Abstract
The insulin peptide B:9-23 is a natural antigen in the non-obese diabetic (NOD) mouse model of type 1 diabetes (T1D). In addition to αβ T cells and B cells, γδ T cells recognize the peptide and infiltrate the pancreatic islets where the peptide is produced within β cells. The peptide contains a cysteine in position 19 (Cys19), which is required for the γδ but not the αβ T cell response, and a tyrosine in position 16 (Tyr16), which is required for both. A peptide-specific mAb, tested along with the T cells, required neither of the two amino acids to bind the B:9-23 peptide. We found that γδ T cells require Cys19 because they recognize the peptide antigen in an oxidized state, in which the Cys19 thiols of two peptide molecules form a disulfide bond, creating a soluble homo-dimer. In contrast, αβ T cells recognize the peptide antigen as a reduced monomer, in complex with the MHCII molecule I-Ag7. Unlike the unstructured monomeric B:9-23 peptide, the γδ-stimulatory homo-dimer adopts a distinct secondary structure in solution, which differs from the secondary structure of the corresponding portion of the native insulin molecule. Tyr16 is required for this adopted structure of the dimerized insulin peptide as well as for the γδ response to it. This observation is consistent with the notion that γδ T cell recognition depends on the secondary structure of the dimerized insulin B:9-23 antigen.
Keywords: Gamma delta T cells, T Cell Receptor, Insulin, Autoreactivity, Autoimmune diabetes, Oxidation
1. Introduction
The adaptive immune system consists of three lymphocyte-types, B cells, αβ T cells and γδ T cells, which share the ability to express diverse antigen receptors encoded by genes that undergo somatic gene rearrangement. Widely distributed in present day vertebrates, all three cell types can be traced back ~ 500 million years through their antigen receptor genes [1], suggesting that they each have essential functions in the survival of the species [2, 3].
Like the other lymphocyte-types, γδ T cells take part in immune responses [2, 3], become mobilized during sterile or infectious inflammation [4, 5], and contribute to host protection, especially early in life [6]. They have been implicated in tissue repair [7], antigen presentation to T cells [8], B cell help [9], the elaboration and control of certain cytokines (e.g. IFN-γ, IL-4, IL-17, IL-22) [5, 10–12], and much evidence suggests that γδ T cells monitor and respond to stressed cells and tissues [13, 14]. Many of these functions are overlapping with those of other lymphocyte populations, which besides conventional T and B cells include innate-like NKT cells, MAIT cells and B1 B cells. However, γδ T cells express antigen receptors that are distinct from the BCRs and αβ TCRs [15–17], suggesting that they recognize antigens differently. Differences in TCR triggering [18] and responsiveness also set γδ T cells apart. Thus, comparison of TCR-mediated signaling showed that γδ TCRs signal more robustly when compared to αβ TCRs [19]. Second, studies of the antigen binding site revealed that the γδ TCR differs from the αβ TCR in that the complementarity-determining regions 3 (CDR3) of the γ and δ chains are more variable in length [20], and from immunoglobulins (Igs) by lesser diversity of CDR1 and 2 [21]. Finally, the antigens recognized, and the mode of antigen recognition, indicate that antigens for γδ T cells are structurally far more diverse than those of αβ T cells [3, 22], and γδ T cells tend to recognize antigens directly, without requirement for processing and presentation [23]. The broad range of molecular moieties stimulating TCR-dependent γδ T cell responses includes intact proteins (cell surface expressed or soluble), protein fragments (peptides), phosphoantigens (e.g. isoprenyl-pyrophosphate), phospholipids and/or complexes between phospholipids or sulfatides and proteins [3, 22]. Furthermore, individual γδ TCRs can mediate responses to several structurally unrelated molecules [24], so that the mode of recognition might change depending on the antigen involved. As a caveat, many of the presumed γδ antigens have not yet been directly shown to bind to the γδ TCR, or to elicit physiological γδ responses in vivo.
Early reports that small synthetic peptides elicit TCR-dependent responses of γδ T cells [25, 26] left unanswered how peptides might be recognized, and whether natural peptides can be antigens for γδ T cells. However, these studies, which involved γδ T hybridomas, already indicated that such responses do not require antigen-presenting cells (APCs) [27], in marked contrast to the peptide antigen-specific, MHC-restricted responses of αβ T cells. This suggested that γδ T cells and αβ T cells recognize peptides by different mechanisms. More recently, we reported a TCR-dependent γδ response to the insulin B chain-derived peptide B:9-23 [28], which is a natural auto-antigen in non-obese diabetic (NOD) mice [29]. NOD mice spontaneously develop a type-1 diabetes (T1D)-like autoimmune disease, which unfolds in several stages, including the early appearance of autoantibodies directed against pancreatic islet antigens, insulitis, and finally β cell destruction and diabetes [30]. γδ T cells appear to play both pathogenic and regulatory roles in this autoimmune disease [31–33], but what triggers their engagement remains unclear. Insulin is an early, prominent and essential auto-antigen in this disease [34, 35]. The naturally occurring insulin B chain-derived peptide B:9-23 is recognized by B cells [29, 36] and CD4+ αβ T cells [37], and by γδ T cells [28]. NOD-derived αβ T cells “see” this peptide antigen in the context of the MHCII molecule I-Ag7, and the molecular parameters of this recognition have been studied in much detail [38, 39]. Here, we provide an initial account of requirements for the recognition of the insulin peptide B:9-23 by γδ T cells, which is not restricted by I-Ag7 but might depend on a distinct secondary structure associated with dimerization of the oxidized peptide.
2. Results
2.1 Oxidizing the insulin peptide B:9-23 improves its capability of stimulating γδ T cell hybridomas
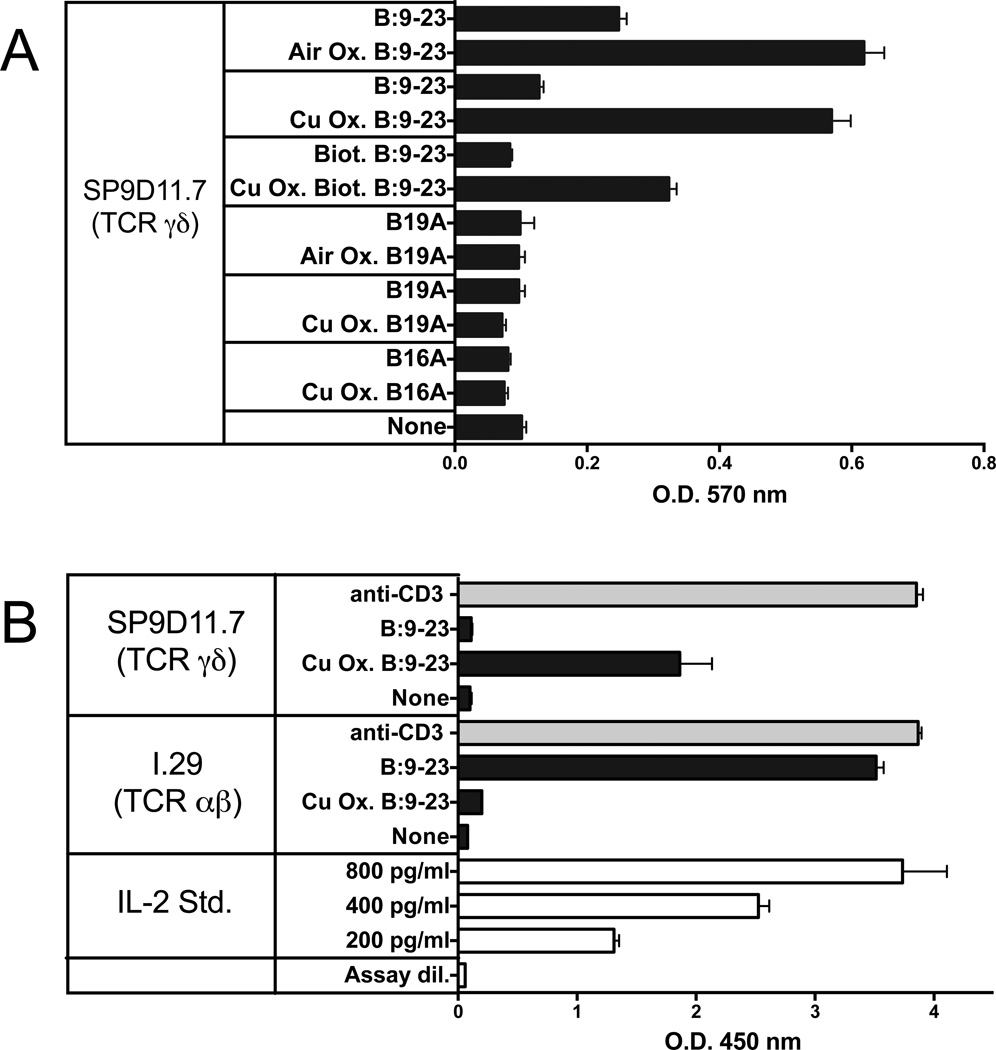
We previously found that a γδ T cell hybridoma derived from a mouse of the non-obese diabetic (NOD) genetic background (hybridoma SP9D11 expressing Vγ4 and Vδ10 TCR-genes) responded specifically to the insulin peptide B:9-23. The response was TCR-dependent. SP9D11 cells also responded specifically to pancreatic islet cells but not to the intact insulin molecule [28]. Specific reactivity to the B:9-23 peptide was also seen with several other NOD-derived γδ TCR-expressing hybridomas, revealing considerable diversity among γδ TCRs capable of supporting this response. Unlike αβ T cells, the γδ hybridomas responding to the insulin peptide did not require APCs, in this regard reminiscent of previously reported peptide responses by γδ hybridomas [25, 27], and even isolated single SP9D11 cells were activated by this soluble peptide [28]. Interestingly, a B:9-23 peptide in which the cysteine in position 19 (Cys19) was replaced with alanine (B:19A) was not stimulatory, suggesting that the cysteine might be required for the γδ response, in marked contrast to B:9-23-reactive αβ T cells, which respond well to the B:19A peptide [40, 41]. In addition, we noted variations between batches of untreated synthetic B:9-23 peptide in terms of their stimulatory capacity. Because cysteine contains a thiol group, which can be oxidized to form disulfides and higher oxidized states [42], we considered the possibility that the peptide must be oxidized to be stimulatory. We therefore compared the stimulatory capacity of fresh B:9-23 peptide preparations that were untreated vs. those that were intentionally oxidized, either by prolonged exposure to ambient air, or by adding copper chloride, which accelerates the oxidative process [43]. We also tested in this manner other peptides, including the previously identified non-stimulatory B:19A peptide [28], and another non-stimulatory B:9-23 peptide in which Tyr16 was replaced with alanine (B:16A). The experiments assembled in Fig.1, panel A show that oxidation substantially enhanced the ability of the wild-type B:9-23 peptide to stimulate SP9D11 cells, but failed to produce responses to the two alanine-substituted non-stimulatory peptides. In contrast, a B:9-23-reactive αβ T cell hybridoma (I 29; [44]) did not respond to the oxidized peptide (Fig.1, panel B). These findings suggested that while oxidation of Cys19 is not required for the αβ response, it might be critical for the γδ response.
Figure 1. Oxidation of the B:9-23 peptide antigen enhances stimulation of antigen-specific γδ T cell hybridomas while reducing stimulation of an antigen-specific αβ T cell hybridoma.
Panel A Responses of hybridoma SP9D11 to untreated and oxidized peptide antigens
3×104 hybridoma cells were cultured overnight either alone (none) or with untreated or oxidized (Air Ox., oxidized by exposure to ambient air, Cu Ox., oxidized with copper (II) chloride) peptide antigens at 100 µg/ml. Cellular responses were measured in triplicate using the LacZ stimulation assay. Bars show mean response values +/− SE.
Panel B Comparison of the responses of the γδ hybridoma SP9D11 and the αβ T cell hybridoma I.29 to untreated and oxidized B:9-23 peptide antigens
Culture conditions and peptide stimulation were as described for panel A, except for the addition to all cultures of 1×105 fixed APCs. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs. Cellular responses were measured in triplicate using ELISA for IL-2. Bars show mean response values +/− SE.
2.2 The oxidized insulin B:9-23 peptide stimulates γδ T cells as a dimer, and without requirement for MHCII
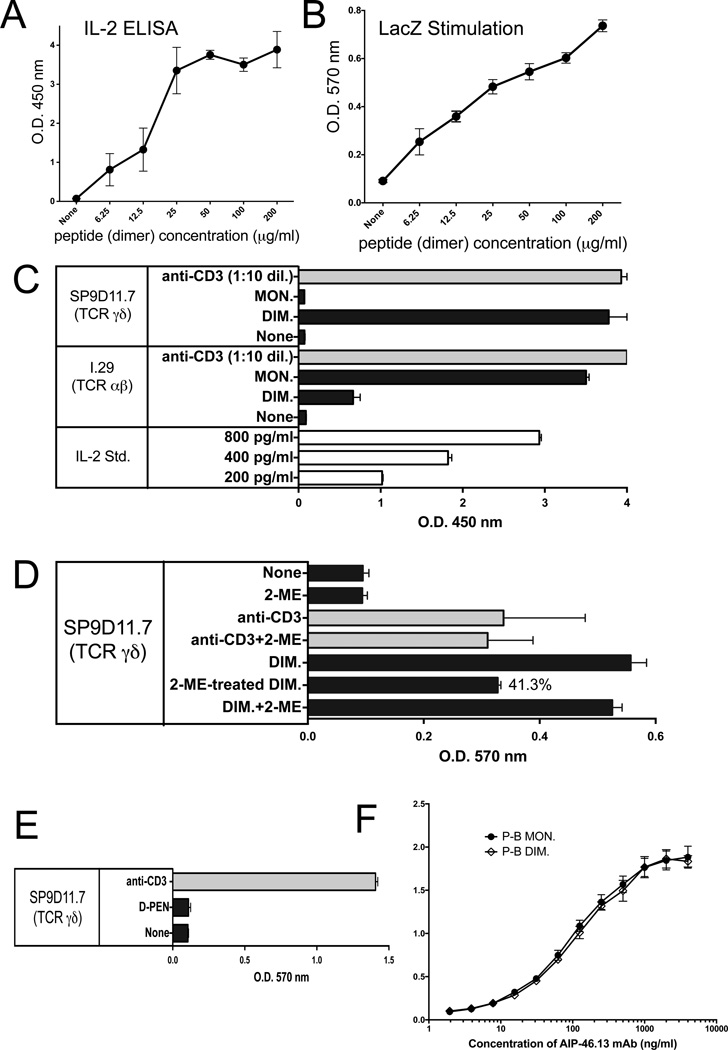
The thiol group in cysteine is readily oxidized to form disulfides, whereas higher oxidized forms require stronger oxidants [43]. Hence, the oxidized B:9-23 peptide might form a dimer and stimulate γδ T cells in this configuration. To examine this possibility, and to exclude other oxidized forms, we again employed the B:9-23 peptide, oxidized it with the comparatively weak oxidant DMSO [45], separated monomers from dimers by HPLC, and finally analyzed the purified fractions for their monomer and dimer content using EMS. Monomeric and dimeric peptide preparations with a purity of > 95% were then tested for their ability to stimulate SP9D11 cells (Fig. 2). SP9D11 hybridoma cells readily responded to the dimer fraction, even at peptide concentrations below 10 µg/ml as indicated in two different stimulation assays (Fig.2, panels A, B). In contrast, there was no response to the monomer fraction (Fig.2, panel C). Treatment of the dimer fraction with the reducing agent 2-ME significantly diminished stimulatory activity of the peptide, at a concentration that did not (yet) affect other cellular responses (Fig.2, panel D). The insulin B:9-23-specific αβ T cell hybridoma I.29, however, responded strongly to the monomer fraction and poorly to the dimer fraction (Fig.2, panel C). Of note, fixed APCs were used to demonstrate this difference between the γδ and αβ T cells (Fig.2, panel C). Because this result raised the possibility that the γδ T cells merely require a disulfide, we also tested a purified penicillamine adduct, in which the B:9-23 peptide was disulfide-linked to D-PEN instead of itself (A.M., unpublished). In contrast to the homo-dimer, this hetero-dimer failed to stimulate the SP9D11 cells, indicating that the monomeric B:9-23 peptide plus a disulfide is not sufficient to elicit the γδ response (Fig.2, panel D). Finally, we compared plate-bound monomeric and homo-dimeric B:9-23 peptides for their ability to immobilize the peptide-specific mAb AIP-46.13. Both forms immobilized the antibody indicating that AIP-46.13 recognizes both monomer and dimers of the B:9-23 antigen (Fig.2, panel E).
Figure 2. The γδ T cell hybridoma SP9D11 specifically recognizes oxidized dimers of the B:9-23 peptide antigen whereas the αβ T cell hybridoma I.29 recognizes monomers.
Panels A, B Response of SP9D11 to titrated amounts of dimeric B:9-23 peptide measured via IL-2 ELISA and LacZ assays, respectively.
Culture conditions and response measurements were as described in Fig.1.
Panel C SP9D11 γδ T cells selectively respond to dimers of the B:9-23-peptide, whereas I.29 αβ T cells respond to monomers.
Culture conditions, peptide stimulation and response measurements as described in Fig.1, panel B, except that HPLC-purified monomeric B:9-23 peptide (MON.) and DMSO-oxidized dimeric B:9-23 peptide (DIM.) were used as peptide antigens. Cellular responsiveness was determined by stimulation with plate-bound anti CD3ε mAbs. Supernatants of cultures with anti CD3ε were diluted 10× prior to response measurements. Bars show mean response values +/− SE.
Panel D SP9D11 Treatment of the dimeric B:9-23-peptide with 2-ME reduces its stimulatory activity.
Culture conditions, peptide stimulation and response measurements as described in Fig.2, panel C. None: no stimulation. 2-ME: 2-ME added to the cultures to a final concentration of 0.25 mM. anti-CD3: soluble anti CD3ε mAb added. Anti-CD3+2-ME: soluble anti CD3ε mAb added as well as 0.25 mM 2-ME. DIM: dimeric B:9-23 peptide added at 100 µg/ml. 2-ME-treated DIM: dimeric B:9-23 peptide pretreated with 5mM 2-ME, then added to stimulation cultures to a final peptide concentration of 100 µg/ml and a final 2-ME concentration of 0.25 mM. DIM+2-ME: dimeric B:9-23 peptide added at 100 µg/ml and 2-ME added at 0.25 mM. Cellular responses were measured in triplicate using the LacZ stimulation assay. Bars show mean response values +/− SE.
Panel E SP9D11 γδ T cells fail to respond to the penicillamin adduct of B:9-23
Culture conditions, stimulation and response measurements were as described in Fig.1, panel A. The B:9-23-penicillamin adduct was added at 100 µg/ml. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs. Responses were measured in triplicate using the LacZ stimulation assay. Bars show mean response values +/− SE.
Panel F Monoclonal antibody AIP-46.13 recognizes monomeric and dimeric B:9-23 peptide (binding assay)
High protein binding ELISA plates were coated with purified monomeric or dimeric B:9-23 peptide at 3 µg peptide/ml. Titrated amounts of AIP-46.13 mAb were added and plate-bound antibody detected by ELISA. Curves show mean absorbance values of triplicate determinations +/− SE.
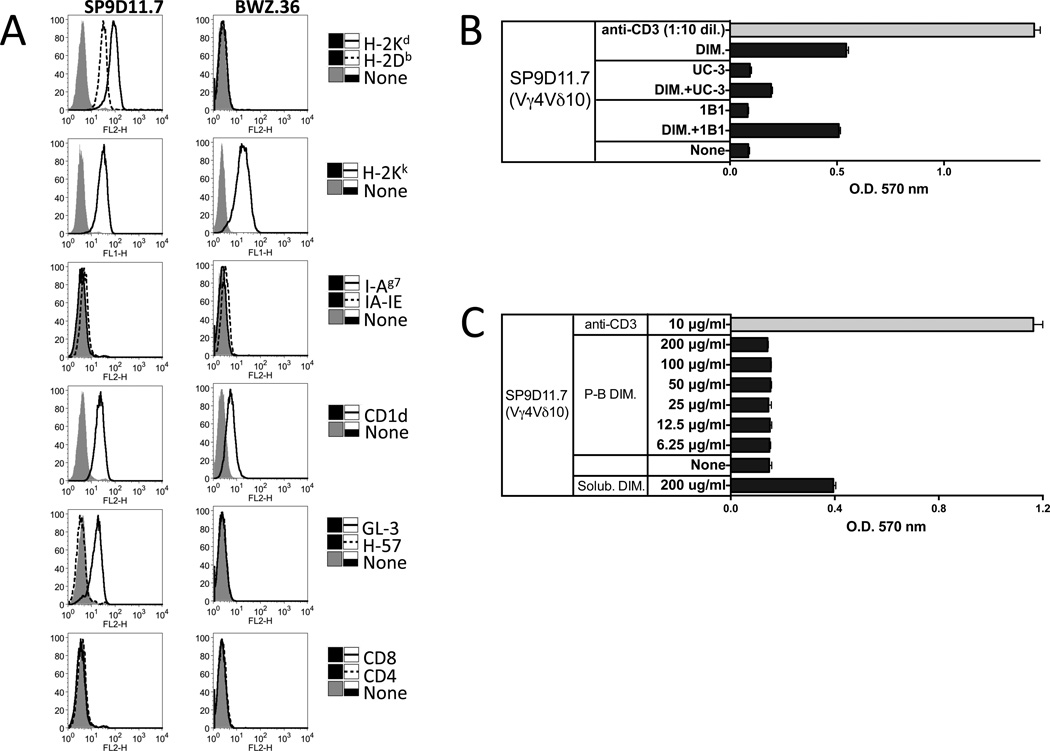
Whereas αβ T cells require APCs for their MHCII-restricted response to the B:9-23 antigen, the γδ response to the B:9-23 antigen was APC-independent ([28] and this study). However, the γδ T cells still potentially might auto-present [8, 46]. We therefore examined the B:9-23-reactive γδ hybridomas themselves for the expression of the NOD-derived MHCII molecule, I-Ag7, using mAb RT1B (clone OX-6) [47], which recognizes this molecule [48, 49] as well as mouse I-Ak and I-As, and mAb M5/114 [50], which recognizes mouse I-Ab,d,q and I-Ed,k. As shown in Fig. 3, panel A, neither I-Ag7 nor other I-A/E molecules were detected on the NOD-derived B:9-23 peptide-reactive SP9D11 hybridoma, nor did we find these or other MHCII molecules on other peptide-reactive or non-reactive γδ T cell hybridomas (77BAS-12, 96BLT-15, 123BLT-27, data not shown). However, hybridoma SP9D11 as well as other γδ T cell hybridomas (not shown) expressed CD1d (Fig.3, panel A for SP9D11). Nevertheless, blocking CD1d with mAb 1B1 known to inhibit CD1d-restricted responses of iNKT cells failed to inhibit the insulin peptide-response of SP9D11 cell, which was readily inhibited by an anti TCR-Vγ4 mAb detecting the SP9D11 TCR (Fig.3, panel B). Hence, the oxidized dimeric insulin peptide that stimulates the γδ response does not appear to be MHCII or CD1d-presented. Finally, we tested whether plate-bound insulin peptide might stimulate the SP9D11 cells but did not observe any response when using tissue culture plates and standard culture conditions (Fig.3, panel C). Although this does not rule out the possibility that immobilized peptide might be recognized by B:9-23-reactive γδ cells, the experiment showed that under the conditions of the stimulation assay used in this study, plate-bound insulin peptide does not substantially contribute to hybridoma stimulation. In view of these findings, and of our earlier observation that single isolated SP9D11 cells can be stimulated by the B:9-23 peptide [28], our data suggest a mechanism of B:9-23 recognition by γδ T cells, which is very different from that of conventional MHCII-restricted or non-conventional NKT-like αβ T cells.
Figure 3. No roles for MHC II or CD1d in the peptide response of the B:9-23-reactive γδ T cell hybridoma SP9D11.
Panel A Cell surface expression of TCR, MHC and CD1d molecules by SP9D11.7 cells
SP9D11.7 hybridoma cells and the fusion line BWZ.36 were stained with mAbs specific for MHC I and II molecules, CD1d, TCR-δ, TCR-β, CD4 and CD8, and analyzed cytofluorimetrically.
Panel B Anti CD1d mAb 1B1 fails to inhibit the peptide response of SP9D11.7 cells
Culture conditions, stimulation and response measurements were as described in Fig.1, panel A. Antibodies specific for TCR-Vγ4 (mAb UC3) or CD1d (mAb 1B1) were added to some cultures at a concentration of 10 µg/ml. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs. Supernatants of cultures stimulated with plate-bound anti-CD3ε were diluted 10× prior to response measurements. Responses were measured in triplicate using the LacZ stimulation assay. Bars show mean response values +/− SE.
Panel C Plate-bound insulin peptide does not substantially contribute to the stimulation of SP9D11 hybridoma cells
Prior to the stimulation cultures, dimeric insulin peptide dissolved in tissue culture medium was added to the wells of a tissue culture plate at the indicated concentrations, and plates were incubated overnight at 37°C. After washing the wells to remove unbound peptide, stimulation cultures were set up. Culture conditions, stimulation and response measurements were as described in Fig.1, panel A. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs. Responses were measured in triplicate using the LacZ stimulation assay. Bars show mean response values +/− SE.
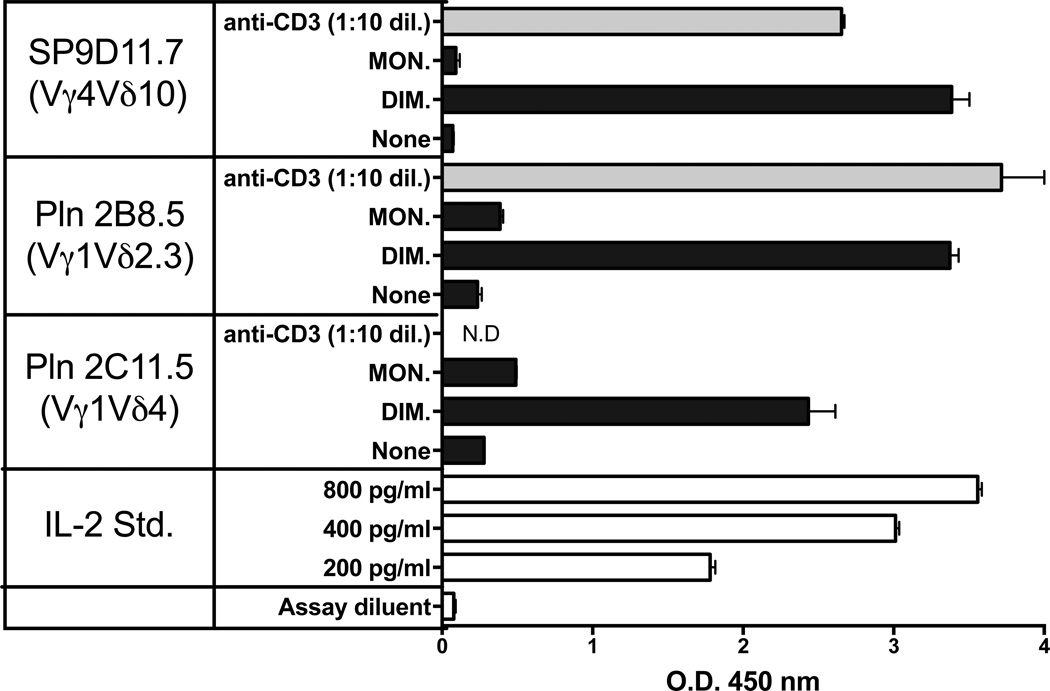
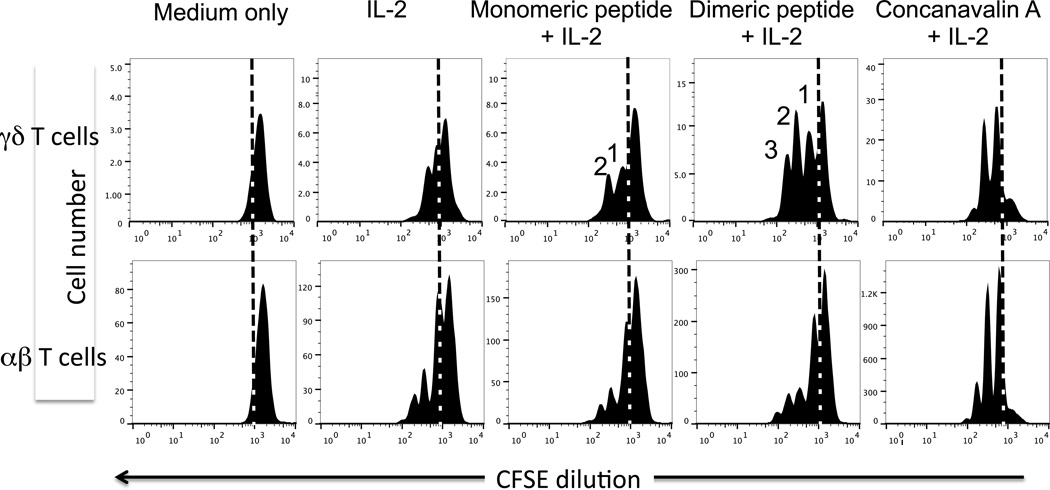
Examination of additional B:9-23 reactive γδ T cell hybridomas derived from NOD mice showed that all clones tested responded to the dimer (Fig. 4). APCs were not present in these assays. The responder cells expressed several different γδ TCRs, including various combinations of Vγ and Vδ genes [28]. With such diversity, it seemed possible that the insulin peptide response would be directly detectable, even among freshly isolated normal γδ T cells with their diverse TCRs. We therefore prepared NOD splenocytes, enriched for T cells via passage through nylon wool, and labeled the non-adherent (NAD) cells with the cytoplasmic vital dye CFSE. These cells then were incubated in vitro either alone or with purified dimeric or monomeric insulin peptide, in the presence of IL-2. NAD cells cultured with either concanavalin A or plate-bound anti-CD3 antibodies plus IL-2 were also included as a positive control. After the culture period, we stained the αβ and γδ T cells within the NAD cell cultures with specific antibodies, and compared their proliferative responses using flow cytometry (Fig.5). As shown by the positive controls, both αβ and γδ T cells were able to divide under these culture conditions, beyond the IL-2-supported background reactivity. The dimeric insulin peptide also stimulated divisions well above background, but this was only seen with γδ T cells and not with αβ T cells. The monomeric insulin peptide failed to elicit substantial responses over the IL-2-supported background of either type of T cell.
Figure 4. APC-independent responses of γδ T cell hybridomas expressing diverse TCRs to the oxidized dimeric B:9-23 antigen.
Culture conditions and peptide stimulation were as described for Fig. 2, panel C. Cellular responsiveness was determined by stimulation with plate-bound anti CD3ε mAbs. Supernatants of cultures with anti-CD3ε were diluted 10× prior to response measurements. Responses were measured in triplicate using an IL-2 ELISA. Bars show mean response values +/− SE.
Figure 5. Proliferation of freshly isolated γδ T cells from NOD spleen in response to stimulation with the oxidized dimeric B:9-23 antigen.
Nylon wool non-adherent lymphocytes from the spleens of 10 wks old NOD females were labeled in vitro with CFSE, then cultured at 5 × 106 cells/ml for two days in medium with 10 units/ml of murine IL-2 plus the peptide indicated at 300 µg/ml. The mitogen Concanavalin A (5 µg/ml) plus IL-2 was used as a positive control. Following a two day culture period, cells were stained with mAbs against either the γδ or αβ TCR. After gating on blasts based on forward/side scatter properties, γδ and αβ T cells were identified and assessed for CFSE levels by fluorescence intensity. Cells left of the dashed vertical line in each histogram have divided, and peaks denoting the expected CFSE levels after 1, 2, or 3 cell divisions among the γδ T cells are indicated.
2.3 The response to the oxidized insulin peptide is linked to certain γδ TCRs
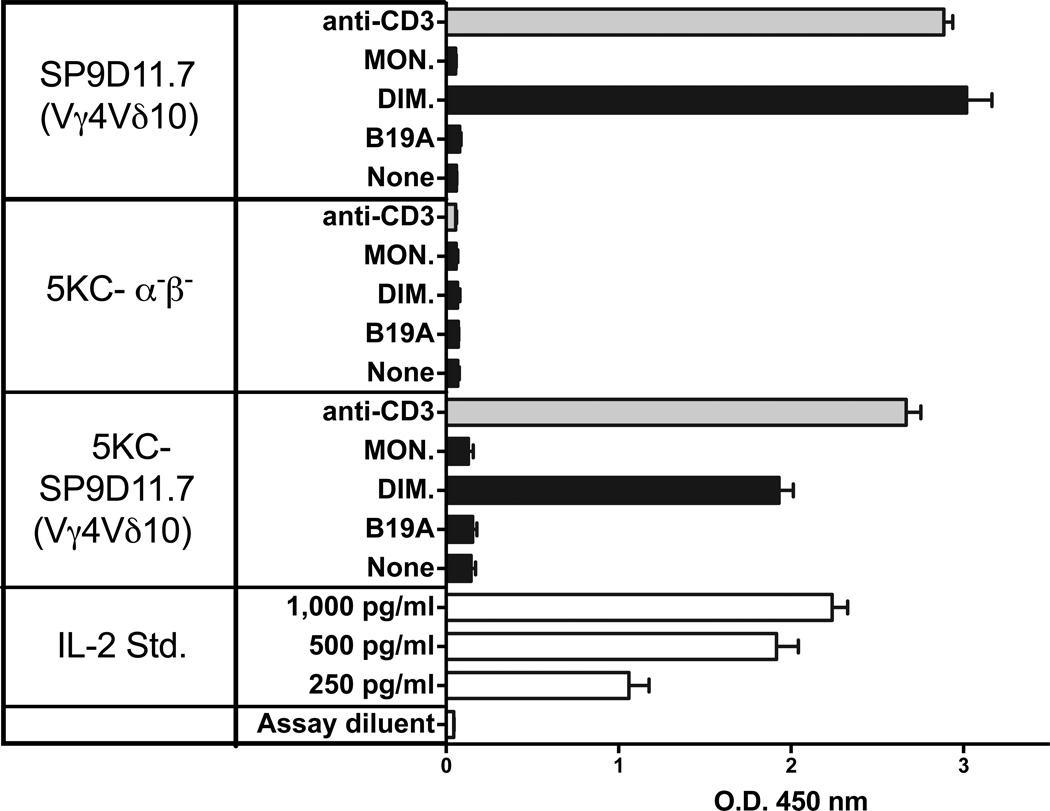
The response of hybridoma SP9D11 to the B:9-23 peptide was TCR-dependent as demonstrated with a TCR transfectoma expressing the SP9D11 γδ TCR [28]. Using the same transfectoma (5KC-SP9D11), we confirmed TCR-dependence of the response to the oxidized dimeric B:9-23 peptide (Fig. 6). 5KC-SP9D11 responded to the purified dimeric peptide whereas non-transfected 5KC cells failed to respond. The purified monomeric peptide did not elicit any responses.
Figure 6. The γδ T cell response to the oxidized dimeric B:9-23 antigen is TCR-dependent.
Hybridoma SP9D11.7 and transfectoma 5KC-SP9D11.7 expressing the SP9D11.7 γδ TCR show similar peptide responses whereas whereas non-transfected cells (5KC-α-β-) are non-responsive. Culture conditions and peptide stimulation were as described for Fig. 2, panel C. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs. Responses were measured in triplicate using ELISA for IL-2. Bars show mean response values +/− SE.
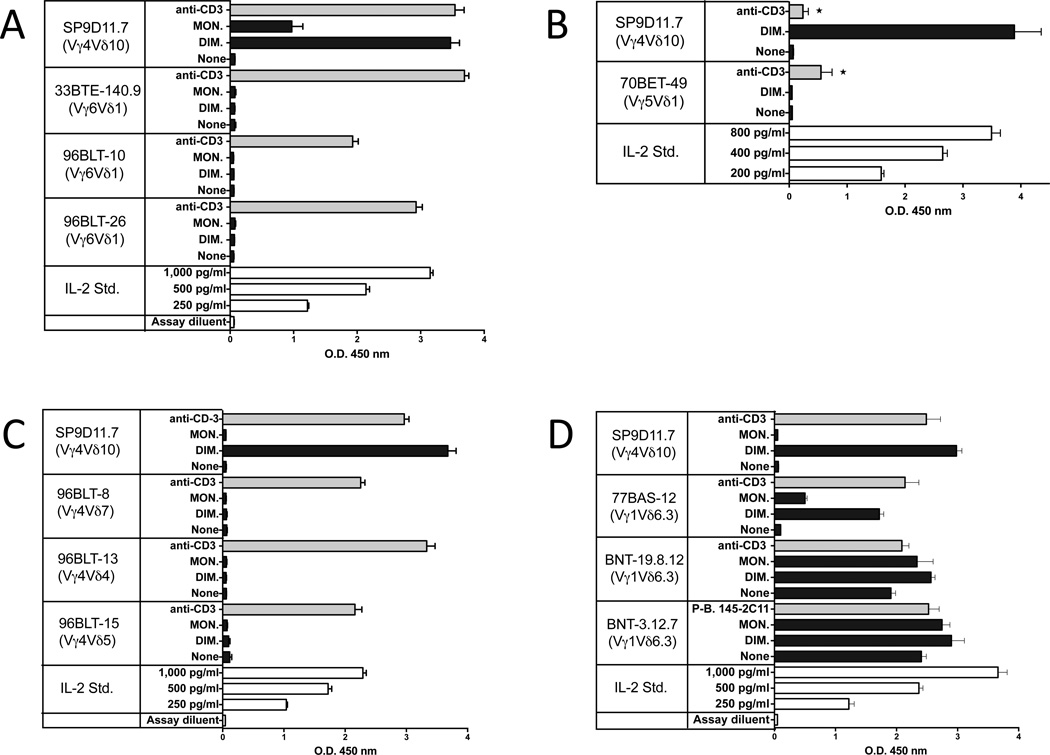
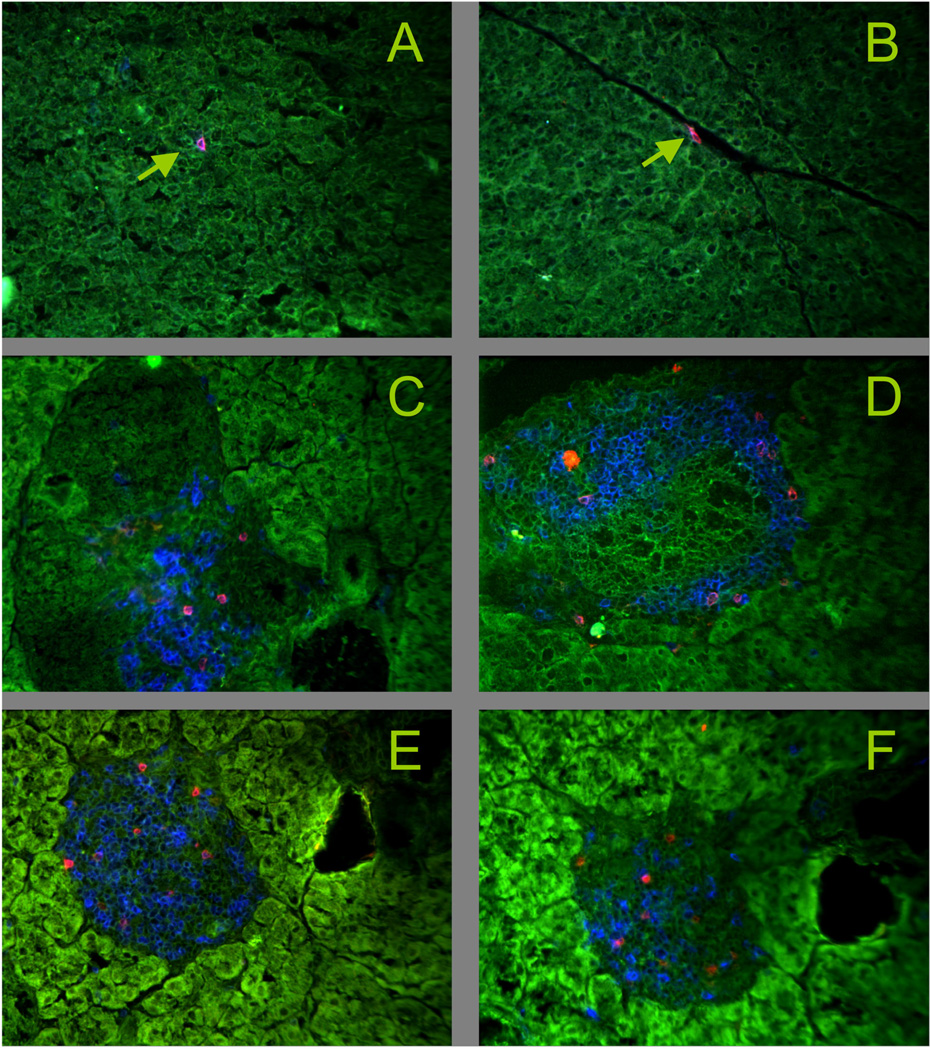
To explore the limits of the B:9-23-specific γδ repertoire, we examined γδ T cell hybridomas corresponding to major populations of γδ T cells in mice (Figure 7). Clones expressing invariant Vγ6Vδ1+ TCRs, representative of the γδ T cells present in the female reproductive tract, in the lung and during various inflammatory responses [2], were not stimulated by the insulin peptide (panel A), and another expressing the canonical invariant Vγ5Vδ1+ TCR, representative of epidermal γδ T cells [2], did not respond either (panel B). Several hybridomas expressing diverse Vγ4+ TCRs, commonly found among γδ T cell populations in the lymphoid organs, the liver and the lung [2] also failed to respond, despite considerable variation in their expression of TCR-Vδ and CDR3 regions (panel C) [51]. However, as shown with the SP9D11 cells and one other previously identified hybridoma expressing Vγ4 that responded to the insulin peptide [28], TCR-Vγ4+ clones can potentially be B:9-23 peptide responders. We also examined hybridomas expressing Vγ1, representative of the largest γδ T cell population in the spleen and other lymphoid tissues, and in the liver (panel D) [2]. Since these cells tend to show TCR-dependent “spontaneous” reactivity [52], it can be difficult to discern antigen-specific responses. Indeed, several hybridomas were highly reactive without any deliberate stimulation, and only small increases in cytokine production were seen when the purified dimeric peptide was added. Whether such clones can recognize the insulin peptide presently remains unclear. However, hybridoma 77BAS-12, derived from a C57BL/10 splenic γδ T cell expressing Vγ1Vδ6.3 [27], had little background reactivity and responded strongly to the insulin peptide. Given that we also found several peptide responders among Vγ1+ hybridomas derived from NOD mice (see Fig.4 and [28]), it is clear that the Vγ1+ γδ T cell subset contains γδ T clones capable of recognizing the oxidized insulin peptide. Moreover, hybridoma 77BAS-12 shows that such clones reach the periphery even in the absence of insulitis/diabetes-development or the particular disease-susceptible genetic background of the NOD mouse strain. In sum, we found multiple clones with specificity for the insulin peptide within two subsets of γδ T cells expressing diverse TCRs but none within the two subsets expressing invariant TCRs. Interestingly, the majority of the insulin peptide reactive, Vγ1+ γδ T cell hybridomas in the NOD-derived collection (3/5), and one hybridoma expressing the very similar Vγ2 gene, shared a distinctive CDR3 motif in the junction of their rearranged γ genes (W-MR-S/T) [28]. We did not find the complete motif in any of a large number of C57BL/6, C57BL/10 or AKR/J-derived Vγ1+ hybridomas (0/96), although numerous cells contained a partial match [27, 51, 53, 54]. Testing just one these cells - the B10-derived hybridoma 77BAS-12 containing a γ CDR3 with the sequence W-R-S – we found a peptide responder (Fig.7, panel D). Thus, CDR3γ might well contribute to insulin peptide recognition in these cells. Finally, because we found insulin peptide responders most readily among Vγ1+ γδ T cells, and because several of these have been derived from pancreatic lymph nodes [28], we examined the pancreas histologically for the presence of Vγ1+ γδ T cells. Indeed, such cells were easily detectable infiltrating the islets of Langerhans during insulitis in NOD mice, although we rarely saw γδ T cells in the pancreas of normal C57BL/6 mice (Fig. 8).
Figure 7. Limitations in the TCR repertoire of γδ T cells responsive to the oxidized dimeric B:9-23 antigen.
Panel A Hybridomas expressing Vγ6Vδ1 fail to respond to dimeric B:9-23
Panel B A hybridoma expressing Vγ5Vδ1 fails to respond to dimeric B:9-23
Panel C Several hybridomas expressing Vγ4 fail to respond to dimeric B:9-23
Panel D High spontaneous reactivity makes it difficult to detect B:9-23 reactivity in most Vγ1+ hybridomas, but hybridoma 77BAS-12 (Vγ1+Vδ6.3+) is a responder.
For all panels, culture conditions and peptide stimulation were as described for Fig.2, panel C. Cellular responsiveness was determined by stimulation with plate-bound anti-CD3ε mAbs, except for panel B (asterisks). Here, soluble anti-CD3ε mAbs were used (10 µg/ml), which provide a weaker stimulus. Responses were measured in triplicate using ELISA for IL-2. Bars show mean response values +/− SE.
Figure 8. Vγ1+ γδ T cells infiltrate the islets of Langerhans in NOD mice.
Snap frozen pancreas tissue was acetone dehydrated and stained with mAbs. Green: tissue autofluorescence.
Panels A, B Control: C57BL/6, blue: CD3ε, red TCR-δ, arrows: individual γδ T cells, not in the islets
Panels C–F NOD, C: 16 wks of age, blue: CD3ε, red: TCR-δ; D: 8 wks of age, blue: CD3ε, red: TCR-Vγ1; E: 12 wks of age, blue: CD3ε, red: TCR-Vγ1; F: 12 wks of age, blue: CD8α, red: TCR-Vγ1
2.4 The oxidized dimeric B:9-23 peptide adopts a secondary structure
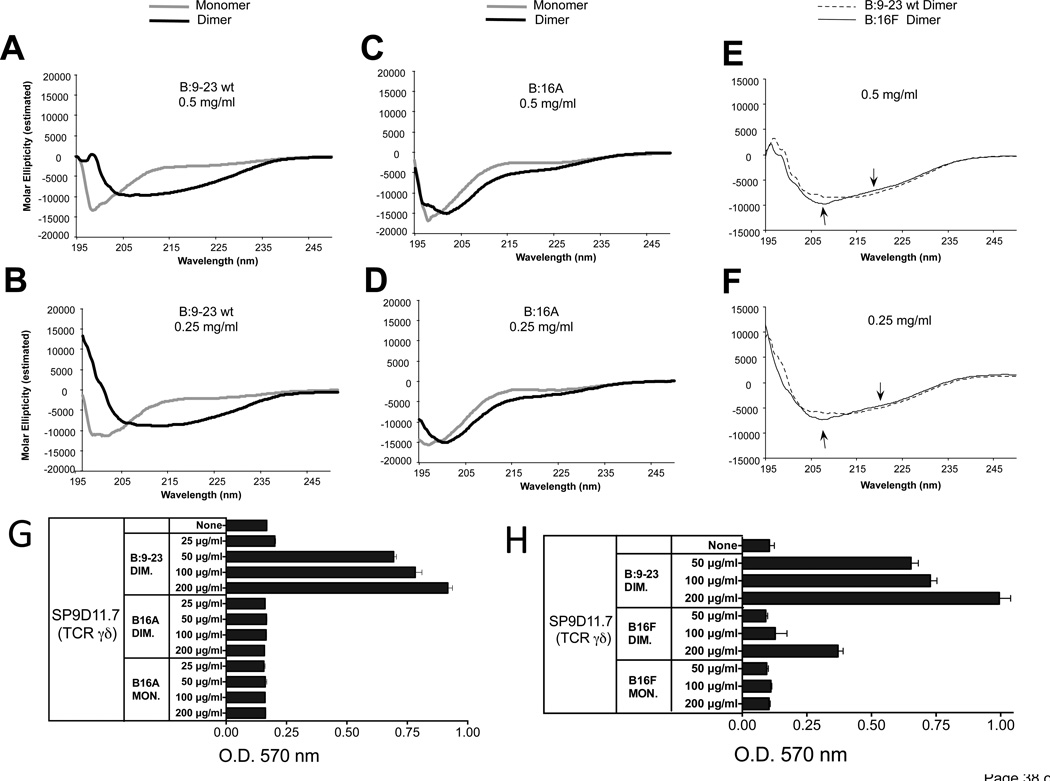
The distinct stimulatory activity of the oxidized dimeric B:9-23 peptide might derive from an ability to cross-link two γδ TCRs or from other distinct properties. In the absence of evidence that the stimulatory peptide must be presented, we considered the possibility that it is “seen” in a manner like intact non-processed protein antigens, which are recognized based on their three-dimensional epitopes. To detect possible structural differences between wild-type monomeric and oxidized dimeric B:9-23 peptides, we measured their circular dichroism (CD) in solution (Fig. 9). As indicated by the CD spectra in panels A and B, the non-stimulatory monomeric peptide had no particular structure as is common for small peptides. In contrast, the stimulatory dimer adopted an organized configuration consistent with a beta-pleated sheet secondary structure. The measurements at two peptide concentrations confirmed that this difference between monomer and dimer was not merely caused by variations in protein concentration. Unlike the wt peptide, the non-stimulatory B16A peptide did not show a substantial spectral shift as a dimer (panels C, D). Instead, monomer and dimer both retained CD spectra indicative of a disorganized structure, similar to the wt monomer. The spectral shift in the dimer was partially recovered when substituting Tyr16 more conservatively, with phenylalanine (B16F dimer) (panels E, F). However, there was still a small difference when compared to the wt peptide (arrows). Comparing these peptides in parallel stimulation assays with the hybridoma SP9D11.7 showed that whereas the B16A dimer had lost all stimulatory activity (panel G), the B16F dimer etained some stimulatory activity but was clearly a weaker stimulator than the wild-type dimeric peptide (panel H). Hence, these data reveal a correlation between the CD spectra and γδ-stimulatory activity of the peptides (see discussion).
Figure 9. As an oxidized dimer, the B:9-23 peptide antigen adopts a distinct secondary structure, which requires Tyr16.
Panels A, B CD spectra of monomeric and oxidized dimeric B:9-23 peptides (wild-type)
Panels C, D CD spectra of monomeric and oxidized dimeric B16A peptides
Panel E, F Comparison of CD spectra of oxidized dimeric wild-type and B16F peptides
CD spectroscopy was performed on HPLC-purified monomeric and dimeric B:9-23 wild-type (wt) (panels A and B) and amino-acid substituted peptides (panels C–F), at two peptide concentrations. Dimerization induces a shift in the circular dichroism of the wt peptide consistent with a change from a random structure of the monomer to a beta-pleated sheet structure of the dimer (panels A, B). In contrast, dimerization does not substantially change the circular dichroism of the B:16A substituted peptides (panels C, D). The dimeric B16F substituted peptide does shift in circular dichroism, but slightly less than the dimeric wt peptide (see arrows in panels E, F).
Panel G The B16A substituted peptide fails to stimulate a response of γδ hybridoma SP9D11
Culture conditions and response measurements using the LacZ assay were as described in Fig.1.
Panel H The B16F substituted peptide elicits a smaller response of γδ hybridoma SP9D11 than the wt peptide
Culture conditions and response measurements using the LacZ assay were as described in Fig.1.
3. Discussion
Although TCR-mediated ligand recognition by γδ T cells remains obscure and controversial, the notion that the mechanism resembles more the binding interaction of BCRs with their cognate antigens than that of αβ TCRs with antigen/MHC complexes [23] has gained considerable support. Thus, the structure of the CDR3s of the γδ TCRs resemble more those of antibodies than those of αβ TCRs [20], and the structures of antigens recognized by γδ T cells are far more diverse than those recognized by conventional MHC-restricted αβ T cells [22]. In particular, several examples of γδ TCR interactions with native proteins have been described [18, 55–58], and evidence for the recognition of structural protein motifs has been found [58]. In contrast, hardly any evidence for recognition of peptides in the context of presenting molecules has been uncovered [59]. Therefore, it would seem counterintuitive that γδ T cells should recognize and respond to small peptides. Small peptide antigens are often devoid of secondary structure or have unstable structures. When stimulating αβ T cells, they are bound to MHC molecules and are recognized as part of this molecular complex [60–62]. Nevertheless, several small peptides have now been reported to stimulate specific and TCR-dependent γδ responses [63]. Among these, the insulin peptide B:9-23 is particularly interesting for several reasons: First, the peptide is generated naturally, during the breakdown of insulin in β cells within the pancreatic islets of Langerhans. Here, the peptide could be detected in situ with a specific antibody [29]. Second, the peptide is a well-known auto-antigen, recognized by diabetogenic αβ T cells in non-obese diabetic (NOD) mice [35]. Early arising antibodies, which play a role in the development of autoimmunity in NOD mice also recognize this peptide [35], and immunization with it elicits anti-peptide autoantibodies and fatal anaphylaxis in NOD mice [36].
That the B:9-23 peptide might not necessarily be “seen” in the fashion of MHC-bound peptides recognized by MHC-restricted αβ T cells became evident in stimulation experiments, where γδ hybridomas responded to it in the absence of APCs [28], reminiscent of previous reports involving other peptides [27]. αβ T cell hybridomas and clones that are reactive with the B:9-23 peptide, require APCs [41]. In contrast, we could show that even isolated individual γδ T cell hybridoma cells expressing a B:9-23-specific γδ TCR responded to the peptide [28]. Furthermore, we determined in the current study that this and other insulin peptide-reactive γδ T cell hybridomas do not themselves express I-Ag7, the restricting element for NOD-derived B:9-23-specific αβ T cells, or any other MHCII molecules. Finally, we confirmed that under the culture conditions (using normal tissue culture plates), plate-bound B:9-23 peptide does not substantially contribute to stimulating the hybridoma response. It thus appears that the peptide, at least in one mode of recognition, can be “seen”, and stimulates responses, as a soluble antigen.
As recognizable soluble antigen, the oxidized insulin peptide might be expected to have a distinct three-dimensional structure. Two sets of data in the current study support this notion. First, we found that the cysteine in position 19 of the B:9-23 peptide is required for the γδ response, because it enables dimerization of the peptide under oxidative conditions. The γδ T cells only responded to the oxidized dimeric peptide, unlike αβ T cells, which responded to the monomeric peptide. Second, CD spectra revealed that the oxidized dimeric B:9-23 peptide adopts a distinct secondary structure, in marked contrast to the disorganized monomer. The presence of this secondary structure was closely correlated with the ability of the peptide to stimulate γδ T cells, because (i) only the dimeric wild-type peptide was stimulatory, (ii) the non-stimulatory modified dimeric peptide (B16A dimer) did not adopt a distinct secondary structure, and (iii) a more conservative substitution of the amino acid in this position (B16F) produced a modified dimeric peptide with a weaker stimulatory activity and a smaller difference in detectable secondary structure. It remains possible that the hydroxyl-group of Tyr16 in the wt peptide, which is the only difference with the modified B16F peptide, is actually recognized, but it seems more likely that differences in secondary structures of the dimeric peptides, and perhaps related physicochemical properties, are critical. More extensive molecular and structural studies will be required to decide this issue. Nevertheless, secondary structure as basis for the recognition of both native proteins and small peptides, would readily reconcile the current controversy over peptide responses by γδ T cells. Some short peptides do have a secondary structure. Those that do tend to recapitulate the structure of the corresponding segment within the native protein [64]. Interestingly, in the native insulin molecule, the B:9-23 peptide overlaps with an α-helical portion of the insulin B chain [65]. Nevertheless, the oxidized dimeric B:9-23 peptide acquired what appears to be a beta-pleated sheet structure, unlike the native protein, and γδ T cells responded to this alternatively folded peptide but not to the intact insulin protein. Conceivably, mis-folding of a given peptide sequence could be specifically recognized by some γδ T cells. Alternatively, mis-folding simply might alter peptide-protein interactions and help in the development of larger antigen complexes providing a stronger stimulatory signal. Finally, a recent study with ovalbumin-peptide specific B cells indicated that although monomers can be recognized, dimers and multimers of a minimal peptide epitope are capable of eliciting stronger and qualitatively different cellular responses [66].
Of note, our earlier study indicated that NOD islets stimulate a response of B:9-23-specific γδ cells [28], and the B:9-23 peptide itself or very similar peptides have been detected in islets of NOD mice [29]. This suggests that the islets are stimulatory because such insulin peptides are present, and in a form that can stimulate γδ T cells. Whether this includes the oxidized dimeric state of B:9-23 remains to be determined. The particular requirements for this response remain to be determined. However, insulitis and type-1 diabetes are known to be associated with oxidative stress [67, 68]. In keeping with other stress-dependent reactivity ascribed to γδ T cells [14], perhaps oxidative stress and associated antigen modification are capable of drawing γδ T cells into the autoimmune response.
4. Conclusions
In this study, we present experimental evidence that insulin peptide B:9-23-reactive γδ T cells recognize this antigen when it forms a homo-dimer due to thiol oxidation. The response to the oxidized B:9-23 antigen is γδ TCR-dependent. We also show that the B:9-23 homo-dimer adopts a distinct secondary structure, and finally, that an amino acid (Tyr16), which is not required for dimerization but essential in secondary structure formation, is also critical for γδ-stimulation, consistent with the notion that γδ T cells recognize the secondary structure of the oxidized peptide. These findings clearly differentiate the mechanisms underlying the responses of γδ and αβ T cells to the same insulin antigen and extend our earlier observation that the γδ response is APC-independent in contrast to the APC-dependent αβ response. The marked difference in stimulation requirements between the two T cell-types suggests that circumstances leading to the activation of these insulin-specific T cells might be very different as well.
5. Materials and Methods
5.1 Animals
NOD.ShiLT/J mice were originally purchased from The Jackson Laboratory (Bar Harbor, ME), and housed and bred in the Biological Resources Center at National Jewish Health, Denver, CO. Animals were used in these experiments at 7–16 wks of age. These studies were approved by Institutional Animal Care and Use Committee.
5.2 Antibodies
Anti-mouse Cd1d (1B1), anti-mouse CD3 (145-2C11), anti-mouse CD16/CD32 (2.4G2), anti-mouse Vγ4 (UC3-10A6), anti-mouse Vγ1 (2.11) and anti-mouse B:9-23 insulin peptide (AIP-46.13) mAbs were purified from the hybridoma culture supernatants in our laboratory by Protein G affinity chromatography. Hybridoma AIP-46.13 [44] was a generous gift from Dr. E. Unanue (Wash. U., St Louis, MO). Anti-mouse CD4 (GK1.5-PE), anti-mouse H-2Db (28-14-8-PE), anti-mouse αβ TCR (H-57-APC), anti-mouse γδ TCR (GL-3-APC), and anti-mouse I-A/I-E (M5/114.15.2-PE) mAbs were purchased from eBiosciences. Anti-mouse H-2Kd (SF1-1.1.1-PE), anti-rat RT1B (OX-6-PE; anti-I-Ag7), anti-mouse CD1d (1B1-PE), and anti-mouse CD8α (53-6.7-PE) were purchased from BD Pharmingen. Anti-mouse H-2Kk (AF3-12.1-FITC) was obtained from BioLegends.
5.3 Histology
Snap frozen pancreas tissue was acetone dehydrated, stained with fluorescent antibodies and analyzed microscopically as previously described in detail [69, 70].
5.4 Culture medium
All hybridomas, transfected cell lines, and freshly isolated cells were cultured in Iscove’s Complete Tissue Culture Medium (ICTM). This medium was prepared by supplementing dissolved Iscove’s Modified Dulbecco’s Medium (IMDM) (Sigma) with D-(+)-glucose, essential and non-essential amino acids, sodium pyruvate, sodium bicarbonate, gentamycin, penicillin G, streptomycin sulfate, 2-ME, and 10% FBS.
5.5 Hybridomas
The NOD-derived γδ T cell hybridomas have been previously described [28]. These hybridomas were generated with the BWZ.36 cell line [71], which is derived from the αβ TCR-deficient BWα-β-T cell fusion line [72] and carries a nuclear factor of activated T cells (NFAT)-LacZ reporter construct. Activation of these cells can be measured by the LacZ enzymatic activity assay (see below).
Production and characterization of non NOD-derived γδ T cell hybridomas expressing Vγ1Vδ6.3, Vγ6Vδ1, Vγ4 with different Vδs, and Vγ5Vδ1 TCRs have been previously published by our group [17, 27, 51, 53]. TCR αβ+ hybridoma I.29 was kindly provided by J. Kappler (National Jewish Health, Denver, CO). This hybridoma was originally produced in the laboratory of Dr. E. Unanue [44].
5.6 Expression of the SP9D11 TCR in TCR-αβ-deficient cells
Expression of the SP9D11 TCR by transduction of the αβ-deficient 5KC-73.8.20 hybridoma cells (5KC-TCR:SP9D11) has been described [28].
5.7 Peptide oxidation and purification
B:9-23 insulin peptide and modified forms of this peptide (B:16A and B:19A) were synthesized at Genemed Synthesis, Inc. (San Antonio,TX). To generate oxidized B:9-23 insulin peptide (Cu-Ox), copper (II) chloride (Sigma Aldrich) was added for a final concentration of 1 mM to solubilized (2 mg/ml) insulin peptide. The mixture then was incubated at room temperature for 40 minutes before use in the activation experiment. For air oxidation (Air-Ox), solubilized B:9-23 insulin peptide (2mg/ml) was dispensed in open Eppendorf vials placed in a still air hood, and exposed to ambient air overnight. After the exposure, evaporation loss of sample volume was replaced prior to the use of the samples in activation experiments.
Wild-type and modified monomeric and dimeric (DMSO-oxidized) insulin peptides were purchased from CPC Scientific (Sunnyvale, CA). These peptide preparations were separated by HPLC to a purity of > 95%, and analyzed by Electrospray Mass Spectrometry (EMS).
Penicillamine disulfide-linked to the B:9-23 peptide (penicillamine adduct) was generated by reacting D-PEN with the peptide in the presence of copper (II) chloride for 30 min at RT. The reaction product was purified by HPLC and molecular mass determined by ESI-TOF-MS (Applied Biosystems BIO-Spec Workstation, Foster City, CA). The final product was lyophilized and >95% pure.
5.8 Solubilization of peptides
To solubilize the peptides, one milligram of lyophilized peptide was resuspended in 480 µl of PBS after which the pH of the mixture was increased by addition of 20 µl of 200 mM NaOH (2.5 µl at a time) while vortexing the sample gently. Solubilized samples were aliquoted in small volumes and stored at −80°C.
5.9 Peptide Activation Experiments
3×104 hybridoma cells in 100 µl of ICTM were incubated overnight (17–21 hrs) in triplicate culture wells, either alone, with plate-bound anti-CD3 antibody, or with the indicated types of insulin peptides. γδ T cells were tested without addition of APCs [28]. Unless otherwise indicated, the peptide concentration used in the activation experiments was 200 µg/ml.
Hybridoma responses were measured by using either a LacZ assay or IL-2 ELISA. When αβ T cell hybridomas were included for comparison in the activation experiments, both αβ and γδ responder cells were plated at 1×105 cells/well, and paraformaldehyde-fixed M12.C3 cells, a mouse B cell line transfected with IAg7 (1×105 cells/well), were added to all cultures because the αβ T cells require antigen presenting cells (APCs).
5.10 LacZ enzymatic activity assay
After overnight incubation, cells were washed 2-times with 200 µl of PBS, and 100 µl of CPRG reagent (91 mg/L chlorophenol red-beta-D-galactopyranoside, 0.125% IGEPAL CA-630, 1.0 mM MgCl2 in 10 mM phosphate buffer) was added onto the cells. The change in the substrate color was measured by reading the absorbancies at 570 nm after 6 and 24 hours of incubation at 37°C. This assay was used only for hybridomas produced with the cell fusion partner BWZ.36 that contains NFAT-LacZ reporter construct [71].
5.11 IL-2 ELISA
The presence of IL-2 in the supernatants of the overnight activation cultures was measured by using the mouse IL-2 ELISA ready-set-go kit from eBioscience (San Diego, CA). The manufacturer’s protocol was followed to perform the assay.
5.12 ELISA for plate-bound monomeric and dimeric insulin peptides
Immulon-2-HB 96-well flat-bottom plates (Thermo Scientific) were coated with 100 µl of peptides (at 2 µg/ml) overnight at 4°C. The wells were washed 5-times with 200 µl of washing buffer (PBS+0.05% TWEEN 20; Sigma Aldrich) and then blocked with 200 µl of blocking buffer (PBS+10% FBS) for 2 hours at RT. The plates were washed and received different concentrations of AIP-46.13 mAb diluted in the blocking buffer. The plates were incubated at RT for 1 hour. After the plates were washed, 100 µl of anti-mouse IgG1-HRPO antibody (Caltag Labs.) diluted 1:2,500 in the blocking buffer was added to the wells, and then the plates were incubated at RT for 45 minutes. Afterwards the plates were washed and 100 ml of TMB single solution (3,3’, 5,5’-tetramethylbenzidine; Life Technologies) were added to the wells. After 10 minutes of incubation at RT, 50 µl of stop solution (2.0 N H2SO4 in distilled water) were added to the wells. The change in the substrate color was measured by reading the absorbancies at 450 nm.
5.13 Cytofluorometric analysis of cellular proliferation
Nylon wool non-adherent (NAD) lymphocytes from the spleens of 10-week-old NOD female mice were suspended at a concentration of 1×107 cells/ml in balanced salt solution (BSS) and labeled with 0.15 µM 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) at room temperature for 5 minutes. Labeled cells were washed twice with BSS+5% fetal bovine serum (FBS) solution and resuspended at a concentration of 5×106 cells/ml in ICTM. CFSE-labeled cells were then plated in 24-well flat-bottom tissue culture plates (5×106 cells/well) and cultured for 50 hours at 37°C in medium alone, or with the peptides as indicated. Conconavalin A (5 µg/ml; Sigma) was used as positive control. Each well also received murine recombinant IL-2 (10 U/ml) in the form of X63 BMG cell culture supernatant. At the end of the culture period, the cells were collected and washed with staining buffer (BSS+2% FBS+0.1% sodium azide), and dispensed into 96-well round-bottom tissue culture plates. Cells were pre-incubated with 2.4G2 mAb (Fc block) for 20 min at 4°C, and washed with cold staining buffer. To identify αβ and γδ T lymphocyte populations, cells were stained with H57-APC and GL-3-APC mAbs, respectively. Stained cells were fixed with 2% paraformaldehyde solution and analyzed on a FACSCalibur flow cytometer (BD Biosciences). Histograms were generated using FlowJo 9.5.2 software (Tree Star).
5.14 Measurement of circular dichroism of peptides in solution
Circular Dichroism (CD) measurements of the peptides were performed at the Biophysics core of University of Colorado Denver School of Medicine (Aurora, CO). The Jasco J-815 spectropolarimeter (Jasco, Inc. Easton, MD) is equipped with a Lauda model RMS circulating water bath (LAUDA-Brinkman, Lauda-Brinkman, Lauda-Konigshofen, Germany) for thermal uniformity for the PFD-452S peltier temperature controller that maintains the temperature control of the optical cell. CD absorbance is expressed as molar ellipticity. Variable wavelength measurements (spectrum scans) of buffer (PBS) and protein solutions (0.5 mg/ml and 0.25 mg/ml concentrations) were scanned at 4°C from 195 nm to 250nm, with data points collected every 0.2 nm, and a scan rate 50 nm per minute. The average of 6 scans was recorded for each experiment and the curves were normalized.
Gamma delta T cells specifically respond to the insulin peptide B:9-23.
The response requires dimerization of the peptide via thiol oxidation.
The oxidized dimeric peptide adopts a distinct secondary structure.
This secondary structure appears to be required for the gamma delta response.
Acknowledgments
The authors would like to thank Drs. John Kappler and Robert Hodges for advice and discussion, Drs. Emil Unanue and Laurent Gapin for the generous gift of reagents, and Shirley Sobus and Joshua Loomis for expert technical assistance in flow cytometry.
Abbreviations
- T1D
type 1 diabetes
- B:9-23
insulin2 B chain peptide (amino acids 9-23)
- TCR
T cell receptor for antigen
- APC
antigen presenting cell
- mAb
monoclonal antibody
- MHC
major histocompatibility complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by grants from the National Institutes of Health (R01DK55969 to G.E. and R21AI097962 to R.L.O.), the NIH Autoimmunity Prevention Center (2U19A1050864), the Diabetes Endocrine Research Center grant from the National Institute of Diabetes and Digestive and Kidney Diseases (P30 DK57516), the American Diabetes Association, the Juvenile Diabetes Foundation (1-2006-16 and 4-2007-1056), and by two separate NIH Autoimmunity Center Pilot Project grants (to W.K.B. and to R.L.O., respectively). Li Zhang was supported by a fellowship grant from the Juvenile Diabetes Foundation (10-2011-138), and an ADA postdoctoral fellowship (7-06-MN-17).
References
- 1.Rast JP, Anderson M, Strong SJ, Luer C, Litman RT, Litman GW. α, β, γ and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 2.Bonneville M, O'Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nature Reviews Immunology. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 3.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nature Reviews Immunology. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukasa A, Lahn M, Pflum EK, Born W, O'Brien RL. Evidence that the same gd T cells respond during infection-induced and autoimmune inflammation. J Immunol. 1997;159:5787–5794. [PubMed] [Google Scholar]
- 5.Simonian PL, Wehrmann F, Roark CL, Born WK, O'Brien RL, Fontenot AP. gammadelta T cells protect against lung fibrosis via IL-22. Journal of Experimental Medicine. 2010;207:2239–2253. doi: 10.1084/jem.20100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsburg E, Tigelaar R, Craft J, Hayday A. Age-dependent requirement for γδ T cells in the primary but not secondary protective immune response against an intestinal parasite. Journal of Experimental Medicine. 2003;198:1403–1414. doi: 10.1084/jem.20030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jameson J, Havran WL. Skin gammadelta T-cell functions in homeostasis and wound healing. Immunological Reviews. 2007;215:114–122. doi: 10.1111/j.1600-065X.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 8.Brandes M, Williman K, Moser B. Professional antigen-presenting function by human gamma delta T cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 9.Wen L, Pao W, Wong FS, Peng Q, Craft J, Zheng B, et al. Germinal center formation, immunoglobulin class switching, and autoantibody production driven by "non α/β" T cells. Journal of Experimental Medicine. 1996;183:2271–2282. doi: 10.1084/jem.183.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roark CL, Simonian PL, Fontenot AP, Born WK, O'Brien RL. Gamma/delta T cells: an important source of IL-17. Current Opinon in Immunology. 2008;20:353–357. doi: 10.1016/j.coi.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-g by gd T cells. J Immunol. 2002;168:1566–1571. doi: 10.4049/jimmunol.168.4.1566. [DOI] [PubMed] [Google Scholar]
- 12.Ferrick DA, Schrenzel MD, Mulvania T, Hsieh B, Ferlin WG, Lepper H. Differential production of interferon-γ and interleukin-4 in response to Th1- and Th2-stimulating pathogens by γδ T cells in vivo. Nature. 1995;373:255–257. doi: 10.1038/373255a0. [DOI] [PubMed] [Google Scholar]
- 13.Janeway CA, Jr, Jones B, Hayday A. Specificity and function of T cells bearing γδ receptors. Immunol Today. 1988;9:73–76. doi: 10.1016/0167-5699(88)91267-4. [DOI] [PubMed] [Google Scholar]
- 14.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Brenner MB, McLean J, Dialynas DP, Strominger JL, Smith JA, Owen FL, et al. Identification of a putative second T-cell receptor. Nature. 1986;322:145–149. [PubMed] [Google Scholar]
- 16.Chien Y-H, Iwashima M, Kaplan K, Elliott JF, Davis MM. A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature. 1987;327:677–682. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- 17.Born W, Miles C, White J, O'Brien R, Freed JH, Marrack P, et al. Peptide sequences of T-cell receptor δ and γ chains are identical to predicted X and γ proteins. Nature. 1987;330:572–574. doi: 10.1038/330572a0. [DOI] [PubMed] [Google Scholar]
- 18.Zeng X, Wei YL, Huang J, Newell EW, Yu H, Kidd BA, et al. Gammadelta T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity. 2012;37:524–534. doi: 10.1016/j.immuni.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes SM, Love PE. Distinct structure and signalling potential of the γδ TCR complex. Immunity. 2002;16:1–20. doi: 10.1016/s1074-7613(02)00320-5. [DOI] [PubMed] [Google Scholar]
- 20.Rock EP, Sibbald PR, Davis MM, Chien Y-H. CDR3 length in antigen-specific immune receptors. Journal of Experimental Medicine. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis MM, Bjorkman PJ. T cell antigen receptor genes and T cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 22.Born WK, Aydintug M, O'Brien RL. Diversity of gammadelta T-cell antigens. Cell Mol Immunol. 2013;10:13–20. doi: 10.1038/cmi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien Y-H, Jores R, Crowley MP. Recognition by γ/δ T cells. Annu Rev Immunol. 1996;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 24.Born WK, Vollmer M, Reardon C, Matsuura E, Voelker DR, Giclas PC, et al. Hybridomas expressing gammadelta T-cell receptors respond to cardiolipin and beta2-glycoprotein 1 (apolipoprotein H) Scand J Immunol. 2003;58:374–381. doi: 10.1046/j.1365-3083.2003.01315.x. [DOI] [PubMed] [Google Scholar]
- 25.Born W, Hall L, Dallas A, Boymel J, Shinnick T, Young D, et al. Recognition of a peptide antigen by heat shock reactive γδ T lymphocytes. Science. 1990;249:67–69. doi: 10.1126/science.1695022. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y-X, Cranfill R, Vollmer M, van der Zee R, O'Brien RL, Born W. In vivo response of murine γδ T cells to a heat shock protein-derived peptide. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:322–326. doi: 10.1073/pnas.90.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien RL, Fu Y-X, Cranfill R, Dallas A, Reardon C, Lang J, et al. Heat shock protein Hsp-60 reactive γδ cells: A large, diversified T lymphocyte subset with highly focused specificity. Proc Natl Acad Sci USA. 1992;89:4348–4352. doi: 10.1073/pnas.89.10.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Jin N, Nakayama M, O'Brien RL, Eisenbarth GS, Born WK. Gamma delta T cell receptors confer autonomous responsiveness to the insulin-peptide B:9-23. J Autoimmun. 2010;34:478–484. doi: 10.1016/j.jaut.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohan JF, Levisetti MG, Calderon B, Herzog JW, Petzold SJ, Unanue ER. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nature Immunol. 2010;4:350–354. doi: 10.1038/ni.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bluestone JA, Herold K, Eisenbarth GS. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markle JG, Mortin-Toth S, Wong ASL, Geng L, Hayday A, Danska JS. Gammadelta T cells are essential effectors of type 1 diabetes in the nonobese diabetic mouse model. J Immunol. 2013;190:5392–5401. doi: 10.4049/jimmunol.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison LC, Dempsey-Collier M, Kramer DR, Takahashi K. Aerosol insulin induces regulatory CD8 gammadelta T cells that prevent murine insulin-dependent diabetes. Journal of Experimental Medicine. 1996;184:2167–2174. doi: 10.1084/jem.184.6.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han G, Wang R, Chen G, Wang J, Xu R, Wang L, et al. Interleukin-17-producing gammadelta+ T cells protect NOD mice from type 1 diabetes through a mechanism involving transforming growth factor-beta. Immunology. 2010;129:197–206. doi: 10.1111/j.1365-2567.2009.03166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Nakayama M, Eisenbarth GS. Insulin as an autoantigen in NOD/human diabetes. Current Opinion in Immunology. 2008;20:111–118. doi: 10.1016/j.coi.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu E, Moriyama H, Abiru N, Miao D, Yu Y, Taylor RM, et al. Anti-peptide autoantibodies and fatal anaphylaxis in NOD mice in response to insulin self-peptides B:9-23 and B:13-23. J Clin Invest. 2002;110:1021–1027. doi: 10.1172/JCI15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simone E, Daniel D, Schloot N, Gottlieb P, Babu S, Kawasaki E, et al. T cell receptor restriction of diabetogenic autoimune NOD T cells. Proc Natl Acad Sci (USA) 1997;94:2518–2521. doi: 10.1073/pnas.94.6.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. Journal of Experimental Medicine. 2011;208:2375–2383. doi: 10.1084/jem.20111502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford F, Stadinski B, Jin N, Michels A, Nakayama M, Pratt P, et al. Specificity and detection of insulin-reactive CD4+ T cells in type-1 diabetes in the nonobese diabetic (NOD) mouse. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16729–16734. doi: 10.1073/pnas.1113954108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alleva DG, Gaur A, Jin L, Wegmann D, Gottlieb P, Pahuja A, et al. Immunological charactrization and therapeutic activity of an altered-peptide ligand, NBI-6024, based on the immunodominant type 1 diabetes autoantigen insulin B-chain (9-23) peptide. Diabetes. 2002;51:2126–2134. doi: 10.2337/diabetes.51.7.2126. [DOI] [PubMed] [Google Scholar]
- 41.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to I-Ag7 in an unexpected, weakly binding register. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Go Y-M, Jones DP. The redox proteome. The Journal of Biological Chemistry. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pecci L, Montefoschi G, Musci G, Cavallini D. Novel findings on the copper catalysed oxidation of cysteine. Amino Acids. 1997;13:355–367. [Google Scholar]
- 44.Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol. 2007;178:6051–6057. doi: 10.4049/jimmunol.178.10.6051. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Zhou L, Chen L, Dastidar SG, Verma C, Li J, et al. Effect of structural parameters of peptides on dimer formation and highly oxidized side products in the oxidation of thiols of linear analogues of human beta-defensin 3 by DMSO. J Pept Sci. 2009;15:95–106. doi: 10.1002/psc.1100. [DOI] [PubMed] [Google Scholar]
- 46.Cheng L, Cui Y, Shao H, Han G, Zhu L, Huang Y, et al. Mouse gammadelta T cells are capable of expressing MHC class II molecules, and of functioning as antigen-presenting cells. J Neuroimmunol. 2008;203:3–11. doi: 10.1016/j.jneuroim.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMaster WR, Williams AF. Identification of Ia glycoproteins in rat thymus and purification from rat spleen. European Journal of Immunology. 1979;9:426–433. doi: 10.1002/eji.1830090603. [DOI] [PubMed] [Google Scholar]
- 48.Rinderknecht CH, Lu N, Crespo O, Tuong P, Hou T, Wang N, et al. I-Ag7 is subject to post-translational chaperoning by CLIP. Int J Immunol. 2010;22:705–716. doi: 10.1093/intimm/dxq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hattori M, Buse JB, Jackson RA, Glimcher L, Dorf ME, Minami M, et al. The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science. 1986;231:733–735. doi: 10.1126/science.3003909. [DOI] [PubMed] [Google Scholar]
- 50.Bhattacharya A, Dorf ME, Springer TA. A shared alloantigenic determinant on Ia antigens encoded by the I-A and I-E subregions: evidence for I region gene duplication. J Immunol. 1981;127:2488–2495. [PubMed] [Google Scholar]
- 51.Roark CE, Vollmer MK, Cranfill RL, Carding SR, Born WK, O'Brien RL. Liver γδ T cells: TCR junctions reveal differences in HSP-60 reactive cells in liver and spleen. J Immunol. 1993;150:4867–4875. [PubMed] [Google Scholar]
- 52.O'Brien RL, Happ MP, Dallas A, Palmer E, Kubo R, Born WK. Stimulation of a major subset of lymphocytes expressing T cell receptor γδ by an antigen derived from Mycobacterium tuberculosis. Cell. 1989;57:667–674. doi: 10.1016/0092-8674(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 53.Happ MP, Kubo RT, Palmer E, Born WK, O'Brien RL. Limited receptor repertoire in a mycobacteria-reactive subset of γδ T lymphocytes. Nature. 1989;342:696–698. doi: 10.1038/342696a0. [DOI] [PubMed] [Google Scholar]
- 54.Kalataradi H, Eyster CL, Fry A, Vollmer MK, Fu Y-X, Born WK, et al. Allelic differences in TCR γ-chains alter γδ T cell antigen reactivity. J Immunol. 1994;153:1455–1465. [PubMed] [Google Scholar]
- 55.Adams EJ, Chien Y-H, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 56.Hampl J, Schild H, Litzenberger C, Baron M, Crowley MP, Chien Y-h. The specificity of a weak γδ TCR interaction can be modulated by the glycosylation of the ligand. J Immunol. 1999;163:288–294. [PubMed] [Google Scholar]
- 57.Sciammas R, Bluestone JA. HSV-1 glycoprotein I-reactive TCRγδ cells directly recognize the peptide backbone in a conformationally dependent manner. J Immunol. 1998;161:5187–5192. [PubMed] [Google Scholar]
- 58.Bruder J, Siewert K, Obermeier B, Malotka J, Scheinert P, Kellermann J, et al. Target specificity of an autoreactive pathogenic human gammadelta-T cell receptor in myositis. The Journal of Biological Chemistry. 2012;287:20986–20995. doi: 10.1074/jbc.M112.356709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vidovic D, Roglic M, McKune K, Guerder S, MacKay C, Dembic Z. Qa-1 restricted recognition of foreign antigen by a γδ T-cell hybridoma. Nature. 1989;340:646–650. doi: 10.1038/340646a0. [DOI] [PubMed] [Google Scholar]
- 60.Wilson IA, Fremont DH. Structural analysis of MHC class I molecules with bound peptide antigens. Sem Immunol. 1993;5:75–80. doi: 10.1006/smim.1993.1011. [DOI] [PubMed] [Google Scholar]
- 61.Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, et al. Peptides presented to the immune system by the murine Class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 62.Grey HM, Sette A, Buus S. How T cells see antigen. Sci Am. 1989;261:56–64. doi: 10.1038/scientificamerican1189-56. [DOI] [PubMed] [Google Scholar]
- 63.Born WK, Zhang L, Nakayama M, Jin N, Chain JL, Huang Y, et al. Peptide antigens for gamma/delta T cells. Cell Mol Life Sci. 2011;68:2335–23343. doi: 10.1007/s00018-011-0697-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho BK, Dill KA. Folding very short peptides using molecular dynamics. PLOS Computational Biology. 2006;2:228–237. doi: 10.1371/journal.pcbi.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaarsholm NC, Ludvigsen S. The high resolution solution structure of the insulin monomer determined by NMR. Receptor. 1995;5:1–8. [PubMed] [Google Scholar]
- 66.Avalos AM, Bilate AM, Witte MD, Tai AK, He J, Frushicheva MP, et al. Monovalent engagement of the BCR activates ovalbumin-specific transnuclear B cells. Journal of Experimental Medicine. 2014;211:365–379. doi: 10.1084/jem.20131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Styskal J, van Remmen H, Richardson A, Salmon AB. Oxidative stress and diabetes: What can we learn about insulin resistance from antioxidant mutant mouse models? Free Radical Biology and Medicine. 2011;52:46–58. doi: 10.1016/j.freeradbiomed.2011.10.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiskopf D, Schwanninger A, Weinberger B, Almanzar G, Parson W, Buus S, et al. Oxidative stress can alter the antigenicity of immunodominant peptides. J Leukocyte Biology. 2010;87:165–172. doi: 10.1189/jlb.0209065. [DOI] [PubMed] [Google Scholar]
- 69.Wands JM, Roark CL, Aydintug MK, Jin N, Hahn Y-S, Cook L, et al. Distribution and leukocyte contacts of gd T cells in the lung. J Leukocyte Biology. 2005;78:1086–1096. doi: 10.1189/jlb.0505244. [DOI] [PubMed] [Google Scholar]
- 70.Cook L, Miyahara N, Jin N, Wands JM, Taube C, Roark CL, et al. Evidence that CD8+ dendritic cells enable the development of gammadelta T cells that modulate airway hyperresponsiveness. J Immunol. 2008;181:309–319. doi: 10.4049/jimmunol.181.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Intl Immunol. 1993;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 72.White J, Blackman M, Bill J, Kappler J, Marrack P, Gold D, et al. Two better cell lines for making hybridomas expressing specific T cell receptors. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]