Abstract
Small, noncoding microRNAs (miRNAs) regulate diverse biological functions in the liver and increasing evidence suggests that they have a role in liver pathology. This Review summarizes advances in the field of miRNAs in liver diseases, inflammation and cirrhosis. MicroRNA-122, the most abundant miRNA in hepatocytes, has well-defined roles in HCV replication, and data indicate that it also serves as a viable therapeutic target. The role of miR-122 is also emerging in other liver diseases. Ample evidence exists for the important regulatory potential of other miRNAs in conditions associated with liver inflammation related to alcohol use, the metabolic syndrome or autoimmune processes. In addition, a broad array of miRNAs have been associated with the development of liver fibrosis both in animal models and human studies. The significance of the function and cellular distribution of miRNAs in the liver and the potential of miRNAs as a means of communication between cells and organs is discussed as well as the emerging utility of circulating miRNAs as biomarkers of different forms of liver damage and as early markers of disease and progression in hepatocellular carcinoma. Importantly, miRNA modulation in the liver represents a new therapeutic approach in the treatment armamentarium of hepatologists in the future.
Introduction
MicroRNAs (miRNAs), discovered by Ambros and colleagues in 1993,1 are small noncoding RNAs, 18–24 nucleotides in length, that regulate gene expression by binding to mRNAs to interfere with the process of translation.2 Genes that encode miRNAs are transcribed from DNA to a primary transcript (pri-miRNAs), which is processed into a short precursor (pre-miRNA) and then exported into the cytoplasm where it is further processed into a mature, single stranded miRNA2,3 (Figure 1). The biogenesis of miRNAs can be regulated at the transcriptional level by specific transcription factors and at the post-transcriptional level by changes in processing. Evidence suggests that single nucleotide polymorphisms in miRNA genes might also modulate miRNA activity and function.4 In most cases, miRNAs repress their targets via interaction with the 3' untranslated region (UTR), and this change is detectable at the RNA level;2,3 however, miRNAs that interact with their targets in a non-3' UTR-dependent or non-seed-dependent fashion cause upregulation of their targets. There are ~1,400 mammalian miRNAs5 and each miRNA can influence hundreds of gene transcripts. More than one miRNA can regulate each specific mRNA, which creates substantial complexity in their capacity to modulate fundamental biological processes.
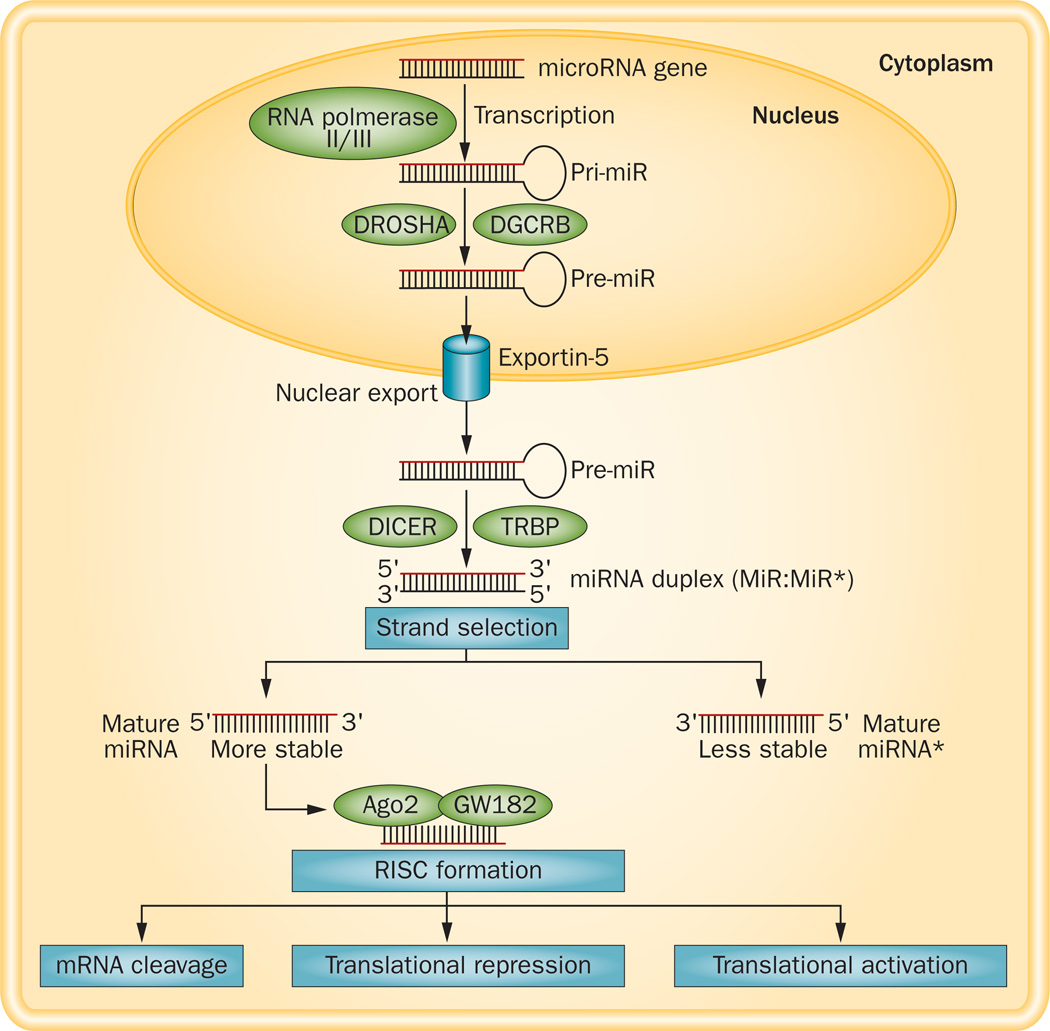
Figure 1.
Biogenesis of miRNAs. miRNAs are transcribed from miRNA genes via RNA polymerase II or III as pri-miRNA and cleaved by DROSHA–DGCR8 complex in the nucleus. The resulting precursor (pre)-miRNA is exported to the cytoplasm via exportin-5 complex. In the cytoplasm, DICER, along with TRBP, cleaves pre-miRNA to form mature miRNA (miRNA duplex). The strand is selected depending upon the stability, and the functional strand is loaded together with Ago2 and GW182 into the RISC. The less stable strand of miRNA gets degraded. Depending on the complementarity of the seed region of mature miRNA to the 3' UTR of the target mRNA gene, target mRNA either undergoes cleavage, translational repression or activation. Abbreviations: RISC, RNA-induced silencing complex; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA.
MiRNAs target and regulate essentially all biological processes and cell types, including those in the liver, and influence complex programmes of gene expression in virtually all cellular processes. Numerous reports have demonstrated that alterations in intracellular miRNAs correlate with various liver diseases including viral hepatitis, alcoholic and nonalcoholic steatohepatitis, drug-induced liver injury, autoimmune liver disease and ischaemia–reperfusion injury. Evidence is also emerging that miRNA expression profiles are distinct between liver diseases with different aetiologies.
This Review summarizes current knowledge on the role of different miRNAs in liver diseases and highlights the functions of the most relevant miRNAs that have specific roles in liver damage, hepatocyte functions, viral hepatitis, alcoholic and nonalcoholic liver disease, liver fibrosis and hepatocellular carcinoma (HCC). In addition, we discuss the emerging utility of miRNAs as potential biomarkers in liver diseases and promising aspects of miRNAs in therapeutic interventions for liver diseases.
Role of microRNAs in the liver
MiRNAs regulate lipid and glucose metabolism
Excess accumulation of hepatic triglycerides and fatty acids is characteristic of several liver diseases including alcoholic liver disease (ALD), NAFLD and NASH. Several lines of evidence suggest that miRNAs have a crucial role in metabolic homeostasis (reviewed extensively elsewhere6).
In the liver, miR-122 affects various genes involved in hepatic cholesterol and lipid metabolism, thereby having a central role in maintaining liver homeostasis. Inhibition of miR-122 using antisense approaches resulted in reduction of plasma cholesterol levels in mice7 and chimpanzees.8,9 A reduction in hepatic miR-122 expression has been reported in both human NASH and animal models of this disease,10,11 as well as in ALD in mice.12 Intriguingly, two studies have demonstrated that deletion of the gene encoding miR-122 in mice leads to the development of steatohepatitis, fibrosis and HCC.13,14 Despite the fact that miR-122-deficient mice (liver-specific knockouts and germ line knockouts) had lower levels of serum cholesterol, LDL and serum triglyceride than wild-type mice, they developed steatohepatitis and HCC.13,14 Reduced levels of HDL were also observed in these knockout mice.13,14
miR-33, miR-34, miR-103, miR-104 and miR-370 also modulate lipid and cholesterol regulatory genes.6 miR-370 affects lipid metabolism by targeting the mitochondrial enzyme, carnitine palmitoyl transferase, which is involved in the transport of long-chain fatty acids across the membrane.15 miR-34a targets hepatic SIRT1, and an increase in levels of miR-34a with a concurrent decrease in Sirt1 levels was found in fatty livers of diet-induced obese mice16 (reviewed elsewhere17). Meng et al.18 demonstrated an induction in miR-34a in ALD both in humans and mice; miR-34a regulates SIRT1 and caspase-2 levels and contributes to hepatic steatosis in this disease (Figure 2). miR-27b has also been described as a regulatory hub in lipid metabolism, potentially affecting both the liver and the heart.19 These findings indicate that dysregulation of miRNAs contributes to metabolic abnormalities; hence miRNA-based therapeutics might deliver promising results in this setting.
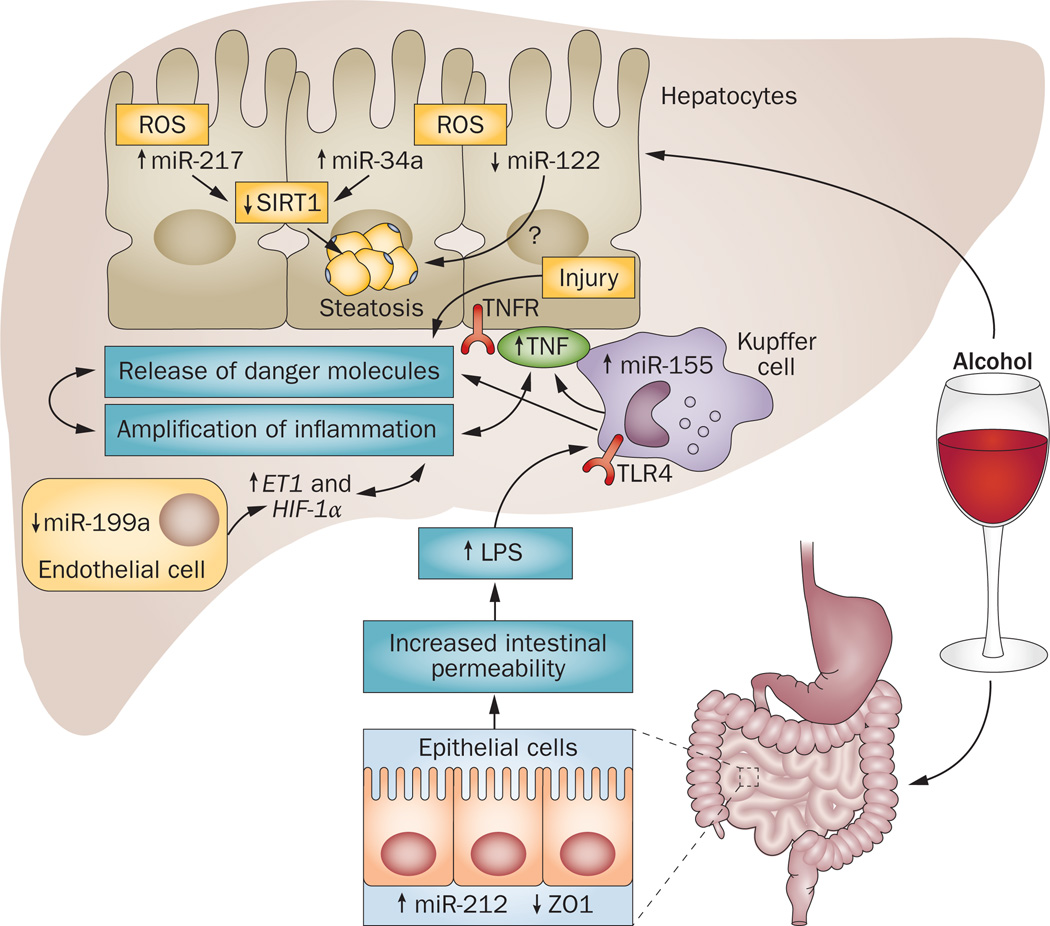
Figure 2.
Role of microRNAs in alcoholic liver disease. Alcohol alone, or along with its metabolites, increases levels of miR-212 in gut epithelial cells, which regulate the tight junction protein, ZO1. Decreases in ZO1 are associated with disruption of gut integrity and thereby increased intestinal permeability and increased bacterial or microbial translocation into the intestinal lumen and a subsequent increase in LPS. The excess of LPS in the liver affects immune (Kupffer cells), parenchymal (hepatocytes), and non-immune cells (endothelial cells). In response, Kupffer cells become activated and induction of miR-155 and release of TNF takes place. TNF causes further injury to hepatocytes and damaged hepatocytes release danger molecules, including miR-122, which are recognized by various immune cells; amplification of inflammation takes place. Alcohol intake also increases oxidative stress, which results in the upregulation of miR-34a and miR-217 and perhaps decrease in miR-122 in hepatocytes. Dysregulation of these miRNAs results in hepatic steatosis via SIRT1 and many other unidentified genes. Oxidative stress downregulates miR-199a in endothelial cells, which results in increase of ET1 and HIF-1α, both of which are genes that contribute to liver inflammation, steatosis and perhaps fibrosis. Abbreviation: LPS, lipopolysaccharide; ROS, reactive oxygen species; TLR4, Toll-like receptor 4; TNFR, tumour necrosis factor receptor.
MicroRNAs regulate inflammation
Various factors contribute to liver inflammation, including viral and bacterial infections, metabolic disorders, toxins and dietary factors. Growing evidence indicates that miRNAs have a fine-tuning role in liver inflammation by targeting various signalling molecules. A plethora of miRNAs has been implicated in inflammatory responses: miR-155, miR-132, miR-125b, miR-146a, miR-150, miR-181, let-7 and miR-21.20,21 miR-155 is involved both in innate and adaptive immune responses.22
miRNA-155 has received attention for its role in various liver diseases. Induction in miR-155 expression has been reported in mouse models of NASH and alcoholic steatohepatits.10,23 miR-155 levels were increased in Kupffer cells after alcohol feeding, and TNF has been identified as a target of miR-155 to promote liver inflammation23 (Figure 2). In mice receiving a choline-deficient and amino acid-defined (CDAA) diet, miR-155 was associated with HCC, in which it regulates C/EBP β levels.10 In addition, we have reported the increased expression of miR-155 in peripheral monocytes from patients with chronic HCV whereas no increase was found in patients who responded to treatment.24 Similarly, two other reports have described the induction of the BIC gene (a precursor of mature miR-155)25 and mature miRNA-155 in peripheral blood mononuclear cells (PBMCs) of patients infected with HCV.26 Interestingly, these reports also found that patients who eliminated HCV RNA successfully after antiviral treatment showed no induction of miR-155.25,26 Remarkably, the hepatic levels of miR-155 were also increased in patients with HCV and were shown to promote HCC through Wnt signalling.27 The role of miRNAs in HCV-induced hepatitis is shown in Figure 3.
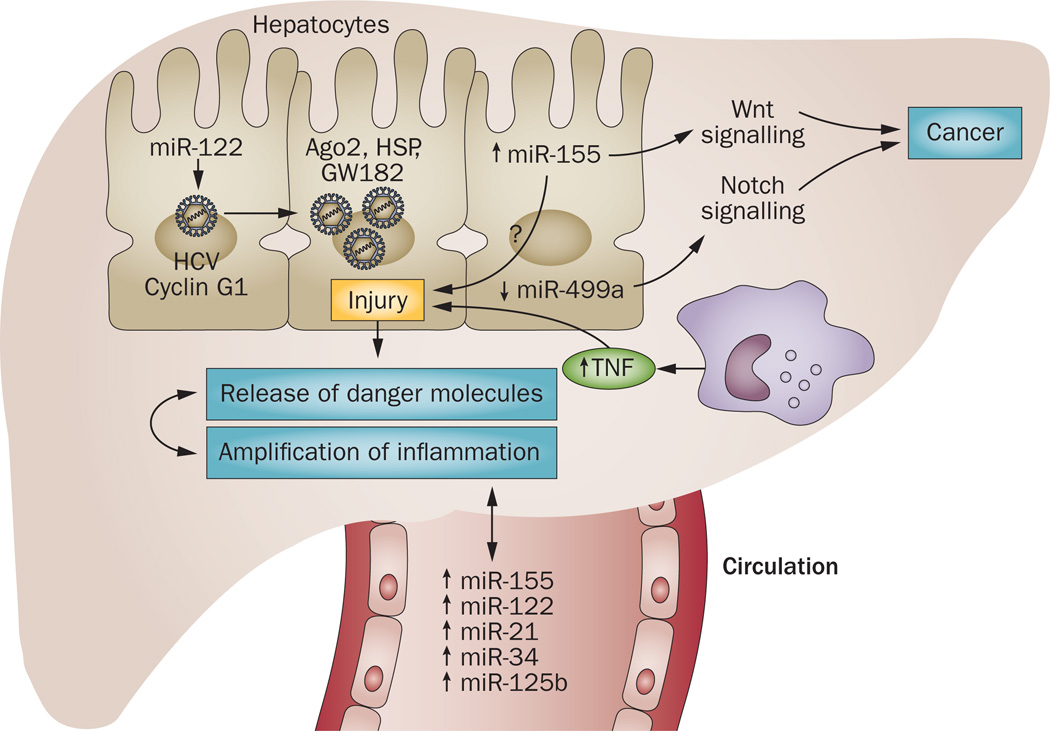
Figure 3.
MicroRNAs in chronic HCV infection. HCV infects hepatocytes and a unique interaction take place between host miR-122 and HCV 5’UTR along with proteins of RISC (Ago2, GW182 and HSPs), which results in enhanced replication of HCV virus. Other host factors, such as cyclinG1 (miR-122 target), are also involved in enhanced HCV replication in the presence of alcohol. HCV infection modulates other host miRNAs, leading to an increase in levels of miR-155 and a decrease in levels of miR-499 in hepatocytes, linking inflammation to cancer via Notch and Wnt signalling. HCV infection causes hepatocyte damage and thereby release of danger molecules, which activate immune cells and enhance local and systemic inflammation. Activation of peripheral monocytes causes an increase in levels of miR-155, which subsequently enhances release of TNF into the circulation. Various miRNAs such as miR-122, miR-155, miR-34a, miR-21, miR-146a and miR-125b are increased in the circulation of patients infected with HCV and might contribute to pathogenesis of the disease. Abbreviations: HSP, heat shock protein; RISC, RNA-induced silencing complex.
Unlike miR-155, miR-146a is a negative regulator of TLR signalling; it dampens proinflammatory responses via TRAF6 and IRAK-1. Consistent with its negative regulatory role in inflammation, mice deficient in miR-146a acquire hyperinflammation and myloproliferative phenotypes owing to hyperactivation of NF-κB.28
miR-132 is known to have a pivotal role in peripheral inflammation.29 Although its role in the brain has been widely studied, limited information is available on the role of miR-132 in immune regulation. Induction of miR-132 has been reported in the liver and Kupffer cells after chronic alcohol administration in mice.21 miR-132 also has a role in innate antiviral immunity by inhibiting expression of the p300 transcriptional coactivator during Kaposi sarcoma-associated herpesvirus infection.30 Intriguingly, miR-132 was shown to promote inflammation in adipocytes during nutrient starvation via a SIRT1–p65 axis,31 suggesting a cell-specific or stimulispecific role. Further studies are warranted to define its role in liver inflammation and immunity.
An unexpected role of miR-122 is emerging in liver inflammation. miR-122-deficient mice showed upregulation of Ccl2, which resulted in the intrahepatic recruitment of CD11bhiGr1+ inflammatory cells. These cells were the source of proinflammatory cytokines, IL-6 and TNF.14 Interestingly, serum miR-122 has been proposed as a marker of necroinflammation in patients with chronic HCV infection.32 Our own study also supports this notion as we found a correlation between serum miR-122 and serum miR-155 levels to be a marker of inflammation in patients with chronic HCV infection.24 These observations suggest that alterations in expression of specific miRNAs in liver inflammation might herald general inflammation or be disease-specific.
MicroRNAs in apoptosis and necrosis
In a mouse model of acute liver failure induced by D-galactosamine/lipopolysaccharide (LPS), miR-15b and miR-16 regulated TNF-mediated apoptosis via Bcl2, an anti-apoptotic gene.33 Inhibition of miR-15b and miR-16 resulted in reduced hepatic apoptosis and TNF production, which indicates a pro-apoptotic role for these miRNAs. miR-1187 is implicated in hepatocyte apoptosis via its targeting of caspase-8 in D-galactosamine/LPS-induced acute liver failure;34 the hepatic expression of miR-1187 was decreased with a concomitant increase in caspase-8 expression.34 In FASLG (CD95) receptor-induced apoptosis, the global loss of miRNAs in hepatic cells was associated with increased cell death, underscoring the importance of miRNAs in hepatocytes.35 miR-221 was highly induced in response to apoptosis, and delivery of miR-221 in vivo by adeno-associated virus serotype 8 (AAV8) resulted in delayed FAS-induced fulminant liver failure in mice.35 miR-221 regulates hepatic expression of BBC3 (a pro-apoptotic gene; also known as PUMA) and protects hepatocytes from apoptosis, suggesting a pleiotropic role of miRNAs in acute liver failure.
MicroRNAs affect cell cycle and proliferation
Various miRNAs control hepatocyte proliferation, and miR-21 has been shown to regulate various genes involved in the cell cycle and DNA synthesis. After performing partial hepatectomy in mice, miR-21 is induced and it inhibits hepatocyte DNA synthesis via its effect on Btg2 and FoxM1 (secondary target).36 miR-21 also has a role in hepatocyte proliferation by promoting cyclin D1 translation.37 Inhibition of induced miR-21 after two-thirds partial hepatectomy impaired the progression of hepatocytes into S phase via RhoB, which is involved in cyclin D1 translation. 37 Contrary to miR-21, miR-26a is downregulated after partial hepatectomy and promotes hepatocyte proliferation by regulating G1/S-specific cyclin-D2 (CCND2) and G1/S-specific cyclin-E2 (CCNE2) cell cycle protein expression.38 miR-217 has an antiproliferative role and methylation of its promoter region results in its decrease during liver regeneration.39 Decreases in levels of miR-127 inversely correlated with levels of B-cell lymphoma 6 protein homolog (Bcl6) and N-lysine methyltransferase SETD8 24 h after partial hepatectomy.39 miR-221 is identified as a pro-proliferative miRNA as it regulates hepatocyte proliferation during liver regeneration by affecting the expression of Arnt.40
MicroRNAs and epithelial–mesenchymal transition
Epithelial–mesenchymal transition (EMT) contributes to liver regeneration and is potentially involved in cancers in the liver. EMT involves the termination of epithelial tight junctions, modulation of adherens junctions, remodelling of the cytoskeleton and loss of cell polarity.41 Little is known about the role of miRNAs in EMT regulation in liver diseases. However, given that different cells undergo EMT in various liver diseases, it is likely that miRNAs have a crucial and unidentified role in this process. miR-155 is highly induced in TGF-β-induced EMT in NMuMG epithelial cells, in which it has a role in cell migration and invasion by targeting RhoA kinase.42 Various studies have demonstrated the critical role of members of the miRNA-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) in EMT by modulating the expression of ZEB1 and ZEB2, the transcriptional repressors of cadherin-1 (E-cadherin).43–45 Consistent with these findings, miR-200a and miR-200b are upregulated in liver fibrosis.46 A common network of miRNAs (miR-106a, miR-106b, miR-18a, miR-18b, miR-17, miR-93, miR-301a and miR-130b) that regulate embryonic stem cell self-renewal are also involved in EMT of hepatocytes via the PTEN–TGF-β axis, implicating their potential to transform normal hepatocytes into neoplastic cells.47 These studies have opened a new avenue of research to explore the dual role of miRNAs in EMT and embryonic stem cell processes.
MicroRNAs and liver fibrosis
The importance of miRNAs in liver fibrosis has been reviewed extensively elsewhere,48 thus here we focus on emerging miRNAs identified in liver fibrosis. miRNAs are involved in regulation of liver fibrosis at multiple levels. Key cytokines and growth factors determining liver fibrosis regulate expression of profibrogenic and antifibrogenic miRNAs (summarized elsewhere49). Hepatic stellate cells (HSCs), the primary cell type that contributes to liver fibrosis, undergo proliferation and differentiation into myofibroblast-like cells upon fibrogenic injury of the liver. A broad variety of miRNAs have been identified in regulating HSC activation (Figure 4). Increased serum levels of miR-571 have been proposed as a potential biomarker of liver fibrosis, and serum levels of miR-542, miR-652 and miR-181b are decreased in cirrhosis50,51 (Figure 4). miR-19b and miR-29 have been identified as antifibrotic genes on the basis of their effects on HSCs.52–54 Interestingly, miR-29 has also been identified as a regulator of disease-associated pathways in biliary atresia in a mouse model, which suggests that individual miRNAs have regulatory potential in different cell types in the liver.55
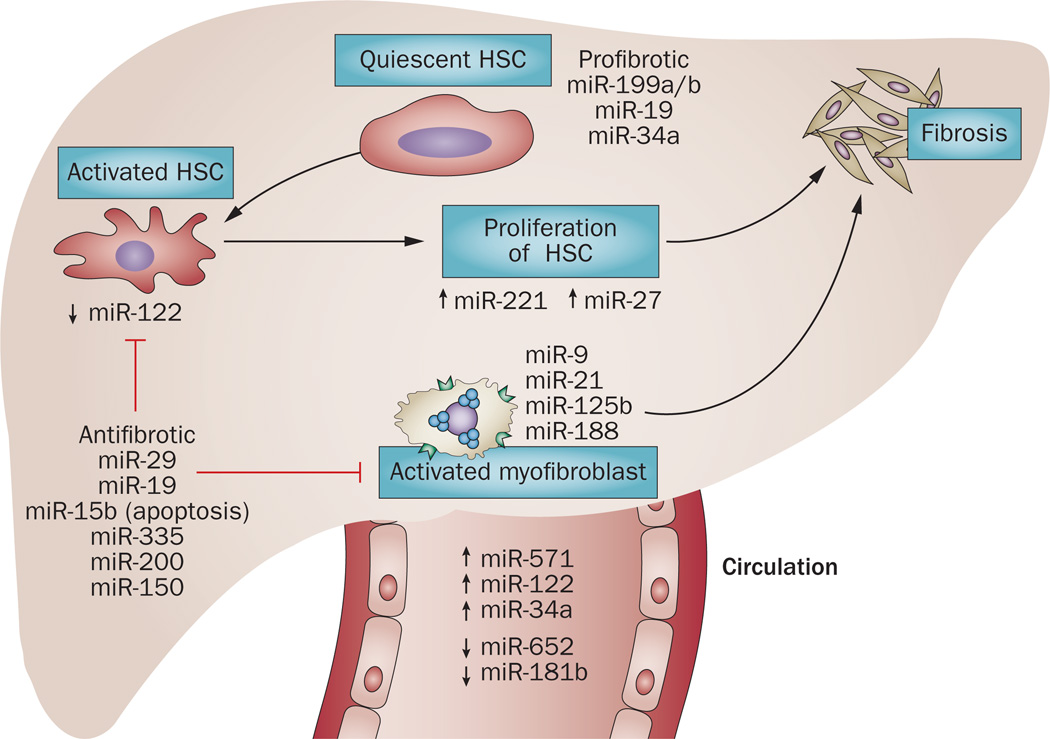
Figure 4.
Regulation of fibrogenetic events by microRNAs. miRNAs can be profibrotic (miR-199a/b, miR-19 or miR-34) and antifibrotic (such as miR-29, miR-19 and miR-150). miR-221 or miR-27 regulate activation and proliferation of HSCs. miRNAs, such as miR-9, miR-21 and miR-188 are involved in myofibroblast activation and synthesis of extracellular proteins and collagen deposition. Fibrotic changes are also associated with dysregulation of miRNAs in the circulation. Both increased levels (miR-122, miR-34a and miR-571) and decreased levels (miR-652 and miR-181b) of miRNAs have been found in the circulation. Abbreviation: HSC, hepatic stellate cell.
TGF-β, a modulator of fibrosis, induces miR-21 in patients infected with HCV, in whom it downregulates SMAD7, a negative regulator of TGF-β signalling. This finding suggests positive feedback regulation of miR-21 and TGF-β.56 miR-199a, miR-199a*, miR-200a and miR-200b levels are highly associated with progression of liver fibrosis both in patients with chronic HCV infection and in mice with CCl4-induced fibrosis.46
A decrease in miRNA-150 and miRNA-194 was found in HSCs of bile-duct-ligated rats; these miRNAs targeted c-Myc and Rac-1, respectively, and thereby inhibited HSC activation.57 miR-199 and miR-200 families have been shown to promote liver fibrosis by enhancing fibrotic genes (such as those that encode α1 procollagen, collagenase 3 and metalloproteinase inhibitor 1).46 miR-146a, which regulates SMAD4, was downregulated in HSCs in CCl4-induced liver fibrosis.58 Overexpression of miR-146a repressed TGF-β-induced HSC proliferation and increased HSC apoptosis.58
miR-122 is required for normal liver homoeostasis and decreased levels of this miRNA have deleterious effects on the liver. In fact, miR-122-deficient mice develop steatohepatitis and fibrosis.13 KLF6, a profibrogenic factor, was identified as a target of miR-122 and a role for the miR-122a–KLF6 axis has been proposed in liver fibrosis.13 We have shown that chronic alcohol use can modulate miR-122 expression, and decreased expression of this miRNA was found in the livers of mice fed with alcohol.12 Given that prolonged chronic alcohol use can lead to liver fibrosis, a decrease in miR-122 after alcohol consumption suggests a direct or indirect role for miR-122 in alcohol-induced fibrosis. Consistent with this notion, a study revealed the role of miR-122 in HSCs; expression of miR-122 was decreased in activated HSCs in a CCl4-induced liver fibrosis mouse model.59 miRNA-122 negatively regulates liver fibrosis by targeting the gene that encodes transmembrane prolyl 4-hydroxylase, which is involved in the process of collagen maturation.59 However, the underlying mechanisms of decreased miR-122 in ALD and its precise role in different cell types is yet to be defined. To conclude, it seems that the majority of miRNAs modulate HSCs and therefore hold promise for therapeutic interventions in liver fibrosis.
Emerging role of organelle-associated microRNAs
Advances in miRNA detection have revealed the unexpected existence of certain miRNAs in various organelles such as mitochondria and the nucleus60,61 (summarized in Table 1). The first study demonstrating the existence of mitochondrial miRNA (mito-miRNA) was carried out in rat livers, and it was postulated that these mito-miRNAs might be involved in processes such as apoptosis, cell proliferation and differentiation.61 Streptozotocin treatment induces type 1 diabetes in mice and results in the alteration of mito-miRNAs.62 In particular, levels of miR-494, miR-202-5p, miR-134 and miR-155 are increased, whereas levels of miR-122 are decreased in the liver mitochondria of these mice.62 Interestingly, Argonaute-2 (Ago2, the effector of the miRNA-induced silencing complex that mediates mRNA repression) was also detected in mitochondrial lysates, raising the possibility that mitochondrial-associated miRNAs might be functional. The finding of the presence of Ago2 and miRNA in mitochondria could be extrapolated to have a role in liver diseases as mitochondria have a major role in liver disease pathogenesis; a functional RISC (RNA-induced silencing complex) along with miRNAs might have a direct role in modulating mitochondrial proteins. In this context, Das and colleagues showed for the first time that miR-181c can translocate to mitochondria where it targets the mitochondria genome via Cox1 resulting in electron transport chain complex IV remodelling and mitochondrial dysfunction.60
Table 1.
Mitochondrial and nuclear miRNAs and their potential roles
| miRNAs | Function | References |
|---|---|---|
| Mitochondrial miRNAs | ||
| Rno miR-21, miR-130a, miR-130b, miR-140*, miR-320, miR-494 and miR-671 | Potential role in apoptosis, cell proliferation and differentiation | Kren et al. (2009)61 |
| miR-705, miR-494, miR-202-5p miR-451-7b, miR-26a, miR-122, miR805, miR-690, miR-155 and miR-134 | Mitochondrial dysfunctions | Bian et al. (2010)62 |
| pre-miR-302a, pre-let-7b, miR-365, miR-720, miR-133b, miR-1974, miR-24, miR-133a, miR-125a-5p and let-7 family members | Potential role in cell proliferation and differentiation | Barrey et al. (2011)135 |
| has-miR-494, miR-1275 and miR-1974 | A possible role in regulating translation in mitochondria | Bandiera et al. (2011)136 |
| miR-181c | Mitochondrial dysfunctions | Das et al. (2012)60 |
| Nuclear miRNAs | ||
| miR-29b | Might regulate transcription or splicing of target transcripts | Hwang et al. (2007)63 |
| miR-320 | Transcriptional gene silencing | Kim et al. (2008)64 |
| miR-671 | Gene silencing of noncoding antisense transcripts | Hansen et al. (2011)65 |
Emerging studies suggest that miRNAs might have biological functions that are distinct from canonical 3' UTR target mRNA repression. miR-29b, miR-320 and miR-671 are mostly abundant in the nucleus and regulate gene transcription.63–65 However, the distribution and function of mitochondrial and nuclear miRNAs in liver diseases is yet to be explored.
Contribution of microRNAs to liver diseases
Viral hepatitis
The discovery of the involvement of miR-122 in the HCV replication process is one of the best examples of the potential importance of miRNAs and targeted miRNA-based therapeutic approaches.66 Inhibition of miR-122 using antisense approaches such as antagomirs and ‘locked nucleic acids’ (LNAs) has reduced HCV replication in mice7 and chimpanzees.9 Early phase IIa human clinical trials indicate that inhibition of miR-122 can inhibit HCV levels in patients with chronic HCV infection.67 miRNAs are usually thought to control the expression of the target gene. However, miR-122 paradoxically increases the replication of HCV by interacting with the 5'-UTR of HCV RNA.68 This unique requirement of miR-122 for HCV replication is not fully understood. The possibility that miR-122 might be involved in HCV RNA degradation, in its localization to functional replication complexes, or in the translation to replication switch have been suggested and are yet to be explored.69 A study showed that Ago2 protein is directly and efficiently recruited to the HCV 5'-UTR by miR-122 but not by an unrelated miRNA; this finding suggests that direct base-pairing is used to guide miR-122 and Ago2 to the HCV RNA, resulting in RNA stability and translation stimulation.70
miRNA-122 can also be the target of disease modifying factors in HCV infection via inhibition of cyclin G1, which has been identified as a host factor in HCV replication. In this context, alcohol consumption is known to enhance HCV replication, and a role for miR-122 in this process has been demonstrated;71 alcohol treatment in HCV-infected hepatoma cells increases levels of miR-122, which in turn modulates cyclin G1 expression.71 In addition, miR-122 upregulation by alcohol was associated with increased levels of the P body protein, GW182 and heat shock protein 90, which contributed to enhance HCV replication.72 An additional consideration is that regulation of HCV replication by miR-122 has been investigated in isolated hepatocytes or hepatoma cells whereas changes in total liver levels of miR-122 expression might reflect miR-122 regulation in other cell types.
The miRNA expression profile in patients with chronic HCV, HBV and HCC was found to be different although a cluster of 19 miRNAs were commonly affected in both HCV and HBV.73 In HBV-infected livers, pathways related to cell death, DNA damage, recombination and signal transduction were activated. On the other hand, in HCV infected livers, pathways related to immune responses, antigen presentation, cell cycle and lipid metabolism were abnormally expressed.73 miR-122-induced downregulation of heme-oxygenase-1 was shown to negatively affect miR-122-mediated suppression of HBV, indicating that host and viral factors might both involve miRNA regulation.74 miRNA-449a has been demonstrated to have a role in modulating expression of CHI3L1 (which encodes chitinase-3-like protein 1, an inflammatory marker [also known as YKL40]), through targeting the components of the NOTCH signalling pathway after HCV infection, meaning that this miRNA might act as a bridge between inflammation and fibrosis75 (Figure 3).
Drug-induced liver injury
Most available data on miRNAs in drug-induced liver injury (DILI) are related to acetaminophen-induced acute liver failure. Levels of miR-122 and miR-192 are decreased whereas levels of miR-710 and miR-711 are increased in the livers of mice administered with acetaminophen.76 The mechanisms causing a decrease in levels of miR-122 and miR-192 after DILI are not clear, but the most probable explanation is that hepatocyte damage releases these miRNAs into the circulation. Tetrachlorodibenzo-p-dioxin (TCDD)-induced liver damage in mice results in a time-dependent decrease in levels of miR-101a and miR-122 in the liver.77 Cox2 (also known as Ptgs2), a miR-101a target gene, was increased in mice exposed to TCDD.77 In general, various miRNAs regulate P450 enzymes and nuclear receptors that are involved in drug metabolism.78 miR-378 has been shown to regulate Cyp2e1, an enzyme that is involved in the metabolism of acetaminophen, alcohol and CCl4,79 again indicating that miRNAs might have an important role in drug metabolism and toxicity. Thus, improved understanding of the role of miRNAs in drug toxicity might facilitate drug development.
Alcoholic steatohepatitis and NASH
Aside from the abnormalities in miR-122 expression described earlier, increased expression of miR-320, miR-486, miR-705 and miR-1224, and decreased expression of several other miRNAs (miR-27b, miR-214, miR-199a, miR-192 and miR-183) was found in mouse livers after chronic alcohol feeding.80 We have also found increased expression of miR-132 in mice fed with alcohol; this miRNA has been identified as a critical regulator of inflammation along with miR-155.21,29,81 In addition, miR-212 is involved in alcohol-induced gut permeability.82 Alcohol consumption resulted in increased miR-212 expression in the gut, which in turn inhibited ZO-1 protein levels (a tight junction protein).82 miR-217 is involved in alcohol-induced steatosis by regulating SIRT1, which affects essential metabolic regulatory transcription factors.83 Alcohol also modulates the expression of miR-199 in rat liver sinusoidal endothelial cells and human endothelial cells.84 A decrease in miR-199 was associated with an increase in endothelin-1 (ET-1) and hypoxia-inducible factor-1α (HIF-1α), which have a crucial role in inflammation and steatosis. These studies suggest that alcohol modulates miRNAs in different cell types of the liver to amplify its effect. Whether similar miRNAs are involved in NASH remains to be determined. Kupffer cell activation is common both in alcoholic steatohepatitis and NASH, thus it is tempting to speculate that the miRNA-155 upregulation that occurs in Kupffer cells in ALD23 might also be present in NASH.
Altered hepatic miRNA expression has been found in patients with NASH.10 miR-122 levels in the liver are substantially decreased in patients with NASH and it has been proposed that these decreased levels in the liver contribute to altered hepatic lipid metabolism.10 These findings have been corroborated in a study in miR-122-deficient mice, in which liver steatosis was found.13,14 These findings are presumably related to reduced levels of miR-122 in hepatocytes; however, a role for miR-122 in inflammatory and other cells is yet to be evaluated. Cell-specific targeting of miR-122 is now possible using recombinant adeno-associated virus (rAAV)-mediated and ‘Tough Decoy’ (TuD)-directed inhibition of this miRNA in hepatocytes in vivo in animal models.85
Hepatocellular carcinoma
HCC is the third leading cause of cancer-related mortality worldwide.86 Key processes in cancer biology, including cell cycle and proliferation, are regulated by miRNAs at multiple levels; therefore, it is not unexpected that alterations in miRNAs contribute to the development of HCC. Human miRNA genome (miRNome) analyses of samples of healthy liver, liver with hepatitis and hepatocellular carcinoma revealed the downregulation of miR-199a/b-3p in HCC, which was correlated with poor survival.87 miR-199a/b-3p is involved in regulation of the PAK4/Raf/MEK/ERK pathway, which suppresses HCC growth. Furthermore, the delivery of miR-199a/b-3p mimics via AAV8 to HCC-bearing mice inhibited tumour growth, indicating that this miRNA is a potential target for HCC treatment.87 Intriguingly, the majority of miRNAs that localize to genomic fragile sites are associated with cancer.88 In line with this notion, miR-151 is frequently localized to the amplified region of chromosome 8q24.3 and associated with intrahepatic metastasis of HCC.89 miR-151 has a vital role in metastasis by activating RAC1, CDC42 and other Rho GTPase genes by targeting RhoGDIA (a metastasis suppressor) and also enhances cell motility and spreading by acting synergistically along with FAK.89
Various studies have suggested the involvement of miR-221 in HCC. miR-221 is upregulated in HCC and controls various genes involved in the cell cycle (such as CDKN1C and CDKN1B90) and DNA damage (DDIT4).91 In a mouse model, inhibition of miR-221 with Chol-anti-miR-221 (a cholesterol-modified isoform of anti-miR-221) resulted in reduction of tumour cell proliferation and increased levels of markers of apoptosis and cell-cycle arrest and, as a consequence, increased survival.92 From these studies it is apparent that antimiR-221 could be exploited as a potential therapeutic target in HCC-related malignancies.
By contrast, miR-29 is downregulated in HCC and targets various anti-apoptotic genes, such as BCL2, MCL1 and MMP2.93,94 It is becoming increasingly evident that miRNAs not only regulate genes involved in tumour angiogenesis, invasion and metastasis but also modulate the expression of other noncoding RNA, hence amplifying their effect. In this regard, miR-29 has been shown to regulate the long noncoding RNA MEG3 by modulating its methylation through effects on DNMT1 and DNMT3b.95
In addition, certain miRNA expression profiles are characteristic of HCC.46,96 Downregulation of miR-24a, miR-26a, miR-15a/b, miR-150 and miR-195 was found in a myc-induced mouse model of HCC.97 Cyclin D2 and E2 are regulated by miR-26a and this miRNA can also induce G1 arrest in human liver cancer cells.97 In human HCC, miR-26a was decreased and AAV-vector-driven overexpression of miR-26a might prevent liver tumours.97 Interestingly, a gender difference in miRNA expression has been observed.98 The expression of miR-26a and miR-26b in nontumour liver tissue was higher in women than in men, whereas HCC tissue regardless of sex had reduced miR-26 expression compared with noncancerous tissues.98 Transcriptome analysis revealed that activation of signalling pathways between NF-κB and IL-6 might be involved in tumour development.98 Other abnormally expressed miRNAs in HCC include an increase in levels of miR-21, miR-22 and miR-517a46,96,99,100 and a decrease in levels of miR-122, miR-20 family, miR-124 and let-7 family members.100 Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain in metastatic properties, which indicates an important role for miR-122 in hepatocarcinogenesis.101
Other liver diseases
Altered hepatic miRNA expression has been found in patients with primary biliary cirrhosis (PBC).102 Interestingly, miR-122a was downregulated along with miR-16a, whereas miR-328 and miR-299-5p were increased.102 These miRNAs are associated with processes such as cell proliferation, apoptosis, inflammation, oxidative stress and metabolism. Upregulation of miR-506 was associated with decreased CI−/HCO3− anion exchange 2 (AE2) expression in the biliary epithelium of patients with PBC.103 AE2 is involved in intracellular pH homeostasis and secretin-stimulated biliary bicarbonate secretion, and common genetic variations in AE2/SLC4A2 are associated with disease susceptibility in patients with PBC.103,104 The important role of AE2 has also been demonstrated in AE2-deficient mice, which showed immunologic and hepatobiliary changes resembling human PBC.105 Similarly, PBMCs from patients with PBC showed dysregulation of various miRNAs that are involved in cell differentiation, cell cycle and MAPK, TGF-β and Wnt signalling pathways.106
Studies suggest that liver-specific miRNA-122 controls systemic iron homeostasis, which contributes to hemochromatosis.107 The role of miRNAs in autoimmune hepatitis deserves further investigation.
Circulating miRNAs as biomarkers of disease
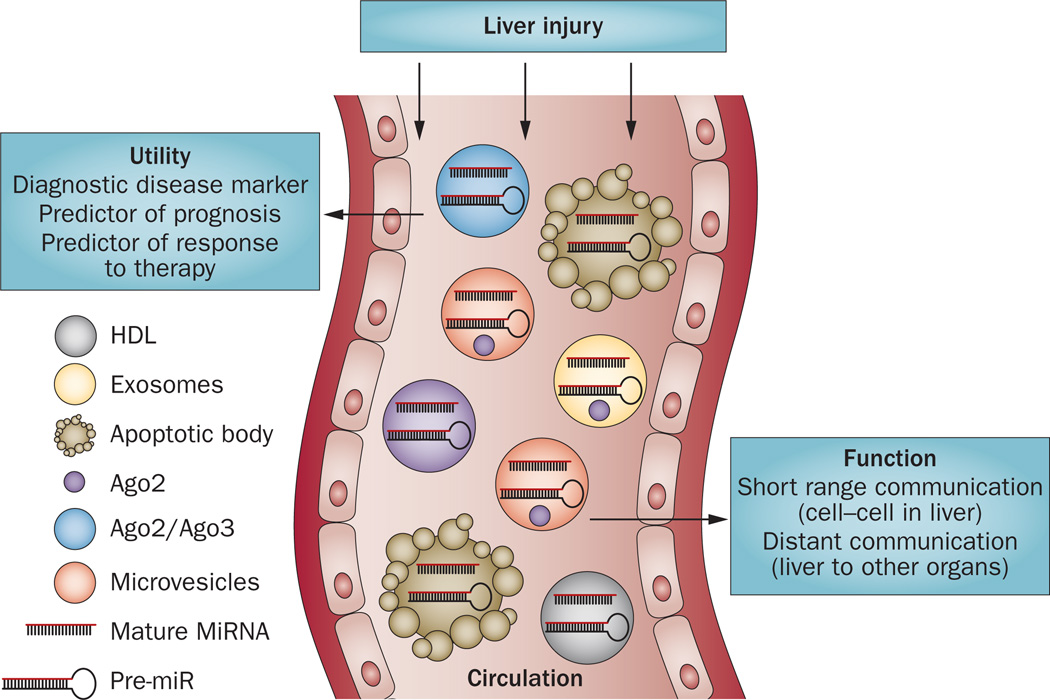
Studies have documented the presence of miRNAs in the circulation, extracellular spaces and in various body fluids, including bile.108–110 Circulating miRNAs are distributed in various forms, including in association with Ago2,111 vesicles such as exosomes,112 macrovesicles or apoptotic bodies113 or bind HDL114 (Figure 5). The cellular release processes of miRNAs and/or factors that might govern these associations are yet to be understood. In addition to miRNAs, the components of RISC proteins such as Ago2 and its interacting partner GW182 have been found in exosomes and microvesicles.115 Moreover, miRNAs are associated with lipid-based carriers and lipidfree proteins and the transfer of miRNAs between cells has a potential role in intracellular communication.116
Figure 5.
Circulating/extracellular microRNAs as biomarkers of liver disease. Liver damage caused by alcohol, acetaminophen, viral or bacterial infection, chemical toxins and diet results in the release of various miRNAs into the circulation. The circulating miRNAs (mature or precursor) can be released inside exosomes, microvesicles, HDL, apoptotic bodies and with proteins (Ago2). The components of RISC proteins such as Ago2 and its interacting partner GW182 have also been found in exosomes and microvesicles. Abbreviations: RISC, RNA-induced silencing complex.
The biodistribution of extracellular miRNAs and their potential functional role on target cells remain to be elicited. It is tempting to speculate that miRNAs in the circulation serve as ‘messengers’ and have the potential to influence functions of cells or tissues distant from their site of origin. In support of this idea, emerging reports indicate that miRNAs in plasma are delivered to recipient cells for uptake.114 Indeed, miRNAs might provide a means of ‘communication’ between tissues, organs or even species (Table 2). Consistent with this notion, a study found that miRNA-168a, which is specifically expressed in rice, was absorbed in the gut of both humans and mice, entered the circulation and had a biological effect on decreasing LDL receptor function.117
Table 2.
Emerging role of circulating miRNAs in cellular processes
| miRNA | Function | References |
|---|---|---|
| Exosomal miRNAs Epstein–Barr virus-encoded miRNAs |
Cell–cell communication | Valadi et al. (2007)112 Pegtel et al. (2010)137 |
| miR-126 | Atherosclerosis | Zernecke et al. (2009)113 |
| miR-150 | Cell migration | Zhang et al. (2010)138 |
| miR-146a | Cell growth inhibition | Kosaka et al. (2010)139 |
| miR-29a | HIV Tat and morphine-mediated neuronal dysfunction | Hu et al. (2012)140 |
| miR-133b | Neurite outgrowth | Xin et al. (2012)141 |
Because of their great stability in the serum and plasma, miRNAs are considered to be attractive biomarkers for liver disease (Figure 5). Numerous studies have reported increased levels of circulating miR-122 in liver diseases with different aetiologies, which suggests that miR-122 in the serum seems to be a potential marker of liver injury in general rather than indicating a specific aetiology of liver disease; increased levels of miR-122 have been found in HCV infection,24,32,118 HBV infection,119 HCC,50,120–123 DILI,76,124 NAFLD/NASH125 and ALD12,126 (Table 3). As miR-122 is abundant in hepatocytes, it is tempting to speculate that miR-122 is released to the circulation after hepatocyte injury through yet to be defined mechanisms.
Table 3.
Circulating miRNA signatures in liver disease
| Liver disease | miRNA signature | Expression | References |
|---|---|---|---|
| ALD | miR-122 (acute alcohol, microsteatosis) miR-122 and miR-155 (chronic alcohol, macrosteatosis) |
Increased Increased |
Zhang et al. (2010)126 Bala et al. (2012)12 |
| NAFLD/NASH | miR-122, miRR-34a and miR-192 | Increased | Cermelli et al. (2011);118 Tryndyak et al. (2012)125 |
| HCV | miR-122, miR-34a, miR-155, miR-125b, miR-146a and miR-21 | Increased | Cermelli et al. (2011);118 Bala et al. (2012);24 Zhang et al. (2012);27 Bihrer et al. (2011)32 |
| HBV | miR-192 and miR-122 | Increased | Ji, et al. (2011);119 Zhang et al. (2010)126 |
| Liver fibrosis/ cirrhosis |
miR-29 and miR-652 miR-513-3p and miR-571 |
Decreased Increased |
Roderburg et al. (2012);52 Roderburg et al. (2011)50 Roderburg et al. (2011)50 |
| HCC | miR-21, miR-16, miR-199a, miR-122, miR-223, and miR-885-5p | Increased | Bihrer et al. (2011);32 Tomimaru et al. (2011);123 Xu et al. (2011);121 Gui et al. (2010)122 |
| Drug overdose | miR-122 and miR-192 | Increased | Wang et al. (2009);76 Starkey Lewis et al. (2011);124 Bala et al. (2012)12 |
Abbreviations: ALD, alcoholic liver disease; HCC, hepatocellular carcinoma.
Apart from miR-122, other miRNA signatures have also been revealed in liver diseases. We have shown the induction of the inflammation-related miRNA, miR-155, in ALD and in TLR9+TLR4-induced liver inflammation in the liver and circulation.12 Chronic HCV infection causes liver inflammation and consistent with this fact, induction of miR-155 was found both in serum and peripheral monocytes of patients with chronic HCV infection.24,27 Another liver miRNA, miR-196 was increased in the serum and plasma of patients with HBV infection,119 NAFLD125 and drug overdose (acetaminophen).124 miRNA-34a, which regulates lipid and cholesterol metabolism, was found to be upregulated in patients with NAFLD and HCV.118 Liver fibrosis and/or cirrhosis was associated with increased levels of miR-513-3p and miR-571 and a reduction in levels of miR-652 and miR-29.50 A number of miRNAs, such as miR-21, miR-199a, miR-1885-5p and miR-16, were increased in the circulation of patients with HCC.32,122,123 In biliary atresia, circulating levels of the miR-200b/429 cluster are elevated in infants and have promising diagnostic clinical potential.127 Interestingly, exosomal miRNA expression pattern is unique to liver diseases and varies between grade and stages of liver disease, which emphasizes their potential as diagnostics biomarkers.128 Moreover, circulating miR-122 could be used as a general marker of liver injury and might have better sensitivity than alanine aminotransferase.12,76,118,124
MicroRNAs as therapeutic targets
The potential therapeutic application of targeting miRNAs is receiving increasing attention in liver diseases and has been reviewed elsewhere.129 Both inhibition and overexpression of miRNAs are therapeutically in reach with current technologies. Perhaps the most advanced therapeutic discovery at this time is related to inhibition of miRNA-122 in chronic HCV infection. On the basis of successful preclinical studies with an anti-miR-122 LNA inhibitor, clinical trials are ongoing. Results of the initial clinical trial with miravirsen, an antisense oligonucleotide targeting miR-122, report prolonged dose-dependent reduction in HCV RNA levels without evidence of viral resistance.130 The long half-life of LNAs and the minimal adverse effect profile promises future development of this approach in human disease.129,131 Delivery systems for small noncoding RNAs can also be approached with AAV delivery of mRNA inhibitors or precursors.
Advances in human AAV modifications have the potential for cell-specific delivery of a target miRNA precursor or antagonist. Delivery of small duplex RNAs can be achieved using cationic lipid-based and nanoparticle delivery systems.131–134 Lipid-based and gold nanoparticles are both under investigation for small RNA-based therapy. miRNA manipulation might be an optimal approach for conditions in which the expression level of a specific miRNA target needs adjustment rather than complete inhibition or upregulation. Given the ‘fine-tuning’ effects of miRNAs on target gene expression and function, incremental modulation of target genes of miRNAs is a realistic therapeutic approach. For example, in liver diseases that involve moderate inflammation in their pathogenesis, such as alcoholic or nonalcoholic liver diseases, targeting inflammatory mediators via miRNA might be sufficient to restore homeostasis instead of fully inhibiting vital signalling pathways. Further investigations will explore possibilities for miRNA targeting in therapeutic intervention for liver diseases.
Conclusions
The biological significance and utility of miRNAs in liver disease is a rapidly growing field. It is increasingly evident that in addition to the ‘traditional’ role of miRNAs in inhibition of target gene expression, miRNAs have more complex and diverse functions that can be specific for individual miRNAs, the cell type or the tissue environment. Although some features of miRNA biology—such as their effects on target genes—are well described, the cell-specific regulatory potential and disease-specific alterations of miRNAs in the liver await further investigation. Another emerging area is the biological significance of extracellular miRNAs. These miRNAs might be particularly important in liver diseases in which some miRNAs enter the systemic circulation in increased quantities during liver damage. Both the quantity and ‘quality’ defined by packaging of miRNAs could also be of importance for circulating miRNAs with respect to their biodistribution, bioavailability and/or target effects.
At this time, little is known about whether the association of miRNAs with Ago2, exosomes, lipid particles or other forms has any bearing on their half-life, biological distribution, cellular uptake and/or target cell or organ effects. It is generally established that circulating miRNAs are stable compared with many other biological markers and that this property has contributed to the rapid explosion of biomarker discovery in circulating miRNAs. Important advances are evolving in HCC-specific miRNA biomarkers; identification of circulating miRNA profiles that are specific for a particular liver disease rather than liver injury in general is more challenging. Finally, understanding of the role and alterations of miRNAs in cellular functions and disease processes in the liver provides a strong basis for targeting miRNAs in treatment development. We believe that it is realistic to expect that miRNA-based therapies will be part of the therapeutic armamentarium for hepatologists in the next decade.
Key points.
-
▪
MicroRNAs (miRNAs) are encoded by genes and exert their intracellular effects by targeting post-transcriptional events on target genes
-
▪
MiRNAs fine-tune all physiological and many pathological processes that are fundamental to normal liver functions and liver disease
-
▪
The distribution and function of some miRNAs is cell-specific, and hepatocytes have the highest abundance of microRNA-122 expression in the body
-
▪
In the liver, miRNAs have been shown to regulate processes such as inflammation, fibrosis, and lipid and glucose metabolism
-
▪
Dysregulation in miRNAs is associated with liver diseases, such as hepatocellular carcinoma, viral hepatitis, alcoholic and nonalcoholic steatohepatitis and drug-induced liver injury
-
▪
Extracellular miRNAs could serve as biomarkers of liver disease, but specificity might be a limitation
Acknowledgements
The work of the authors is supported by NIAAA grant RO1-AA020744 (to G. Szabo).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun G, et al. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. MicroRNAs in cancer treatment and prognosis. Am. J. Cancer. Res. 2012;2:414–433. [PMC free article] [PubMed] [Google Scholar]
- 6.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 8.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung O, et al. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bala S, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai WC, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu SH, et al. Essential metabolic, antiinflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010;2:527–534. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng F, et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 2012;181:804–817. doi: 10.1016/j.ajpath.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers KC, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2012;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 21.Bala S, Szabo G. MicroRNA signature in alcoholic liver disease. Int. J. Hepatol. doi: 10.1155/2012/498232. http://dx.doi.org/10.1155/2012/498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 23.Bala S, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bala S, et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidorkiewicz M, et al. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from Hepatitis C patients after antiviral treatment. Acta Virol. 2010;54:75–78. doi: 10.4149/av_2010_01_75. [DOI] [PubMed] [Google Scholar]
- 26.Grek M, et al. Coordinated increase of miRNA-155 and miRNA-196b expression correlates with the detection of the antigenomic strand of hepatitis C virus in peripheral blood mononuclear cells. Int. J. Mol. Med. 2011;28:875–880. doi: 10.3892/ijmm.2011.748. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Hepatitis C Virus-induced upregulation of miR-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 28.Zhao JL, et al. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl Acad. Sci. USA. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaked I, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Lagos D, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 31.Strum JC, et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bihrer V, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 33.An F, et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis. 2012;17:702–716. doi: 10.1007/s10495-012-0704-7. [DOI] [PubMed] [Google Scholar]
- 34.Yu DS, et al. The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 2012;29:663–668. doi: 10.3892/ijmm.2012.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma AD, et al. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology. 2011;53:1651–1661. doi: 10.1002/hep.24243. [DOI] [PubMed] [Google Scholar]
- 36.Song G, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51:1735–1743. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Invest. 2012;122:1097–1108. doi: 10.1172/JCI46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, et al. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS ONE. 2012;7:e33577. doi: 10.1371/journal.pone.0033577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan C, et al. Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS ONE. 2012;7:e39151. doi: 10.1371/journal.pone.0039151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan Q, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299–310. doi: 10.1002/hep.25984. [DOI] [PubMed] [Google Scholar]
- 41.Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 44.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelialmesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami Y, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CJ, et al. Human ESC self-renewal promoting microRNAs induce epithelial-mesenchymal transition in hepatocytes by controlling the PTEN and TGFβ tumor suppressor signaling pathways. Mol. Cancer. Res. 2012;10:979–991. doi: 10.1158/1541-7786.MCR-11-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front. Physiol. 2012;3:49. doi: 10.3389/fphys.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, et al. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268–2272. doi: 10.1016/j.cellsig.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 50.Roderburg C, et al. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS ONE. 2012;7:e32999. doi: 10.1371/journal.pone.0032999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang B, et al. miR-181b promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem. Biophys. Res. Commun. 2012;421:4–8. doi: 10.1016/j.bbrc.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 52.Roderburg C, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 53.Kwiecinski M, et al. Expression of plateletderived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Lab. Invest. 2012;92:978–987. doi: 10.1038/labinvest.2012.70. [DOI] [PubMed] [Google Scholar]
- 54.Mannaerts I, et al. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS ONE. 2013;8:e55786. doi: 10.1371/journal.pone.0055786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hand NJ, et al. MicroRNA profiling identifies miR-29 as a regulator of disease-associated pathways in experimental biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2012;54:186–192. doi: 10.1097/MPG.0b013e318244148b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marquez RT, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 57.Venugopal SK, et al. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;298:G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y, et al. MicroRNA-146a modulates TGF-β1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell. Signal. 2012;24:1923–1930. doi: 10.1016/j.cellsig.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Li J, et al. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J. Hepatol. 2013;58:522–528. doi: 10.1016/j.jhep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das S, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kren BT, et al. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bian Z, et al. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- 63.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 64.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 67.US National Library of Medicine. ClinicalTrials.gov [online] 2013 http://www.clinicaltrials.gov/ct2/results?term=hepatitis+and+miravirsen+&Search=Search. [Google Scholar]
- 68.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jopling C. Liver-specific microRNA-122: biogenesis and function. RNA Biol. 2012;9:137–142. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conrad KD, et al. microRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS ONE. 2013;8:e56272. doi: 10.1371/journal.pone.0056272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hou W, Bukong TN, Kodys K, Szabo G. Alcohol facilitates HCV RNA replication via upregulation of miR-122 expression and inhibition of cyclin G1 in human hepatoma cells. Alcohol. Clin. Exp. Res. 2012;37:599–608. doi: 10.1111/acer.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bukong TN, Hou W, Kodys K, Szabo G. Ethanol facilitates HCV replication via upregulation of GW182 and HSP90 in human hepatoma cells. Hepatology. 2012;57:70–80. doi: 10.1002/hep.26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ura S, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 74.Qiu L, et al. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 75.Sarma NJ, et al. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS ONE. 2012;7:e50826. doi: 10.1371/journal.pone.0050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl Acad. Sci. USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoshioka W, Higashiyama W, Tohyama C. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicol. Sci. 2011;122:457–465. doi: 10.1093/toxsci/kfr130. [DOI] [PubMed] [Google Scholar]
- 78.Yokoi T, Nakajima M. microRNAs as mediators of drug toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 79.Mohri T, et al. Human CYP2E1 is regulated by miR-378. Biochem. Pharmacol. 2010;79:1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 80.Dolganiuc A, et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol. Clin. Exp. Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang Y, et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol. Clin. Exp. Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 83.Yin H, et al. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J. Biol. Chem. 2012;287:9817–9826. doi: 10.1074/jbc.M111.333534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1α and microrNA-199. J. Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15(Suppl. 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 87.Hou J, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 88.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ding J, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat. Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 90.Fornari F, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 91.Pineau P, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl Acad. Sci. USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park JK, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiong Y, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 94.Fang JH, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 95.Braconi C, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 97.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ji J, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl J. Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of microRNAs in human liver cancers. Semin. Oncol. 2011;38:752–763. doi: 10.1053/j.seminoncol.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang J, et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Padgett KA, et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J. Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Banales JM, et al. Up-regulation of microRNA 506 leads to decreased Cl-/HCO3- anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology. 2012;56:687–697. doi: 10.1002/hep.25691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Poupon R, et al. Genetic factors of susceptibility and of severity in primary biliary cirrhosis. J. Hepatol. 2008;49:1038–1045. doi: 10.1016/j.jhep.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 105.Salas JT, et al. Ae2a, b-deficient mice develop antimitochondrial antibodies and other features resembling primary biliary cirrhosis. Gastroenterology. 2008;134:1482–1493. doi: 10.1053/j.gastro.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 106.Qin B, Huang F, Liang Y, Yang Z, Zhong R. Analysis of altered microRNA expression profiles in peripheral blood mononuclear cells from patients with primary biliary cirrhosis. J. Gastroenterol. Hepatol. 2013;28:543–550. doi: 10.1111/jgh.12040. [DOI] [PubMed] [Google Scholar]
- 107.Castoldi M, et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Invest. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shigehara K, et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE. 2011;6:e23584. doi: 10.1371/journal.pone.0023584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 113.Zernecke A, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 114.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 116.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ji F, et al. Circulating microRNAs in hepatitis B virus-infected patients. J. Viral Hepat. 2011;18:e242–e251. doi: 10.1111/j.1365-2893.2011.01443.x. [DOI] [PubMed] [Google Scholar]
- 120.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 2011;45:355–360. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 121.Xu J, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 122.Gui J, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. (Lond.) 2010;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tomimaru Y, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J. Hepatol. 2011;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 124.Starkey Lewis PJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 125.Tryndyak VP, et al. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 2012;262:52–59. doi: 10.1016/j.taap.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 127.Zahm AM, Hand NJ, Boateng LA, Friedman JR. Circulating microRNA is a biomarker of biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2012;55:366–369. doi: 10.1097/MPG.0b013e318264e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Murakami Y, et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Szabo G, Sarnow P, Bala S. MicroRNA silencing and the development of novel therapies for liver disease. J. Hepatol. 2012;57:462–466. doi: 10.1016/j.jhep.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. doi: 10.1056/NEJMoa1209026. http://dx.doi.org/10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 131.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu XQ, Song WJ, Sun TM, Zhang PZ, Wang J. Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol. Pharm. 2011;8:250–259. doi: 10.1021/mp100315q. [DOI] [PubMed] [Google Scholar]
- 133.Su J, Baigude H, McCarroll J, Rana TM. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 2011;39:e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barrey E, et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bandiera S, et al. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 139.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hu G, et al. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 2012;3:e381. doi: 10.1038/cddis.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xin H, et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–1564. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]