotes
This work was supported by R01 DK067180 and R01 DK058727 (BJ) and AI068029 (LLL).
Authorship: GAO, FT, JS, BM, and CC performed experiments; GAO provided input into the conceptual development and execution of the studies; JCG and LBB performed the genetic analysis; GAO and BS analyzed the results and made the figures; LLL provided constructs and cell lines; LBB and LLL participated in discussion and review of the manuscript; BS and BJ wrote the manuscript; BJ devised the research and supervised all investigations.
The authors declare no competing financial interests.
References
- 1.Tassi I, Klesney-Tait J, Colonna M. Dissecting natural killer cell activation pathways through analysis of genetic mutations in human and mouse. Immunological reviews. 2006;214:92–105. doi: 10.1111/j.1600-065X.2006.00463.x. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nature immunology. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meresse B, Jabri B. NKG2 receptor-mediated regulation of effector CTL functions in the human tissue microenvironment. Springer; 2006. [DOI] [PubMed] [Google Scholar]
- 4.Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nature immunology. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 5.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nature reviews Immunology. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scalzo AA, Yokoyama WM. Cmv1 and natural killer cell responses to murine cytomegalovirus infection. Current topics in microbiology and immunology. 2008;321:101–122. doi: 10.1007/978-3-540-75203-5_5. [DOI] [PubMed] [Google Scholar]
- 7.Schleinitz N, Vely F, Harle JR, Vivier E. Natural killer cells in human autoimmune diseases. Immunology. 2010;131:451–458. doi: 10.1111/j.1365-2567.2010.03360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Botet M, Bellon T, Llano M, Navarro F, Garcia P, de Miguel M. Paired inhibitory and triggering NK cell receptors for HLA class I molecules. Human immunology. 2000;61:7–17. doi: 10.1016/s0198-8859(99)00161-5. [DOI] [PubMed] [Google Scholar]
- 9.Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, Spies T. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 10.Kim DK, Kabat J, Borrego F, Sanni TB, You CH, Coligan JE. Human NKG2F is expressed and can associate with DAP12. Molecular immunology. 2004;41:53–62. doi: 10.1016/j.molimm.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 12.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty DE. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A [see comments] Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomasello E, Vivier E. KARAP/DAP12/TYROBP: three names and a multiplicity of biological functions. European journal of immunology. 2005;35:1670–1677. doi: 10.1002/eji.200425932. [DOI] [PubMed] [Google Scholar]
- 14.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 15.Le Drean E, Vely F, Olcese L, Cambiaggi A, Guia S, Krystal G, Gervois N, Moretta A, Jotereau F, Vivier E. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. European journal of immunology. 1998;28:264–276. doi: 10.1002/(SICI)1521-4141(199801)28:01<264::AID-IMMU264>3.0.CO;2-O. published erratum appears in Eur J Immunol 1998 Mar;28(3):1122. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. 2000;12:109–119. doi: 10.1006/smim.2000.0213. [DOI] [PubMed] [Google Scholar]
- 17.Meresse B, Curran SA, Ciszewski C, Orbelyan G, Setty M, Bhagat G, Lee L, Tretiakova M, Semrad C, Kistner E, Winchester RJ, Braud V, Lanier LL, Geraghty DE, Green PH, Guandalini S, Jabri B. Reprogramming of CTLs into natural killer-like cells in celiac disease. The Journal of experimental medicine. 2006;203:1343–1355. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park KS, Park JH, Song YW. Inhibitory NKG2A and activating NKG2D and NKG2C natural killer cell receptor genes: susceptibility for rheumatoid arthritis. Tissue antigens. 2008;72:342–346. doi: 10.1111/j.1399-0039.2008.01110.x. [DOI] [PubMed] [Google Scholar]
- 19.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 20.Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nature immunology. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser BK, Barahmand-Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 22.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. The Journal of experimental medicine. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang M, Orr MT, Spee P, Egebjerg T, Lanier LL, Sigal LJ. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34:579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabri B, Selby JM, Negulescu H, Lee L, Roberts AI, Beavis A, Lopez-Botet M, Ebert EC, Winchester RJ. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity. 2002;17:487–499. doi: 10.1016/s1074-7613(02)00427-2. [DOI] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Molecular biology and evolution. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 29.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annual review of biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 30.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. The Journal of experimental medicine. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS biology. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


