Abstract
Retinoic acid-inducible protein I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) are cytosolic viral RNA sensors that induce type I interferon production (IFN). In this study, we found that MDA5 undergoes inducible SUMOylation by small ubiquitin-like modifier-1 (SUMO-1) in response to polyI:C stimulation. Enhanced SUMOylation of MDA5 by exogenously expressed SUMO-conjugating enzyme Ubc9 correlated with upregulation of IFN expression and repressed virus replication. Conversely, overexpression of a SUMOylation-deficient mutant of Ubc9 or knockdown of endogenous Ubc9 reduced IFN production. Furthermore, we showed that PIAS2β, a SUMOylation E3 ligase, could specifically interact with and enhance the SUMOylation of MDA5. Consequently, PIAS2β knockdown reduced the SUMOylation of MDA5 and the IFN production. Collectively, these findings suggest that SUMO-1 modification of MDA5 possibly via PIAS2β may play a role in the MDA5-mediated IFN response to viral infections.
Keywords: SUMOylation, IFNs, MDA5, Ubiquitylation
1. Introduction
Type I interferon (IFN) plays a key role in mediating antiviral innate immunity. Mammalian cells have developed two distinct pathways to recognize viral nucleic acids and trigger the production of IFN. One pathway is mediated by Toll-like receptors (TLRs), which mainly recognize extracellular viral nucleic acids. The other pathway utilizes the retinoic acid-inducible gene I (RIG-I)-like helicase (RLH), which contains RIG-I and melanoma differentiation-associated gene 5 (MDA5, also referred as helicard or IFNH1) to recognize intracellular nucleic acids (Andrejeva et al., 2004; Perry et al., 2005; Uematsu and Akira, 2006; Yoneyama et al., 2004). Both RIG-I and MDA5 are RNA helicases, consisting of two N-terminal caspase-recruiting domains (2CARD), a central DExD/H box RNA helicase domain, and a C-terminal regulatory domain (Kang et al., 2004, 2002; Kato et al., 2005). The two CARD domains act as a signaling domain, which interacts with the adaptor protein Cardif/IPS-1/MAVS/VISA to initiate signal cascades (Kawai and Akira, 2006; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Despite the similarity in domain structure and signal pathway, RIG-I and MDA5 are not redundant in function. They are involved in specific RNA binding in addition to recognising different subsets of viruses (Kato et al., 2006). It seems that RIG-I is responsible for short double stranded RNA and 5′ppp single stranded RNA, while MDA5 is activated by long double stranded RNA (Gitlin et al., 2006; Hornung et al., 2006; Yoneyama and Fujita, 2008).
The majority of proteins involved in the IFN induction process, such as RIG-I, MDA5, TRAF3, TBK1 and IRF3, have been reported to be ubiquitinated (Friedman et al., 2008; Kayagaki et al., 2007; Wang et al., 2009; Zhang et al., 2008; Zheng et al., 2008). Particularly, RIG-I is ubiquitinated by four different E3 ligases TRIM25, RNF125, RNF135 and REUL to activate IFN signaling (Arimoto et al., 2007; Gack et al., 2007; Gao et al., 2009; Kim et al., 2008; Oshiumi et al., 2009). As a homolog of RIG-I, however, MDA5 is only reported to be ubiquinated by RNF125 in the negative regulation of IFN signaling (Arimoto et al., 2007).
Similar to ubiquitylation, SUMOylation is a multi-step reaction that covalently conjugates a 12 kDa small ubiquitin-like modifier (SUMO) to target proteins by a single E1-activating enzyme (Aos1/Uba2, also called SAE1/2), a unique E2 conjugating enzyme (Ubc9), and an array of different E3 ligases (e.g., PIAS family and RanBP2), so as to regulate their activity, stability and subcellular localization (Hay, 2005; Hershko and Ciechanover, 1998). SUMOylation is believed to regulate the signaling pathway of IFNs. It has been reported that virus-mediated IRF3 and IRF7 SUMOylation attenuates activation of IFNs (Kubota et al., 2008). In this work, we have demonstrated that SUMOylation of MDA5 increased markedly after cells were transfected with polyI:C, a mimic of double stranded RNA virus recognized by MDA5. This was correlated with elevated IFN-β induction and inhibition of virus replication. We have further shown that PIAS2β directly associates with MDA5 and functions as a putative E3 ligase to SUMOylate MDA5. Over-expression and gene-silencing experiments showed that PIAS2β modulates MDA5-driven IFN-β expression. Based on these results we hypothesize that SUMOylation of MDA5 by PIAS2β participates in the regulation of type I IFN signaling.
2. Materials and methods
2.1. Plasmids
The cDNAs encoding full length SUMO-1 and Ubc9 were sub-cloned between the EcoRI/XhoI or KpnI/XhoI restriction sites of pCMV-HA (Clontech, Mountain View, CA) and pcDNA3.1(+)Myc/His (Invitrogen), respectively, by add-on PCR amplification of a human fetal cDNA library. The primers used were SUMO-1 forward: 5′-GAA TTC GGA TGT CTG ACC AGG AGG CAA AAC C-3′; reverse: 5′-CTC GAG CTA AAC TGT TGA ATG ACC CCC CG-3′; Ubc9 forward: 5′-GCG GTA CCA TGT CGG GGA TCG CCC TCA G-3′; reverse: 5′-GCC TCA GTG AGG GCG CAA ACT TCT TGG-3′. Plasmids encoding Flag-MDA5, Flag-MDA5-N (1–294) and Flag-MDA5-C (295–1025) were kindly provided by Dr. T Fujita (Kyoto University, Japan). pIFN-β-luc was a gift from Dr. G. Cheng (UCLA, USA). Ubc9 (C93S) was kindly provided by Dr. G. Sui (Wake Forest University School of Medicine, USA). pDEF-Flag-SUMO-1, pDEF-GST-SUMO2, pDEF-Flag-SUMO3, pDEF-Myc-PIAS1, pDEF-Myc-PIAS2α, pDEF-Myc-PIAS2β, and pDEF-Myc-PIAS4 were gifts from Dr. X. Peng (Chinese Academy of Medical Science, China). The C362S point mutation in the zinc-finger or SP-RING motif of PIAS2β was constructed using a QuickChange site-directed mutagenesis kit (Stratagene, CA), yielding the SUMOylation-defective PIAS2β-C362S mutant (Schmidt and Muller, 2002). HA-Ub-K48 was kindly provided by Dr K. Lim (National Neuroscience Institute, Singapore).
2.2. RNAi
The lentiviral vector LTV1 expressing shRNA that targets Ubc9 was kindly provided by Dr. G. Sui (Sui and Shi, 2005). Production of the virus and infection of cells were performed as previously described (Rubinson et al., 2003). The shRNA expression cassette was used as the scrambled control. RNAi sequences that target human PIAS2β: 5′-AAG ATA CTA AGC CCA CAT TTG-3′ (Yang and Sharrocks, 2005) was synthesized (RiboBio, China) and transfection was performed as previously described (Yang et al., 2003).
2.3. Reagents and cell lines
Monoclonal antibody to Flag (M2) was purchased from Sigma (St Louis, MO). Antibodies to SUMO-1 and Myc were from Santa Cruz Biotech (San Diego, CA) and polyclonal antibody for Ubc9 was purchased from Abgent (San Diego, CA). Monoclonal antibodies to MDA5 were purchased from Santa Cruz Biotech (San Diego, CA) for immunoprecipitation, and Alexis (San Diego, CA) for Western blotting. HEK293T, HeLa, and A549 cells were routinely maintained in DMEM (Hyclone, UT, USA) supplemented with 10% FCS (PAA, Pasching, Austria) and 1% penicillin and streptomycin (Hyclone, UT).
2.4. Co-immunoprecipitation
Constructs were transiently transfected as indicated either into 293T cells by a standard calcium phosphate method or into HeLa and A549 cells by jetPEI according to the manufacturer’s instructions (Polyplus, NY). Co-immunoprecipitation and Western blotting were performed as described previously (Desterro et al., 1998).
2.5. PolyI:C pull down assay
HEK293T cell lysates transfected with the plasmids indicated were mixed with 20 μL (50% slurry) polyI:C-agarose beads (Sigma, St Louis, MO) at 4 °C for 2 h, and washed extensively with lysis buffer. Pulldown proteins were analyzed by Western blotting with antibodies as indicated.
2.6. Luciferase assays
IFN-β luciferase reporter assays were performed as previously described (Guo and Cheng, 2007; Zheng et al., 2008). HEK293T cells were seeded on 24-well plates and were transfected the next day by standard calcium phosphate precipitation. A Renilla luciferase plasmid was co-transfeced as an internal control for transfection efficiency. Luciferase activities were measured and normalized according to the manufacturer’s instructions (Promega, Madison, WI). All assays were repeated at least three times. Data represent the average of three independent experiments (mean ± SD).
2.7. RT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen, CA, USA) and RT-PCR was performed as previously described (Weng et al., 2005; Zheng et al., 2008). The primer pairs were: Ubc9 forward: 5′-CGG AAT TCG ATG TCG GGG ATC GCC CTC-3′; reverse: 5′-CGG GGT ACC TTA TGA GGG CGC AAA CTT C-3′; PIAS2β forward: 5′-GAG GAA GAC CCT CCT GCC AAA AGG-3′; reverse: 5′-TTA GTC CAA TGA GAT GAT GTC AGG-3′ (Yang and Sharrocks, 2005); β-actin forward: 5′-CAT GGA GTC CTG TGG CAT CCA CGA AAC T-3′; reverse: 5′-ATC TCC TTC TGC ATC CTG TCG GCA AT-3′; GAPDH forward: 5′-AAG CGC ACG GGC ATG GCC T T-3′; reverse: 5′-AGG AGA CCA CCT GGT GCT CAG-3′.
2.8. Plaque assays
VSV (strain New Jersey) was expanded in L929 cells and titrated with plaque assays in HeLa cells as described elsewhere (Huang et al., 2005; Zheng et al., 2008). HEK293T cells were transfected with the plasmids indicated for 16 h. Cells were then infected with VSV (MOI = 0.1) for 1 h and excess virus was washed off with PBS. Supernatants were collected at the times indicated and used to determine VSV replication in HeLa cells. Experiments were repeated three times and each experiment was performed in duplicate. Data represent the average of three independent experiments (mean ± SD).
3. Results
3.1. MDA5 is SUMOylated at the C-terminal region
An initial bioinformatic analysis (SUMOplot Analysis Program, Abgent) revealed that MDA5 possesses several potential SUMOylation acceptor sites (ψKXE, ψ representing hydrophobic amino acids) which are scattered throughout the coding sequence (Rodriguez et al., 2001; Sampson et al., 2001; Song et al., 2004, 2005). To examine the SUMOylation of MDA5, we overexpressed Flag-MDA5 together with Myc-Ubc9 and HA-SUMO-1 in 293T cells. Immunoprecipitation with anti-Flag antibody showed that higher molecular weight band-shifts which were absent in cells transfected with SUMO-1 and Ubc9 alone were associated with exogenous Flag-MDA5 as detected by SUMO-1 antibody (Fig. 1A). In fact, these higher molecular weight bands were also present in whole cell lysates when probed directly with anti-Flag antibody (Fig. 1A). When we co-transfected the enzymatically inactive mutant of SUMO E2-conjugating enzyme Ubc9, Ubc9 (C93S) (Guo et al., 2008; Hahn et al., 1997; Poukka et al., 1999), the specific band shifts disappeared (Fig. 1B). To determine which domain(s) of MDA5, 2CARD, helicase or RD were SUMOylated, we co-expressed Myc-Ubc9 and HA-SUMO-1 in 293T cells with the Flag-tagged full length MDA5, or its N-terminal fragment containing 2CARD (MDA5-N) or C-terminal without 2CARD (MDA5-C) (Saito et al., 2007). Immunoprecipitation results showed that both the full length MDA5 and MDA5-C, but not MDA5-N, were conjugated with SUMO-1 (Fig. 1C). These results suggest the potential involvement of MDA5-C in SUMO-1 modification. To determine whether endogenous MDA5 can be SUMOylated, we expressed Myc-Ubc9 and HA-SUMO-1 in A549 cells in which MDA5 is expressed abundantly. Whole cell extracts were immunoprecipitated with anti-MDA5 antibody, and precipitates were probed with MDA5 or SUMO-1 antibodies, showing that endogenous MDA5 could also be SUMOylated (Fig. 1D).
Fig. 1.
MDA5 is SUMOylated at its C-terminus. (A) MDA5 is SUMO-1 modified. HEK293T cells were transfected with plasmids for Flag-MDA5, HA-SUMO-1 and Myc-Ubc9 by standard calcium phosphate precipitation. Whole cell lysates (WCL) were prepared 48 h post transfection, and immunoprecipitated with anti-Flag M2 antibody. SUMO-conjugated proteins were immunoblotted with anti-SUMO-1 antibody. The whole cell lysates was immunoblotted using anti-Flag as a loading control. (B) Ubc9 is dependent on MDA5 SUMOylation. HEK293T cells were transfected with plasmids for Flag-MDA5, HA-SUMO-1 and Myc-Ubc9 or Ubc9 (C93S) as indicated. Immunoprecipitation was performed exactly as in (A) except that the input was immunoblotted with anti-Flag and anti-Ubc9. (C) The C-terminus of MDA5 is the SUMO-1 acceptor site. HEK293T cells were co-transfected with Flag-tagged full length MDA5, MDA5-N (aa 1–295) or MDA5-C (aa 295–1025) with SUMO-1-HA and Myc-Ubc9 as indicated. SUMOylation of MDA5 was measured exactly as in (A). (D) Endogenous MDA5 is SUMOylated. A549 cells (1 × 107) were transfected with HA-SUMO-1 and Myc-Ubc9 plasmids, or with control plasmids using Lipofectamine 2000. Cell lysates were prepared 48 h after transfection and were immunoprecipitated with anti-MDA5 antibody (Santa Cruz, sc-48031), and detected with anti-MDA5 (ENZO Life Science, ALX-804-863) or anti-SUMO-1 antibody.
3.2. SUMOylated MDA5 still binds to polyI:C
Since the C-terminus of MDA-5 is responsible for RNA recognition and binding (Gitlin et al., 2006; Kang et al., 2002; Kato et al., 2006; Yoneyama et al., 2004), we speculated that the SUMOylation of this region (Fig. 1C) would affect its ability to bind to RNA. polyI:C, a synthetic dsRNA mimic, is a substrate of MDA5 (Gao et al., 2009). When Flag-MDA5 was co-expressed with or without Myc-Ubc9 and HA-SUMO-1 in 293T cells, agarose beads conjugated with polyI:C were able to pull down both unmodified MDA5 (lower bands) and SUMOylated MDA5 (upper bands) from the cell lysate, in proportion to their expression levels (Fig. 2A). This result suggests that SUMOylation of MDA5 might not affect its RNA binding capability. To test the effect of polyI:C on MDA5 SUMOylation, 293T cells co-transfected with Flag-MDA5, Myc-Ubc9 and HA-SUMO-1 were treated with polyI:C for various lengths of time. Compared with mock transfection, the level of SUMOylation of MDA5 kept increasing after polyI:C treatment (Fig. 2B). Taken together, these results indicate that both exogenous and endogenous MDA5 can be SUMOylated, probably in a polyI:C inducible manner.
Fig. 2.
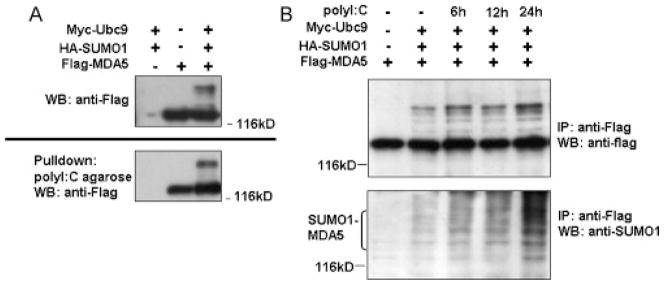
polyI:C enhances the modification of MDA5. (A) SUMOylation of MDA5 does not affect its RNA binding activity. HEK293T cells were transfected with the plasmids for Flag-MDA5, HA-SUMO-1, and Myc-Ubc9 as indicated. Cell lysates were incubated with polyI:C beads and pull-downed proteins were resolved by SDS-PAGE and immunoblotted with anti-Flag antibody. (B) poly I:C induces SUMOylation of MDA5. HEK293T cells were co-transfected with Flag-MDA5, Myc-Ubc9 and HA-SUMO-1. At each indicated time point after transfection, cells were further transfected with polyI:C for an additional 6, 12 or 24 h. SUMOylation status was analyzed as described in Fig. 1A.
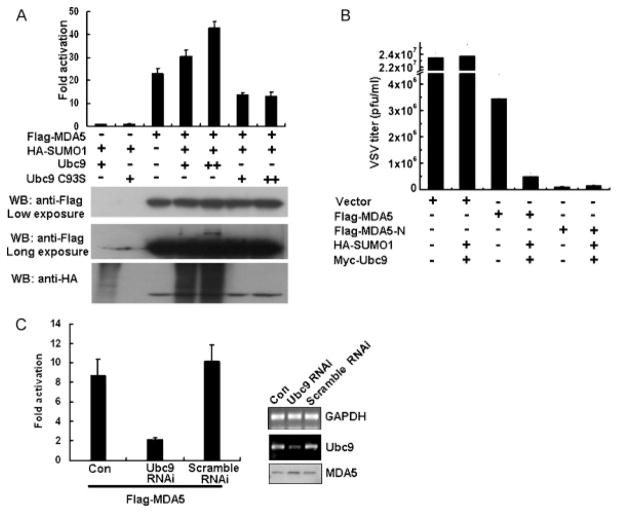
3.3. SUMOylation regulates the MDA5-mediated antiviral response
To determine whether SUMOylation of MDA5 might be involved in type I IFN signaling, Flag-MDA5, Myc-Ubc9 and HA-SUMO-1 were co-transfected into HEK293T cells together with an IFN-β luciferase reporter. In agreement with previous reports that MDA5 overexpression leads to the activation of the IFN-β reporter (Yoneyama et al., 2004), MDA5-driven IFN-β promoter activity increased in a Ubc9-dose dependent manner, probably due to the significantly increased SUMOylation level of MDA5 (Fig. 3A). On the other hand, this enhancement decreased gradually when cells were co-transfected with increasing amounts of the Ubc9-C93S mutant, in accordance with the reduced SUMOylation level of MDA5 (Fig. 3A). Since increased IFN production should inhibit virus replication, we tested whether Ubc9 can influence the MDA5 mediated anti-viral effect by performing plaque formation assays using VSV, an IFN-sensitive viral strain (Huang et al., 2005). As expected, overexpression of MDA5 weakly inhibited VSV propagation, but co-expression of Ubc9 with MDA5 significantly suppressed VSV replication (Fig. 3B). As a control, overexpression of SUMO-1 and Ubc9 per se had no effect on VSV replication. This result suggests that SUMOylation is specifically involved in the MDA5-mediated antiviral response. The 2CARD domain is a dominant-active mutant that can constitutively activate IFN signaling and repress viral replication when transiently expressed in cells (Fig. 3B). However, co-expression of Ubc9 failed to enhance the antiviral activity of MDA5-N (Fig. 3B). This is not surprising because MDA5-N was not subjected to SUMOylation. Therefore, SUMOylation of MDA5 might be necessary for its antiviral function to work at full capacity.
Fig. 3.
SUMOylation activates the MDA5-mediated IFN response. (A) Ubc9 increases MDA5-driven IFN-β reporter activity. HEK293T cells were transfected with a Renilla luciferase internal control plasmid, IFN-β reporter, Flag-MDA5, HA-SUMO-1, and increasing amounts of Myc-Ubc9 or Ubc9 (C93S) as indicated for 24 h. Luciferase activities were determined and normalized against renilla luciferase activity. Fold activation expressed for the ratio of relative luciferase activity (RLU) in the presence or absence of Myc-Ubc9 and HA-SUMO-1. Data represent the average of three independent experiments (mean ± SD). (B) Ubc9 expression inhibits VSV replication. HEK293T cells were transfected with the plasmids indicated for 16 h in 24-well plates. Cells were then infected with VSV (MOI = 0.1) for an additional 1 h. Supernatants were collected 9 h post infection and standard plaque assays in HeLa cells were used to determine virus titers. The data represent at least 3 independent experiments. (C) Downregulation of endogenous Ubc9 expression inhibits IFN production. HeLa cells (1 × 106) were infected with sham lentivirus, lentivirus containing Ubc9-shRNA or scrambled Ubc9 shRNA (MOI = 2) for 12 h. Cells were then co-transfected with a Renilla luciferase internal control plasmid, IFN-β reporter and Flag-MDA5 or a control vector. Dual luciferase activities were determined and normalized as in (A), and fold activation was derived by RLU in the presence or absence of Flag-MDA5 and HA-SUMO-1. Data represent the average of three independent experiments (mean ± SD). Efficiency of gene silencing was detected by RT-PCR, with GAPDH gene expression as an internal control (right panel).
To further confirm that Ubc9 regulates IFN-β reporter activity via modification of MDA5, HeLa cells were infected with a lentivirus expressing shRNA against Ubc9 gene expression (Fig. 3C, insert). Luciferase reporter assays showed that Flag-MDA5 driven IFN-β reporter activities decreased by ~5-fold when Ubc9 expression was reduced (Fig. 3C). Correlation of Ubc9 knockdown with reduced IFN production suggested that endogenous Ubc9 is specifically involved in the MDA5-mediated IFN response. Taken together, overexpression and gene-silencing experiments showed that Ubc9 enhanced IFN-β expression, and SUMOylation of MDA5 could be a target in the cellular antiviral response.
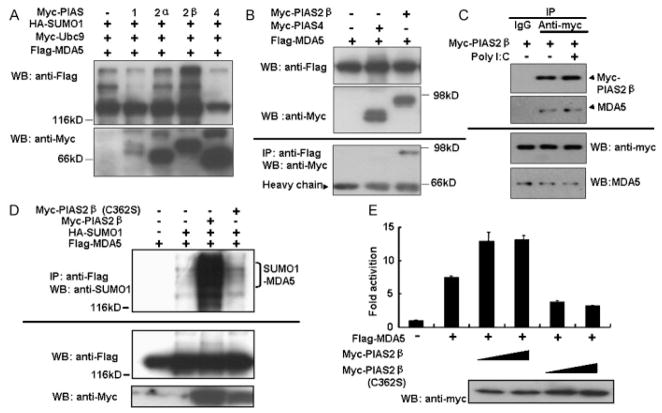
3.4. PIAS2β interacts with MDA5 and stimulates the SUMOylation of MDA5
While E1 and E2 were shown to be sufficient for SUMOylation of various substrates, the majority of SUMO targeting reactions requires E3 activity (Hay, 2005; Hay, 2007). Of the few SUMO E3 ligases discovered thus far (Hay, 2007), PIAS proteins (Rytinki et al., 2009; Schmidt and Muller, 2002) play an important role in innate immunity (Liu et al., 1998; Shuai and Liu, 2005; Tahk et al., 2007; Ungureanu et al., 2005; Zhang et al., 2004). This prompted us to determine whether PIAS proteins are involved in the SUMO conjugation of MDA5. We assessed SUMO conjugation of MDA5 in the presence of PIAS 1, PIAS 2α, PIAS 2β or PIAS 4. Immunoprecipitation showed that exogenously expressed PIAS2β, but not the other PIAS proteins, significantly increased SUMOylation of MDA5 (Fig. 4A). Since E3 ligase usually forms a complex with SUMO targeting proteins for efficient SUMO conjugation (Hay, 2005, 2007), we examined the interaction between MDA5 and PIAS2β. When Flag-MDA5 and Myc-PIAS2β were co-expressed in 293T cells, MDA5 immunoprecipitated with PIAS2β, demonstrating that these two proteins interact in cells (Fig. 4B). This inter-molecular association seems to be specific since MDA5 did not interact with PIAS4 in the same experiments (Fig. 4B) or with other PIAS proteins (data not shown). Moreover, when Myc-PIAS2β was over-expressed in A549 cells, endogenous MDA5 could also be immunoprecipitated with PIAS2 (Fig. 4C). However, this interaction was not influenced by the polyI:C stimulation (Fig. 4C). Taken together, PIAS2β is likely to be a SUMOylation E3 ligase for MDA5.
Fig. 4.
PIAS2β is associated with and enhances the SUMOylation of MDA5. (A) MDA5 is SUMO-modified by overexpressed PIAS2β. HEK293T cells were transfected with plasmids expressing Flag-MDA5, SUMO-1-HA, Ubc9-myc and Myc-tagged PIAS1, PIAS 2α, PIAS 2β, or PIAS 4 as indicated. Cell lysates were prepared 36 h post transfection and SUMO modification of MDA5 was detected by immunoblotting with anti-Flag for the characteristic band shifts. Protein expression levels of different PIAS were detected by immunoblotting with by anti-Myc. (B) MDA5 associates with PIAS2β. Cell lysates of 293T cells co-expressing Flag-MDA5 and Myc-PIAS2β or myc-PIAS4 were immunoprecipitated with anti-Flag antibody and immunoblotted by anti-Myc. (C) Endogenous MDA5 associates with PIAS2β. Cell lysates of A549 (1 × 107) cells expressing Myc-PIAS2β were immunoprecipitated with IgG or anti-Myc antibody and immunoblotted by anti-MDA5 and anti-Myc antibody. polyI:C transfection was done at 24 h post Myc-PIAS2β transfection. (D) Mutant PIAS2β does not SUMO-modify MDA5. Whole cell lysates of HEK293T cells co-expressing Flag-MDA5, HA-SUMO-1 and Myc-tagged PIAS2β or its mutant PIAS2β (C362S) were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-SUMO-1 antibody. Protein expression levels were detected with anti-Flag or anti-Myc, respectively. (E) MDA5 driven IFN-β reporter activities were enhanced by PIAS2β. HEK293T cells were co-transfected with IFNβ reporter, along with plasmids for Flag-MDA5, Myc-PIAS2β or PIAS2β (C362S) as indicated. One day post transfection, dual luciferase activities were measured and the fold activation for each sample was determined by dividing the basal RLU activity. Data represent the average of three independent experiments (mean ± SD). Target protein expression levels are shown in the bottom panels.
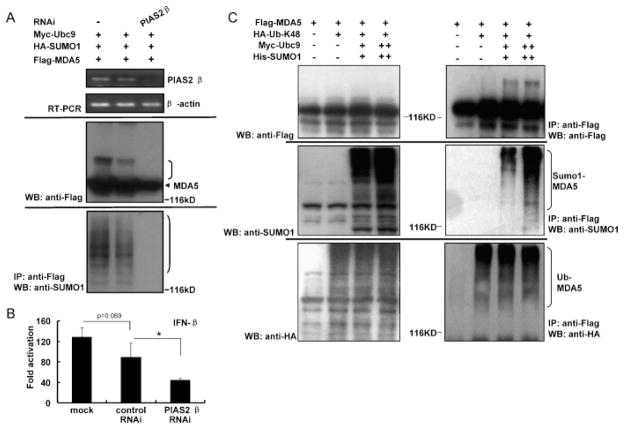
We went further to determine the relationship between SUMOylation by PIAS2β and the MDA5-mediated IFN response. Immunoprecipitation experiments showed that overexpression of wild type Myc-PIAS2β increased the SUMOylation of Flag-MDA5, whereas its defective mutant derivative, PIAS2β-C362S (Schmidt and Muller, 2002), failed to do so (Fig. 4D). Moreover, overexpression of PIAS2β increased IFN-β reporter activity driven by MDA5 in 293T cells, whereas PIAS2β-C362S suppressed it (Fig. 4E). To substantiate this finding, we knocked-down endogenous PIAS2β in 293T cells using siRNA one day prior to the co-transfection of Flag-MDA5, Myc-Ubc9 and HA-SUMO-1 expression plasmids. Down-regulation of PIAS2β gene expression by siRNA (Fig. 5A, top panel) caused a reduction in the SUMOylation level of MDA5 (Fig. 5A, middle panel). This interference in endogenous PIAS2β expression correlated with a decrease in FLAG-MDA5 driven IFN-β reporter activities (Fig. 5B). Collectively, these results strongly suggest that PIAS2β serves as an E3 ligase in the SUMOylation of MDA5, which positively regulates MDA5-mediated IFN induction.
Fig. 5.
PIAS2β knockdown inhibits the SUMOylation of MDA5 and the IFN response. (A) Downregulation of PIAS2β inhibits SUMOylation of MDA5. HEK293T cells were transiently transfected with siRNA sequence targeting the human PIAS2β gene or its scrambled control. Twelve hours later, cells were transfected with plasmids encoding Flag-MDA5, Myc-Ubc9 and HA-SUMO-1 and incubated for 36 h. RT-PCR analysis (left panel) shows the knockdown efficiency of endogenous PIAS2β. β-Actin was detected as an internal control. In parallel, SUMOylation of MDA5 was assessed as in Fig. 1A (middle and right panels). (B) Downregulation of PIAS2β inhibits MDA5-driven IFN-β reporter activities. Twenty-four hours after PIAS2β knockdown as in (A), cells were transfected with plasmids for renilla plasmid, IFN-β reporter and Flag-MDA5 plasmids for an additional 24 h. Calculation of fold activation and statistical analysis were as in Fig. 4D. (C) SUMOylation does not affect the Lys48-linked poly-ubiquitination of MDA5. HEK293T cells were transiently transfected with plasmids encoding Flag-MDA5, Myc-Ubc9, His-SUMO-1 and HA-Ub-K48 for 48 h. Whole cell lysates were immunoprecipitated with anti-Flag antibody followed by immunoblotted with anti-Flag, anti-SUMO1 or anti-HA antibody. Protein expression levels were detected with anti-Flag, anti-SUMO1 or anti-HA antibody, respectively.
3.5. SUMOylation of MDA5 does not affect its ubiquitylation
Lys 48-linked ubiquitylation leads to 26S proteasomal degradation against target protein, whereas Lys 63-linked ubiquitylation is involved in protein–protein interactions, recruitment, and assembly of signaling complexes. According to the reports, RIG-I has both the Lys 63-linked and Lys 48-linked ubiquitylation. However, MDA5 is only ubiquitinated on Lys 48, which contribute to the degradation of MDA5 and suppression of the MDA5 signal pathway. To further investigate the mechanism of SUMOylation of MDA5, we designed the experiment to observe the interplay between SUMOylation and ubiquitylation. As the result shown, the SUMOylation of MDA5 had no influence on its Lys 48-linked ubiquitylation (Fig. 5C).
4. Discussion
RIG-I and MDA5 play a pivotal role in sensing intruding viral RNA and initiating the type I IFN response (Akira et al., 2006). Like many other cytokine signaling pathways, RIG-I and MDA5 are tightly regulated by post-translational modifications to guarantee a timely and appropriate cellular response (Akira et al., 2006; Komuro et al., 2008). It has been well documented that RIG-I undergoes ubiquitylation and ISGylation in response to infection (Arimoto et al., 2007; Gack et al., 2007; Gao et al., 2009; Kim et al., 2008; Oshiumi et al., 2009). Thus far, however, the only report on ubiquitylation of MDA5 is by RNF125, which results in inhibition of IFN signaling (Arimoto et al., 2007). We report that MDA5 can also be SUMOylated, most likely by the E3 ligase, PIAS2β, which upregulates MDA5-mediated type I IFN signaling. This opposing regulation of MDA-5 functionality by SUMOylation and ubiquitylation is reminiscent of a similar dual modification of proliferating cell nuclear antigen (PCNA) that regulates yeast DNA synthesis (Haracska et al., 2004). It remains unclear how such a bipartite modulation of MDA5 coordinates initiation and termination of the antiviral response, and/or keeps the anti-viral response in homeostasis to avoid tissue damage (Carter and Vousden, 2008; Hay, 2004; Hunter and Sun, 2008). At least, in our experiments, the SUMOylation of MDA5 had no influence on its Lys 48-linked ubiquitylation.
MDA5 contains two CARD domains, a helicase domain and a repression domain. The C-terminal domain of MDA5, without the 2CARD domain, is responsible for RNA binding (Saito et al., 2007). Our data suggest, however, that SUMO-1 modification within this region does not affect its binding to polyI:C. Conversely, polyI:C enhances the SUMOylation of MDA5. We therefore speculate that MDA5 may undergo a conformational change upon polyI:C binding, turning MDA5 into a more favorable SUMO acceptor and more potent activator of IFN signaling. This hypothesis, including whether SUMO-1 expression per se is IFN inducible, requires further investigation.
SUMOylation is a versatile post-translational modification involved in various cellular functions including innate immunity. IRF3 and IRF7 can be SUMOylated and this modification down-regulates IFN production (Kubota et al., 2008). In contrast to this finding, we have shown that PIAS2β contributes to the SUMO-1 conjugation of MDA5, and positively regulates MDA5-driven IFN-I induction, thus inhibiting virus replication. These seemly conflicting results can be reconciled because MDA5 functions upstream of IRF3/7, and SUMOylation of MDA5 may override any downstream modifications. All the studies rely on overexpression assay systems. Further dissection of the modification and role of endogenous counterparts should be able to address this issue.
Our findings also point to a new role of PIAS family proteins, since previous studies have shown that the PIAS family of E3 ligases is mainly suppressive in cytokine signaling. For example, PIAS1 and PIAS2α can modify STAT1 to inhibit the JAK1/STAT1 innate immune pathway (Liu et al., 2004; Ungureanu et al., 2005). A recent study suggests that PIAS4 can suppress TRIF-induced IFN activation (Zhang et al., 2004). Therefore, our data echo the notion that different subsets of PIAS members participate in the specific regulation of cellular function (Bergink and Jentsch, 2009), and in this specific case, that PIAS2β can participate in the SUMOylation of MDA5 that positively modulates its role in IFN-I signaling.
Acknowledgments
We thank Drs. T. Fujita (Kyoto University, Japan), G. Sui (Wake Forest University School of Medicine, USA) and X. Peng (Chinese Academy of Medical Science, China) for the gift of plasmids and Dr. G. Gao (Institute of Biophysics, CAS) for providing VSV. This work was supported in part by grants from the Chinese Academy of Sciences (KSCX1-YW-10), and the Ministry of Science and Technology of China (2006CB910901, 2007DFC30190, 2008ZX10001-002, 2009CB522506) to H.T.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Carter S, Vousden KH. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle. 2008;7:2519–2528. doi: 10.4161/cc.7.16.6422. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, Xavier R, Ting AT. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gao D, Yang YK, Wang RP, Zhou X, Diao FC, Li MD, Zhai ZH, Jiang ZF, Chen DY. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One. 2009;4:e5760. doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Cheng G. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J Biol Chem. 2007;282:11817–11826. doi: 10.1074/jbc.M700017200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Yang MC, Weissler JC, Yang YS. Modulation of PLAGL2 transactivation activity by Ubc9 co-activation not SUMOylation. Biochem Biophys Res Commun. 2008;374:570–575. doi: 10.1016/j.bbrc.2008.07.064. [DOI] [PubMed] [Google Scholar]
- Hahn SL, Wasylyk B, Criqui-Filipe P. Modulation of ETS-1 transcriptional activity by huUBC9, a ubiquitin-conjugating enzyme. Oncogene. 1997;15:1489–1495. doi: 10.1038/sj.onc.1201301. [DOI] [PubMed] [Google Scholar]
- Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. Modifying NEMO. Nat Cell Biol. 2004;6:89–91. doi: 10.1038/ncb0204-89. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. Embo J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Sun H. Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found Symp Proc. 2008:1–16. doi: 10.1007/2789_2008_098. [DOI] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, Fisher PB. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23:1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, Zhang Z, Arnott D, Dixit VM. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15:407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- Poukka H, Aarnisalo P, Karvonen U, Palvimo JJ, Janne OA. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA. 2002;99 doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- Sui G, Shi Y. Gene silencing by a DNA vector-based RNAi technology. Methods Mol Biol. 2005;309:205–218. doi: 10.1385/1-59259-935-4:205. [DOI] [PubMed] [Google Scholar]
- Tahk S, Liu B, Chernishof V, Wong KA, Wu H, Shuai K. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci USA. 2007;104:11643–11648. doi: 10.1073/pnas.0701877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and innate immunity. J Mol Med. 2006;84:712–725. doi: 10.1007/s00109-006-0084-y. [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Gronholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- Weng C, Li Y, Xu D, Shi Y, Tang H. Specific cleavage of Mcl-1 by caspase-3 in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Jurkat leukemia T cells. J Biol Chem. 2005;280:10491–10500. doi: 10.1074/jbc.M412819200. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. Embo J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu LG, Han KJ, Wei X, Shu HB. PIASy represses TRIF-induced ISRE and NF-kappaB activation but not apoptosis. FEBS Lett. 2004;570:97–101. doi: 10.1016/j.febslet.2004.05.081. [DOI] [PubMed] [Google Scholar]
- Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- Zheng D, Chen G, Guo B, Cheng G, Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]