Abstract
Nanopores have attracted much interest for nucleic acid sequencing, chemical sensing, and protein folding at the single molecule level. The outer membrane protein OmpG from E. coli stands out because it forms nanopores from single polypeptide chains. This property affords one to separately engineer each of the seven extracellular loops that control access to the pore. The longest of these loops, loop 6, has been recognized previously as the main gating loop that closes the pore at low pH and opens it at high pH. In the present work, we devised a method to pin each of these loops to the embedding membrane and measure the single pore conductances of these constructs. The electrophysiological and complementary NMR measurements show that the pinning of individual loops alters the structure and dynamics of neighbouring and distant loops in a correlated fashion. Pinning of loop 6 generates a constitutively open pore and patterns of concerted loop motions control access to the OmpG nanopore.
Keywords: nanopore, porin, molecular engineering, NMR, electrophysiology
Engineered protein nanopores have attracted interest to detect rare metal ions and neurotransmitters in solution,[1] to sequence at low cost small numbers of molecules of DNA and RNA,[2] and to measure the folding and unfolding kinetics of single proteins.[3] Most of these studies used engineered versions of the heptameric α-hemolysin nanopore although other proteins have also been used.[4] Single molecule detection in these systems has been mostly achieved through electrophysiological experiments with nanopores that were reconstituted into planar lipid bilayers. As an alternative, optical detection schemes have also been devised.[5] Although α-hemolysin has been so successful in these studies, a major limitation is its heptameric seven-fold symmetric structure[6] requiring the assembly and purification of heteroheptamers. The E. coli outer membrane protein OmpG offers an attractive alternative because it forms a nanopore from a single polypeptide chain that folds into lipid bilayers as a 14-stranded β-barrel.[7] Pore access is controlled by seven extracellular loops that have been shown to be highly dynamic and regulated by pH.[7a, 8] Despite the promise of OmpG for further nanopore engineering, a drawback has been its loop dynamics, which give rise to flickering single channel currents. This problem has been overcome partially by the engineering of the so-called quiet OmpG mutant, in which strands 12 and 13 were disulfide cross-linked and the irregularly structured Asp215 in strand 11 was deleted.[9] In the present work, we have devised a new strategy to quiet OmpG by pinning the loops to the membrane by long hydrocarbon-chain alkylation (Figure 1A). Moreover, we have used NMR in lipid micelles to characterize changes in loop structure and dynamics as a consequence of pinning each of the seven loops. Dodecylation of loop 6 had the largest effect on permanently opening the channel, but other loops are shown to also contribute to the channel closing in to various degrees and in cooperation with loop 6. Neighboring loops are particularly affected by pinning of individual loops and loops 1 and 5 were found to cooperate synergistically to control access to the nanopore.
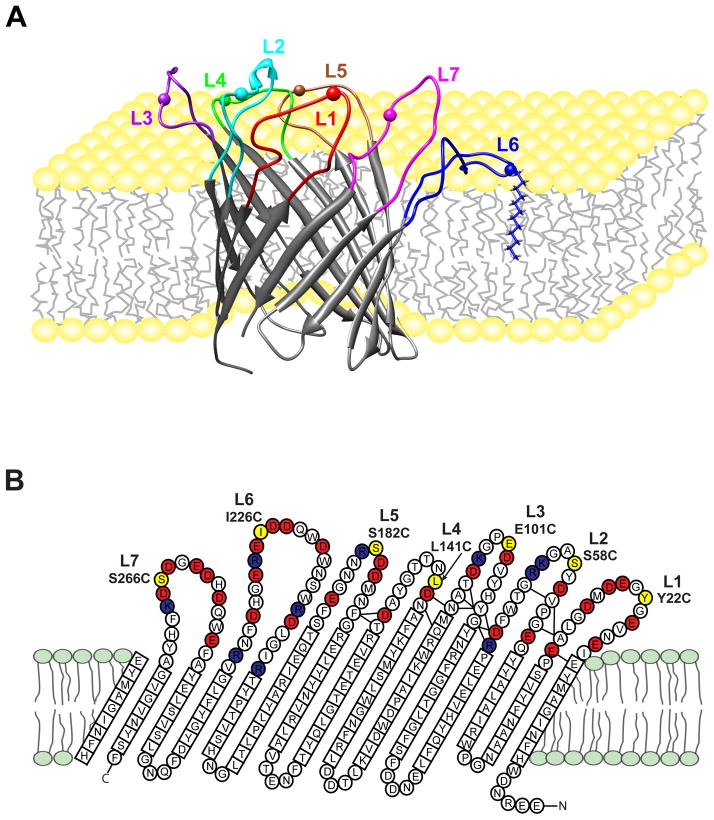
Figure 1.
Pinning of loops of OmpG nanopore to the membrane by dodecylation of engineered cysteines. A. A conformer of the ensemble of solution structures of OmpG[8b] is shown with loop 6 pinned to the membrane by dodecylation at Cys226 (pinned L6, mutant I226C). Similar constructs were made for pinned L1 (mutant Y22C), pinned L2 (mutant S58C), pinned L3 (mutant E101C), pinned L4 (mutant L141C), pinned L5 (mutant S182C), and pinned L7 (mutant S266C). The spheres represented the single cysteine sites. B. Topology of OmpG determined from the solution NMR structure (PDB code 2JQY). 14 transmembrane β-strands are connected by 7 extracellular loops with an excess of negatively charged residues (red) over positively charged residues (dark blue). The residues that were changed to single cysteines for alkylation are shown in yellow in each loop.
To immobilize individual loops of OmpG and pin them to the membrane, we engineered single cysteines into each of the loops. The following sites were chosen that preserve the predominantly negative charge characters of each of the loops, which we figured might be important for their function: Y22C (L1), S58C (L2), E101C (L3), L141C (L4), S182C (L5), I226C (L6), and S266C (L7) (Figure 1B). Introduction of these mutations caused little or no structure perturbation as monitored by 1H,15N-TROSY NMR of OmpG in dodecylphosphocholine (DPC) micelles (not shown). However, dodecylation of these cysteines caused major shifts and broadening of cross-peaks of nearby residues as exemplified by Ile226 in loop 6 (Figure S1). Residues forming the β-barrel pore structure were not significantly changed by dodecylation and the resulting pinning of the loops to the lipid surface. For simplicity, we call the dodecylated pinned loop mutants of OmpG pL1, pL2, … pL7, for the respective seven loop mutants.
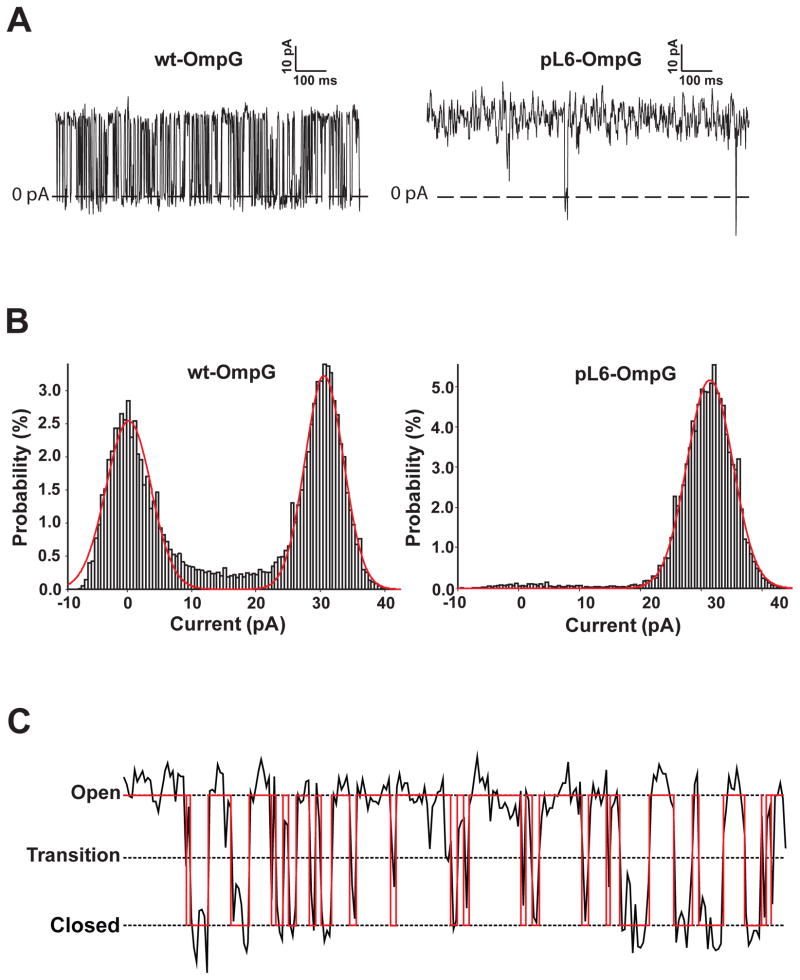
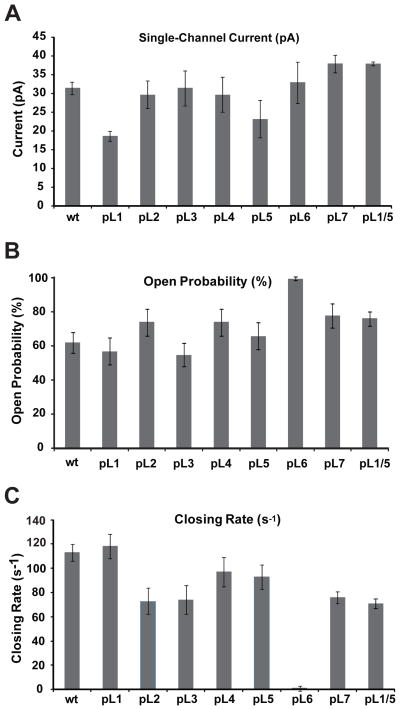
Figure 2 compares typical single-channel current traces of wild-type OmpG (wt) and pL6 OmpG flickering between open and closed states at pH 6.0, i.e. the pH that shows about equal populations of the two states in the wt.[10] Single channel activities (10–20 s recording time for each condition) were analyzed in terms of open current, open probability, and closing rate. Gaussian fits of the open probability histograms reveal that wt OmpG was open 62%, but pL6 was open nearly 100% of the time. The open currents at 50 mV and 1 M KCl were 31 and 33 pA, respectively, and the closing rate of wt was 113 s−1. Pinning of loops 2, 3, 4, and 6 did not significantly change the open currents of OmpG, but the open currents of pL1 and pL5 were reduced to 19 and 23 pA, respectively (Figure 3). Similarly, the open probabilities of pL1, pL3, and pL5 were not changed much, but pL2, pL4, and pL7 were significantly more open than wt. Besides pL6, which was always open, the closing rates of pL2, pL3, and pL7 were most reduced. Since pL1 and pL5 produced the largest current reductions, we also prepared the double-pinned mutant pL1/5. Very interestingly, the open current and open probability of this mutant surpassed that of wt (Figure 3), strongly suggesting that loops 1 and 5 cooperate with each other and likely also with loop 6 in closing the OmpG channel. Only when both loops were immobilized, the channel became 75% open. This double mutant is presumably closed only by the single remaining major pore gating loop 6.
Figure 2.
Illustration of single-channel current recordings and definition of their open-closed state transitions. A. Typical current recordings of wild-type and pL6 OmpG nanopores at pH 6.0. B. Current histograms of wild-type and pL6 OmpG recordings. The red curves are fits of the data to Gaussian distributions. C. Definition of state transitions. Nanopores are considered open/closed when the current is >50%/<50% of the open current. The open probability is defined as the fraction of the total recording time that the pore is open. The closing rate is defined as the number of closings per second. The red trace is the binary analysis of the experimental recording.
Figure 3.
Pinning of individual loops changes the electrophysiological activity of the OmpG nanopore. Changes of the open current (A), open probability (B), and closing rate (C) upon pinning of loops 1 through 7 to the membrane by dodecylation. Data for the double-pinned mutant pL1/5 are also shown. All measurements are averages from at least 5 independent lipid bilayer measurements.
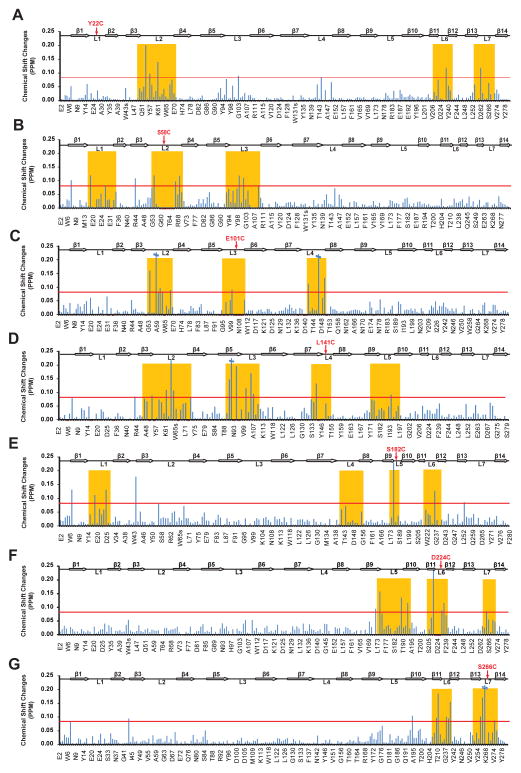
The electrophysiological results suggest that loops 1 and 5 can partially substitute for each other and cooperate with loop 6 in closing the OmpG nanopore. Previous NMR ensemble calculations already suggested that some loop motions are correlated.[8b] To more directly demonstrate structural cross-talk between different loops, we recorded 1H,15N-TROSY NMR spectra of each of the single-pinned loop constructs of OmpG and compared the combined 1H-15N chemical shifts to those of the corresponding unreacted free Cys mutant proteins. The chemical shift perturbations show that the pinned loops mostly affect the structures of the neighbouring loops (Figure 4). For example, pL3 mostly affects the chemical shifts of loops 2 and 4 (Figure 4C), pL2 affects loops 1 and 3 (Figure 4B), and pL6 affects loops 5 and 7 (Figure 4F). Some other pinned loop perturbations reach farther to the second nearest neighbours. For example, pL1 affects the structures of loops 6, 7, and 2 (Figure 4A) and pL4 affects the structures of loops 2, 3, and 5 (Figure 4D). pL5 is the only exception: in addition to affecting its neighbouring loops 4 and 6, it also reaches across and affects the structure of loop 1. This structural evidence nicely supports the synergistic effect of loops 1 and 5 that we observed electrophysiologically. In summary, the loops of OmpG communicate with each other structurally and undergo concerted motions that all contribute to the electrophysiological behavior of the OmpG nanopore. The coupling between neighbouring loop motions is stronger than distant coupling across the pore, which however occurs in the case of a structural cooperation between loops 1 and 5.
Figure 4.
NMR chemical shift perturbations suggest concerted motions of extracellular loops in pinned OmpG loop mutants. Chemical shift changes between dodecylated OmpG mutants and their unreacted counterparts are plotted for all seven pL mutants. A red line is drawn at 0.08 ppm, which is twice the estimated average measuring error. Chemical shifts were obtained from 1H,15N-TROSY spectra of ~0.5 mM 2H,15N-labeled protein samples in 150 mM DPC micelles at pH 7.0 and 600 MHz proton frequency.
If the threshold for structural perturbation is set somewhat lower than the current line at 0.08 ppm in Figure 4, one also finds that pL1 and pL5 affect more loops than the other pL mutants, suggesting that L1 and L5 may have a role in scaffolding the other loops. This possible structural scaffold function of loops 1 and 5 could explain their synergism in opening pore access that we observed electrophysiologically.
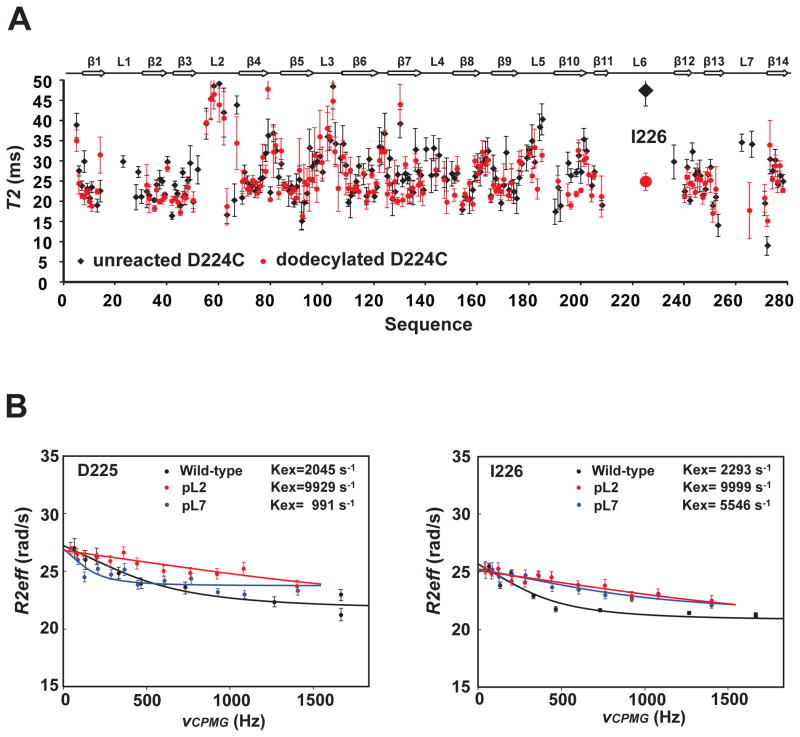
To examine how pinning affects the motions of the different loops, we measured 15N NMR transverse relaxation times T2 of the pL6 mutant D224C before and after dodecylation. Amide 15N T2 times are inversely proportional to and report on segmental ns timescale correlation times τc of the polypeptide backbone. A comparison of the T2 times of unreacted and dodecylated D224C mutant shows that the segmental motions of most loops on the ns timescale are not significantly affected by the pinning of loop 6 (Figure 5A). However, the pinning of loop 6 at residue 224 resulted in a significant decrease of the T2 time from 47 to 25 ms of nearby Ile226. This corresponds to an increase of a rough estimate of the segmental loop τc from 12 to 28 ns when this loop becomes partially immobilized on the lipid surface.
Figure 5.
Nanosecond and sub-millisecond dynamics of pinned and unpinned loops of OmpG nanopore. A. 15N NMR transverse relaxation times (T2) of 2H,15N-labeled D224C-OmpG unreacted (black) or reacted (red) with a dodecyl chain. The T2 times were obtained by fitting the relaxation profiles using exponential decays. Error bars represented the fitting errors. Only residues with T2 times between 0–50 ms are displayed. B. NMR relaxation dispersion experiments indicating correlated sub-millisecond loop motions. 15N CPMG dispersion profiles of Asp225 (left) and Ile226 (right) of wild-type (black), pL2 (red) and pL7 (blue) OmpG. The relaxation dispersion profiles were fitted by a two-state fast exchange model and the resulting exchange rates are indicated in each panel. All data were acquired with ~0.5 mM protein samples in DPC micelles at pH 7.0, 800 MHz and 40 °C.
To assess the stability of dodecyl chain insertion into the membrane, we monitored the accessibility of the disulfide bond by DTT in DPC micelles and DMPC liposomes (Figure S2). I226C was labeled through a disulfide bond with either a nitroxide or a dodecyl chain. Nitroxide-labeled I226C was readily cleaved by DTT, suggesting exposure to solvent as expected. Dodecylated I226C was completely protected from disulfide reduction and cleavage by DTT, strongly suggesting its burrial in DPC micelles and DMPC bilayers. There are previous reports that acyl chains of lipidated peptides and proteins are inserteted into lipid model membranes[11] and anchoring of the loops at their two ends to the βbarrel should enhance this interaction. Together with the reduced mobilities of these loops measured by NMR, these experiments provide strong support for the notion that the lipidated loops are indeed pinned to the membrane.
The opening and closing of the OmpG nanopore by the main gating loop 6 occurs on the low ms timescale (Figure 3C). This and faster timescales can be accessed by NMR relaxation dispersion experiments. We performed 15N Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments to probe sub-ms motions of selected loop 6 residues before and after pinning of the neighbouring loop 7 and the more distant loop 2 (Figure 5B). The relaxation profiles show exchange rates of ~2050 and 2300 s−1, respectively, for the two loop 6 residues Asp225 and Ile226. When loop 2 was pinned, the exchange rates of the loop 6 residues increased to almost 10,000 s−1. However, when loop 7 was pinned, the exchange rate of Asp225 decreased to ~1000 s−1 and that of Ile226 increased to ~5,500 s−1. An increase of exchange rates upon pinning of neighbouring or more distant loops may be rationalized by restraining the motions of other loops (loops 2 and 7 in this case) on the sub-ms timescale that would otherwise hinder loop 6 motions. The increase in motional frequency of loop 6 does not implicate anything about the open probabilities, which happen to be slightly increased in pL2 and pL7 (Figure 3B). The closing rate is slightly decreased on the ms timescale for pL2 and pL7 (Figure 3C), whereas the increased exchange rates measured by NMR occur on the sub-ms timescale and could reflect opening rates. The reduced exchange rate of Asp225 in pL7 is more difficult to explain and probably reflects some more complex motions of this charged residue in the field of many other charged loop residues (see Figure 1B).
Our new approach of loop immobilization has revealed patterns of cross-talk between different loops in controlling access to the OmpG nanopore. As expected from previous studies,[7a, 8b, 12] loop 6 performs the major gating function. When it was pinned to the membrane, a constitutively open pore was found with properties quite similar to those of quietOmpG (Figure S3).[9] pL6 OmpG should therefore be a promising candidate for further nanopore engineering and future practical applications in chemical sensing or sequencing. Structural, dynamical, and functional analysis also revealed important new roles of the other loops of OmpG. For example, we discovered concerted motions between loops 1 and 5, which may form a scaffold coordinating the motions of the other loops. Besides the cooperation of loops 1 and 5, any loop affects the motions of its next and second next neighboring loops. Collective loop motions have been described before by NMR paramagnetic relaxation enhancement data and derived structural ensemble calculations,[8b] but the current study assigns specific patterns of ordered motions to each of the loops, which may be driven by the high (negative) charge density in extracellular loops and hydrogen-bonding networks at the bases of the loops (Figure 1B). The loop dynamics of no other porin has been described in this much detail. Our approach relies on the immobilization of the flexible loops, which usually can be predicted from the amino-acid sequence, thus not requiring prior knowledge of the three-dimensional structure. Therefore, we expect the approach to be general and useful to analyze the structures and functions of other membrane pores and to guide OmpG nanopore engineering for a range of future practical applications.
Experimental Section
Mutagenesis was performed and proteins were prepared as previously described.[8b] Samples were refolded in octyl β-glucoside and transferred into DPC. Refolded single Cys mutants were reacted with a 10-fold excess of dodecyl methanethiosulfonate (Toronto Research Chemicals, Toronto, CA) over night and purified by Superdex 200 size-exclusion chromatography. Completeness of dodecylation was confirmed by MALDI-TOF mass spectrometry and NMR. Electrophysiological recordings were obtained at room temperature and an applied voltage of 50 mV as previously described [8a]. Symmetric buffer conditions with 25 mM Bistris, pH 6.0, 1 M KCl in both chambers were used. 1H,15N-TROSY NMR spectra, T2 relaxation time, and CPMG relaxation dispersion experiments of ~0.5 mM 2H,15N-labeled protein samples in 150 mM DPC micelles were obtained at 40 °C on Bruker Avance 600 or 800 MHz spectrometers in 25 mM Hepes, pH 7.0, 50 mM NaCl, 0.1 mM EDTA, 0.05% NaN3. The unreacted single Cys mutant proteins also contained 5 mM DTT. Detailed experimental methods are available in Supporting Information.
Supplementary Material
Footnotes
This work was funded by grant R01 GM52329 from the National Institutes of Health. We thank Drs. Binyong Liangand Hagan Bayley for critically reading the manuscript.LKT would also like to thank Dr. Bayley for his hospitality when this manuscript was written during a sabbatical stay in his laboratory at the University of Oxford.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx. ((Please delete if not appropriate))
References
- 1.a) Choi LS, Mach T, Bayley H. Biophys J. 2013;105:356–384. doi: 10.1016/j.bpj.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boersma AJ, Brain KL, Bayley H. ACS Nano. 2012;6:5304–5308. doi: 10.1021/nn301125y. [DOI] [PubMed] [Google Scholar]
- 2.a) Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Proc Natl Acad Sci USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ayub M, Hardwick SW, Luisi BF, Bayley H. Nano Lett. 2013;13:6144–6150. doi: 10.1021/nl403469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Larrea D, Bayley H. Nat Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Nat Biotechnol. 2012;30:349–U174. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heron AJ, Thompson JR, Cronin B, Bayley H, Wallace MI. J Am Chem Soc. 2009;131:1652–1653. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 6.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 7.a) Yildiz O, Vinothkumar KR, Goswami P, Kuhlbrandt W. EMBO J. 2006;25:5240–5240. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Subbarao GV, van den Berg B. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 8.a) Liang BY, Tamm LK. Proc Natl Acad Sci USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhuang TD, Chisholm C, Chen M, Tamm LK. J Am Chem Soc. 2013;135:15101–15113. doi: 10.1021/ja408206e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Khalid S, Sansom MSP, Bayley H. Proc Natl Acad Sci USA. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlan S, Zhang Y, Cheley S, Bayley H. Biochemistry. 2000;39:11845–11854. doi: 10.1021/bi001065h. [DOI] [PubMed] [Google Scholar]
- 11.Brunsveld L, Waldmann H, Huster D. BBA-Biomembranes. 2009;1788:273–288. doi: 10.1016/j.bbamem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 12.a) Damaghi M, Bippes C, Koster S, Yildiz O, Mari SA, Kuhlbrandt W, Muller DJ. J Mol Biol. 2010;397:878–882. doi: 10.1016/j.jmb.2010.02.023. [DOI] [PubMed] [Google Scholar]; b) Damaghi M, Koster S, Bippes CA, Yildiz O, Muller DJ. Angew Chem. 2011;123:7560–7562. doi: 10.1002/anie.201101450. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2011;50:7422–7424. doi: 10.1002/anie.201101450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.