Abstract
The impact of environmental stressors on the magnitude of specific chemokine gene expression was examined in mouse bone marrow-derived macrophages stimulated through various Toll-Like Receptors (TLRs). Levels of TLR-stimulated CXCL1 and CXCL2 but not CXCL10 or CCL5 mRNAs were selectively enhanced (>10 fold) in stressed macrophages. The amplification was also manifest for other pro-inflammatory cytokines including TNFα, IL-1α, and IL-6. Responses through TLR3 and TLR4 exhibited the greatest sensitivity, reflecting a requirement for TRIF, the adaptor protein selectively associated with these TLRs. IRF3, a transcription factor that is downstream of TLR4/TRIF signaling, was not required for sensitivity to stress-induced chemokine amplification. CHOP and XBP-1 have been reported to enhance inflammatory cytokine responses but are not required for amplification of TLR3/4-induced CXCL1 expression. Rather, RIPK1, a kinase also linked with TLR3/4/TRIF signaling, is required and involves a stress-dependent increase in its abundance and ubiquitination. While NFκB activation is necessary for TLR-induced chemokine gene transcription, this factor does not appear to be the primary mechanistic target of environmental stress. The application of stress also enhanced chemokine expression in macrophages infiltrating the peritoneal cavity but was not observed in the resident peritoneal cells nor in the liver. These findings identify novel mechanisms for modulating the magnitude and duration of selective TLR-induced chemokine and cytokine gene expression and further establish the importance of cell stress pathways in coordinating the outcomes of cellular and tissue injury.
Introduction
Inflammation following tissue injury involves the careful orchestration of inflammatory cytokine production by infiltrating leukocytes that traffic to such sites. These inflammatory cell populations are exposed not only to pathogens and endogenous inflammatory signals but also to conditions that promote cellular stress responses and indeed, the two processes are often coincident in both time and space (1-4). Indeed the nature and magnitude of inflammatory response can be significantly influenced by response to environmental stressors (5-7).
The production of chemokines and cytokines is triggered through signaling events downstream of pattern recognition receptors (e.g., TLRs) that recognize and respond to molecular markers of injury or infectious challenge (8-10). Signaling downstream of TLRs engages multiple pathways coupling directly and indirectly to altered transcriptional and post-transcriptional mechanisms that govern the magnitude and duration of chemokine and cytokine production (11-13). Likewise environmental conditions that disrupt cellular homeostasis (e.g., nutrient deprivation, redox disturbance, viral and bacterial infection, genotoxic damage), also initiate intracellular signaling pathways culminating in altered patterns of gene expression that are collectively known as cellular stress responses (14-17). The products of stress-induced genes serve to correct deficiencies, prevent further cellular damage, restore normal structure and function, or, if necessary, engage programmed cell death pathways. While the pathways that define cell stress and inflammatory responses are largely distinct from one another, there is now evidence demonstrating substantial crosstalk (5-7).
Recent reports establish that intracellular stress responses often occur coincident with inflammation following infection or sterile injury and the interface between these processes has important physiologic and pathophysiologic consequences in vivo (18-22). Cell stress has been reported to markedly amplify TLR-induced inflammatory cytokine production in myeloid cells, exhibiting substantial target gene specificity (23-27). For example TLR-induced production of IFNβ, IL-6, and IL-23p19 can all be markedly amplified in cells that are undergoing endoplasmic reticulum stress, also known as the unfolded protein responses (UPR). Moreover, individual changes have been linked to specific components of the UPR, particularly c/EBP homologous protein (CHOP) and X box binding protein 1 (XBP1). Nevertheless, the mechanisms mediating cooperative interaction between cell stress and TLR-mediated inflammatory cytokine expression remain poorly defined.
We have observed that engagement of cell stress at the time of TLR stimulation can produce a marked (≥5-50 fold) enhancement of inflammatory chemokine gene expression in mouse bone marrow derived macrophages (BMDM). The magnitude and duration of the effects vary substantially with the nature of the cell stress inducer but neither CHOP nor spliced XBP1 appear to be the major stress sensor mechanisms. Responses also vary with the specific TLR utilized and those initiated through TLR3 or TLR4 exhibit the most sensitivity. This TLR specificity reflects the important contribution of the TRIF adaptor and is further substantiated by a requirement for RIPK1, a stress kinase linked with TLR3/4/TRIF signaling (28, 29). Moreover, the altered pattern of chemokine gene expression in response to cell stress is mediated by elevated and prolonged transcription from specific subsets of chemokine and cytokine genes. Finally, cell stress appears to impact a subset of myeloid cell populations as infiltrating inflammatory macrophages but not resident macrophages exhibit sensitivity. The findings identify novel mechanistic features of the interaction between stress responses and inflammation signaling pathways that shows dramatic impact on the magnitude and duration of selective TLR-induced chemokine and cytokine gene expression and further establishes the importance of environmental stressors in coordinating the outcomes of cellular and tissue injury.
Materials and Methods
Reagents
DMEM, RPMI 1640, lysine-leucine-methionine deficient DMEM, Dulbecco's PBS, and antibiotics were obtained from Central Cell Services of the Lerner Research Institute. FBS was purchased from ATLAS Biologicals. Dialyzed FBS and random primers were obtained from Invitrogen. The Amaxa Nucleofector Kit V was obtained from Lonza. Tri-Reagent was purchased from Molecular Research Center and Nylon transfer membrane was purchased from GE Healthcare Life Sciences. Tunicamycin (Tm), thapsigargin (Tg), formamide, MOPS, salmon sperm DNA and diethyl pyrocarbonate were obtained from Sigma-Aldrich. TLR agonists (Pam3CSK4, LPS, and polyIC) were obtained from Invivogen. Recombinant mouse TNFα (Cat. No. GF027) and antibody specific for TNFα (Cat# 05-168) were purchased from EMD Millipore. Necrostatin 1 (NEC1) was obtained from TOCRIS. The smac mimetic inhibitor of cIAP1/2 (AT 406) was obtained from Selleck Chemicals (Cat. No. S2754). RNase inhibitor was from Roche and MMLV reverse transcriptase was from Promega. SYBR Green PCR Master Mix (2×) was acquired from Applied Biosystems. Brewer's thioglycollate broth was obtained from BD Biosciences. PerkinElmer Life Sciences was the source of [α-32P]-dCTP. CXCL1 ELISA kit was purchased from R&D Systems. Nuclear extract kit and TransAM™ NFκB p65 transcription factor assay kit were purchased from Active Motif. Mouse anti-receptor interacting protein kinase 1 (RIPK1) antibody (Cat. No.610458) was purchased from BD Transduction Laboratories; Rabbit anti-p65 monoclonal antibody (Cat. No. 8242) was purchased from Cell Signaling; anti-ubiquitin antibody (Cat. No. SC 8017) was obtained from Santa Cruz; monoclonal antibody specific for GAPDH was purchased from Millipore (Cat. No. MAB374); antibody specific for CD95 was obtained from BD Biosciences (Cat. No.554255). The siRNAs (5′-CCACUAGUCUGACUGAUGA-3′ for RIPK1 (30) and 5′-GGGAUUCAUGAAUGGCCCUUA-3′ for XBP1 (31)) were purchased from Thermo Scientific Dharmacon RNAi Technologies.
Mice, Cell culture, and siRNA transfection
C57BL/6, CHOP-/-, TRIF-/-, and GCN2-/- mice on C57Bl/6 background were obtained from The Jackson Laboratory. TTP-/- deficient mice were kindly provided by Dr. Perry Blackshear (NIH). IRF3-/- mice (C57BL/6 background) were obtained from RIKEN Bio Resource Center, Japan. Mouse bone marrow derived macrophages (BMDM) were prepared from bone marrow cells that were collected from femurs by flushing with cold Hank's buffered salt solution. The cell suspensions were then passed through a 100 μm nylon cell strainer (BD Falcon), were collected by centrifugation and resuspended in DMEM containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 50 ng/ml M-CSF (R&D). 5 × 106 cells were seeded in 100 mm dishes and cultured at 37°C with 5% CO2 for 7 days. Non-adherent cells were removed by washing and the BMDM were treated as indicated in the text. Thioglycollate elicited peritoneal macrophages were prepared as described previously (32). RAW 264.7 mouse macrophage cells were maintained in DMEM containing penicillin, streptomycin, and 10% fetal bovine serum in humidified 5% CO2. Transient transfections of siRNA oligonucleotides in BMDM and RAW264.7 cells were done using Amaxa Nucleofector Kit V. 5 × 106 cells were transfected with 15 μg of siRNA oligonucleotide duplex and cultures were rested for 48 to 96 hrs following transfection prior to further experimental treatments. For amino acid restriction experiments, cells were washed 3× with PBS, cultured with lysine-, leucine-, and methionine-deficient DMEM supplemented with 10% dialyzed FBS, penicillin and streptomycin (AAR medium) for 1 hr, washed again and fresh AAR medium was added for 16 hrs prior to further treatments. Promoter reporter plasmids were constructed by linking either 5 copies of the mouse CXCL10 κB1 sequence element (33, 34) or 290 nucleotides from the 5′ flanking region of the mouse CXCL1 gene (35, 36) into the multiple cloning site of the pGL2-basic plasmid (Promega). Plasmids were transfected in RAW264.7 cells using Superfect (Qiagen) and rested for 24 hrs prior to further experimental treatment. Luciferase activity was measured using the Luciferase Assay System (Promega) according to the manufacturer's instruction.
Preparation of liver non-parenchymal cells
Non-parenchymal cells were isolated from the mouse liver and prepared as described (37). Briefly, the liver was perfused with collagenase IV (Sigma-Aldrich, St. Louis, MO) solution (1.0 mg/ml) via the portal vein, minced and digested with collagenase for 30 minutes at 37°C with agitation. The digests were filtered through 200 μm nylon mesh and the non-parenchymal cells enriched by Percoll density gradient centrifugation, collected and resuspended in culture medium in polypropylene tubes for treatment as described in the text. Myeloid cell content was assessed by flow cytometry following immunostaining with anti-CD11b.
mRNA Determination
Total RNA was prepared using Tri Reagent and analyzed either by northern blot hybridization as described previously (38) or by reverse transcription and real-time PCR using 0.5-1 μg total RNA as previously described (39). Autoradiographs were quantified by image analysis using the ImageJ software. CXCL1 mRNA levels were normalized to levels of GAPDH mRNA measured in the same RNA sample. Specific primers used for PCR as described below were obtained from Eurofins MWG Operon.:
CXCL1 (40):
Forward: 5′-CACAGGGGCGCCTATCGCCAA-3′
Reverse: 5′-CAAGGCAAGCCTCGCGACCAT-3′
CXCL2 (41):
Forward: 5′-CGCTGTCAATGCCTGAAG-3′
Reverse: 5′-GGCGTCACACTCAAGCTCT-3′
CCL5 (42):
Forward: 5′-GAATACATCAACTATTTGGAGAT-3′
Reverse: 5′-TAGAGCAAGCAATGACAG-3′
CXCL10 (43):
Forward: 5′-AGGACGGTCCGCTGCAA-3′
Reverse: 5′-CATTCTCACTGGCCCGTCAT-3′
TNFα (44):
Forward:5′-CATCTTCTCAAAATTCGAGTGACAA-3′
Reverse: 5′TGGGAGTAGACAAGGTACAACCC-3′
IL6:
Forward: 5′-GGACCAAGACCATCCAAT-3′
Reverse: 5′ACCACAGTGAGGAATGTC-3′
IL1α (45):
Forward: 5′-GTGTTGCTGAAGGAGTTGCCAGAA-3′
Reverse: 5′-GTGCACCCGACTTTGTTCTTTGGT-3′
IL1β (45):
Forward: 5′-AAGAGCTTCAGGCAGGCAGTATCA-3′
Reverse: 5′TAATGGGAACGTCACACACCAGCA-3′
TLR2:
Forward: 5′-AGATGTCGTTCAAGGAGGTGC-3′
Reverse: 5′- CTGTTATGGCCACCAAGATCC-3′
TLR4:
Forward: 5′-TTCTTCTCCTGCCTGACACC-3′
Reverse: 5′-ATCCAGCCACTGAAGTTCTG-3′
spliced XBP1 (46):
Forward: 5′-GAGTCCGCAGCAGGTG-3′
Reverse: 5- GTGTCAGAGTC-CATGGGA-3′
CHOP:
Forward: 5′-TCCCTGCCTTTCACCTTG-3′
Reverse: 5′-GCCCTGGCTCCTCTGTCA-3′
BIP:
Forward: 5′-CATGGTTCTCACTAAAATGAAAGG-3′
Reverse: 5′-GCTGGTACAGTAACAACTG-3′
GAPDH (47):
Forward: 5′-ATCACCATCTTCCAGGAGCGAGAT-3′
Reverse: 5′-GTTGGTGGTGCAGGAGGCATTGCT-3′;
CXCL1 primary transcript:
Forward: 5′-CAATGAGCTGCGCTGTCAGTG-3′
Reverse: 5′-GGAAGTGGCAGAAGCTAACCG-3′.
Immunoprecipitation and western blot analysis
Cell lysates were prepared by incubating cells on ice for 30 mins in RIPA buffer (50 mM Tris-HCL pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.05% SDS, 1mM EDTA, 150 mM NaCl, and protease inhibitor cocktail (Sigma)). Lysates were pre-cleared by incubation with Protein G beads for 2 hr and subsequently incubated with antibody-Protein G beads overnight at 4°C. Samples were washed in RIPA buffer, eluted by boiling in SDS sample buffer, separated by SDS-PAGE and transferred onto PVDF membrane (Millipore). Membranes were blotted with the antibodies and samples were visualized with Western Lightning Plus ECL system (PerkinElmer). For determination of RIPK1 ubiquitination, treated cell cultures were lysed on ice in 50 mM HEPES pH 7.5, 1% NP-40, 20 mM β-glycerophosphate, 250 mM NaCl, 2 mM DTT, 1 mM NEM and protease inhibitor cocktail (Sigma) for 30 min. Cell debris was removed by centrifugation and the supernatant was supplemented with 0.1 volume of 10% SDS to give a final concentration of 1% SDS. Then lysates were boiled for 5 min, placed on ice, lysis buffer added to achieve a final SDS concentration of 0.1%, and incubated with beads and antibodies overnight at 4°C prior to analysis as described above.
NFκB DNA binding ELISA
BMDM cells were fractionated using Nuclear Extract Kit according to the manufacturer's instruction. The nuclear fractions were used to analyze the DNA binding activity of p65 using the TransAM™ NFκB p65 kit.
Results
Cellular stress amplifies expression of a subset of TLR-induced chemokine and cytokine genes
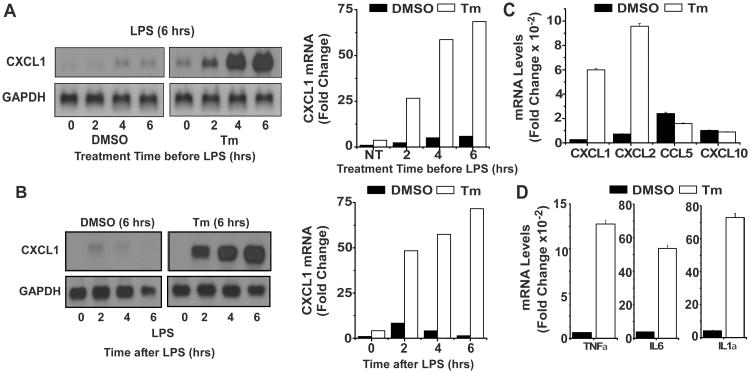
Cellular stress has been shown to promote elevated expression of a limited selection of TLR-induced inflammatory cytokines in macrophages including IL-6, the p19 subunit of IL-23, and IFNβ (23-26). In addition, though more modest effects on other TLR target genes have been reported, early chemokine expression has not been well characterized (24, 27). To examine this in more detail BMDM were exposed to tunicamycin (Tm), a protein glycosylation inhibitor known to promote the UPR, for the indicated times followed by stimulation with LPS for 6 hours. The level of mRNA encoding the chemokine CXCL1 was markedly elevated (>20 fold) in cultures exposed to Tm for 4 hrs prior to LPS (Fig 1A) while only modest change was observed in cells undergoing simultaneous exposure to Tm and LPS (Tm treatment time = 0). Moreover, CXCL1 mRNA expression was also prolonged in cultures exposed to Tm for 6 hours prior to LPS treatment as compared to unstressed macrophages where the response was transient and levels of CXCL1 mRNA returned to baseline by 6 hrs after LPS (Fig 1B). A similar pattern of enhanced expression is also observed with CXCL2 mRNA while levels of mRNAs encoding CXCL10 and CCL5, though induced by LPS, were not enhanced (Fig 1C). These findings suggest a selective effect of stress on the expression of chemokines with the signature gly-leu-arg cys-x-cys (ELR-CXC) motif known to target neutrophils. The effects of stress engagement on TLR response were not limited to chemokines, however, as comparable alterations in levels of TNFα, IL-1α, and IL-6 were also observed (Fig 1D).
Figure 1. Cellular stress amplifies expression of a subset of TLR-induced chemokine and cytokine genes.
A. BMDM treated with DMSO or Tm (1 μg/ml) for the indicated times were subsequently stimulated with LPS (100 ng/ml) for 6 hrs. Levels of CXCL1 and GAPDH mRNA were determined by northern hybridization and autoradiographs were quantified using ImageJ software. CXCL1 levels were normalized to GAPDH levels in the same samples and are presented as fold change relative to samples treated with DMSO and LPS for 6 hrs. Results are representative of more than 4 separate experiments. B. BMDM were treated with DMSO or Tm for 6 hrs followed by LPS for the indicated times prior to determination and quantification of CXCL1 mRNA levels as in A. Data are presented as the fold change relative to samples treated with DMSO but without LPS. C. BMDM were treated for 6 hrs with DMSO or Tm prior to addition of LPS for 6 hrs. Levels of CXCL1, CXCL2, CCL5 and CXCL10 mRNA were measured by real time PCR as described in Materials and Methods and normalized to levels of GAPDH mRNA. Values presented are the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. D. RNA from the experiment described in C was employed to determine levels of TNFα, IL6, and IL1α mRNAs.
Characteristics of stress amplified chemokine expression
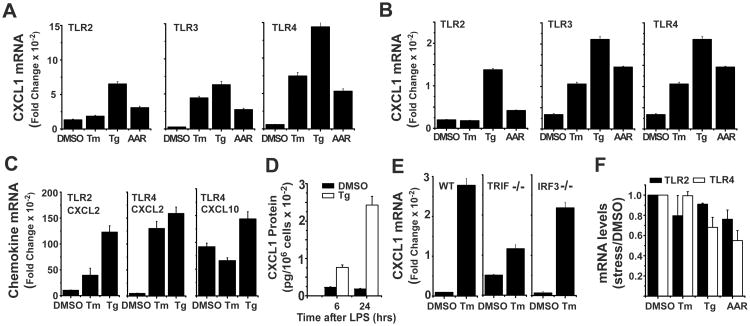
We compared several routes of inducing cell stress including tunicamycin (Tm), an inhibitor of protein glycosyltransferase (48), thapsigargin (Tg), an inhibitor of the ER Ca++ ATPase (49), or essential amino acid restriction (AAR) for their ability to amplify CXCL1 expression in BMDM treated with three different TLR specific ligands (Pam3CSK4 (P3C) for TLR2, polyIC for TLR3, LPS for TLR4). Cultures were exposed to Tm or Tg for 6 hrs or deprived of essential amino acids (leu, met, lys) for 16 hrs followed by stimulation for 6 hrs with the individual TLR specific ligands (Fig 2A). A similar experimental design was used to examine chemokine expression in the RAW264.7 cell line (Fig 2B). All three routes of inducing cell stress resulted in elevated expression of LPS-stimulated CXCL1 mRNA, though the magnitude of response varied with the stress inducer and TLR ligand employed. In both cell types treatment with Tm elevated CXCL1 mRNA when stimulated through TLR3 or TLR4 but not TLR2 while cells exposed to Tg or AAR exhibited amplification for responses to all three TLRs. Though the magnitude of response to TLR4 was generally greater than for other TLRs, particularly in BMDM, this does not reflect greater total chemokine production but rather greater sensitivity to the stress mechanism. CXCL2 expression was modulated comparably to CXCL1 and revealed a similar distinction between TLR2 and TLR3/TLR4 in Tm-stressed cells (Fig 2C). Consistent with the findings in Fig.1, levels of CXCL10 (Fig 2C) and CCL5 (not shown) mRNAs exhibited only modest differences between stressed and non-stressed macrophages. CXCL10 is only poorly induced by TLR2 as it depends on the IRF3 component of the TRIF pathway that is specifically engaged by TLR3 and TLR4 (50, 51). The levels of secreted CXCL1 protein are also selectively increased in LPS-stimulated macrophages pre-exposed to Tg (Fig 2D). The greater sensitivity of TLR3 and TLR4 for stress-enhancement of chemokine expression and the target gene selectivity (CXCL1/2 versus CXCL10/CCL5) suggested that stress might operate with some selectivity for the TRIF signaling pathways and hence we evaluated the role of the TRIF adaptor protein. LPS-induced CXCL1 mRNA expression was markedly enhanced by Tm treatment in wild type macrophages while BMDM from TRIF-/- mice exhibited little change (Fig 2E). In contrast, stress amplified TLR4-induced CXCL1 mRNA expression comparably in macrophages from wild type and IRF3-/- mice (Fig 2E). Finally, the enhanced sensitivity of TLR stimulation to different cell stress conditions does not reflect stress-mediated alteration in TLR expression (Fig 2F).
Figure 2. Characteristics of stress amplified chemokine expression.
A. BMDM were cultured with Tm (1 μg/mL), Tg (50 nM), or under conditions of AAR (-leu, -met, -lys) for 6 hrs (Tm, Tg), or 16 hrs (AAR) prior to stimulation with ligands for TLR2 (Pam3Cys), TLR3 (polyIC), or TLR4 (LPS) for an additional 6 hrs. Levels of CXCL1 and GAPDH mRNA were determined by real time PCR as in Fig 1C. Values presented are the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. B. RAW264.7 cells were treated and analyzed as in A. C. RNA from the experiments described in A was employed to determine levels of CXCL2 mRNA in cultures stimulated through TLR2 or TLR4 and CXCL10 mRNA in cultures stimulated through TLR4. D. BMDM were treated with DMSO or Tg for 6 hrs prior to stimulation with LPS for 6 or 24 hrs. Supernatants were collected and used to determine protein levels of CXCL1 by ELISA. Results are presented as the mean of duplicate determinations +/- ½ range. E. BMDM from wild type, TRIF-/-, or IRF3-/- mice were treated with DMSO or Tm for 6 hrs followed by LPS for 6 hrs. Levels of CXCL1 and GAPDH mRNAs were determined and quantified as in A. F. RNA from the experiments described in A was employed to determine levels of TLR2 and TLR4 mRNAs.
UPR pathways are not required for stress-enhanced CXCL1 expression
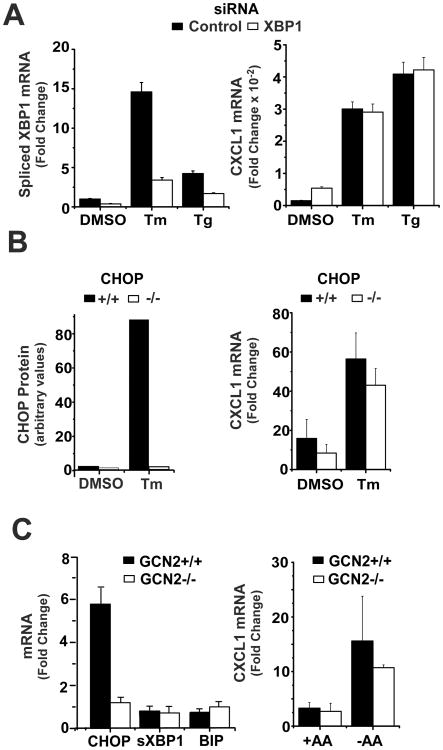
Prior studies have reported important roles for several components of the UPR in the selective enhancement of cytokine expression in inflammatory macrophages (23, 24, 26, 27). For example, the enhanced expression of both IL-6 and the p19 component of IL23 have been shown by several laboratories to be dependent on CHOP, a transcription factor activated via the UPR (24-26, 52). Spliced XBP1, the primary target of the UPR sensor inositol requiring kinase and endonuclease 1 (IRE1), has been demonstrated to regulate the magnitude of IFNβ gene transcription as well as to participate in the enhanced expression of other cytokines including IL-6 and TNFα (26, 27, 53). We used several experimental strategies to test the possible role for these factors in the Tm-mediated amplification of LPS-stimulated CXCL1 expression. Transfection of BMDM with siRNA targeting XBP1 reduced the levels of spliced XBP1 mRNA by more than 70% as compared to control siRNA-treated cultures but did not alter the effects of stress on LPS-induced CXCL1 mRNA levels (Fig 3A). To evaluate the possible participation of CHOP we measured CXCL1 mRNA levels in unstressed or Tm-treated, LPS-stimulated BMDM from wild type and CHOP-/- mice. Exposure to Tm enhanced chemokine expression comparably in both cell populations (Fig 3B). In a third approach we compared the response of BMDM from WT or GCN2-/- mice to AAR (GCN2 is an eIF2α kinase that is activated during AAR and initiates cell stress by inducing ATF4 and CHOP (54)). While CHOP mRNA is strongly induced during AAR in wild type macrophages, this is, as expected, abrogated using BMDM from GCN2-/- mice (Fig 3C). Also noteworthy is the finding that AAR did not activate other UPR pathways (e.g., XBP1 splicing or BIP expression) (Fig 3C). Importantly, the effects of AAR on elevation of CXCL1 mRNA expression were similar in WT and GCN2-/- macrophages (Fig 3C). Hence, deficiency of CHOP or XBP1 does not compromise stress-amplified chemokine expression. Moreover other components of the UPR are not induced during AAR-mediated stress and thus are not required, at least in this context. These findings are consistent with a recent report showing that cell stress can amplify inflammasome activity independently of the UPR (55).
Figure 3. UPR pathways are not required for stress-enhanced CXCL1 expression.
A. BMDM were transfected with control or XBP1 specific siRNAs and exposed to DMSO, Tm, or Tg for 6 hrs followed by LPS for 6 hrs. Levels of CXCL1 and spliced XBP1 mRNAs were analyzed by real-time PCR and normalized to GAPDH mRNA levels. Values presented are the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. B. CHOP+/+ and -/- BMDM were treated with DMSO or Tm for 6 hrs followed by LPS for 6 hrs. CXCL1 mRNA levels were determined by northern hybridization and quantified as described in the legend to Fig 1A and are presented as fold change relative to samples treated with DMSO alone for 12 hrs. Values represent the mean of triplicate determinations +/- 1 S.D. CHOP protein levels were measured by western blot. Results are representative of 4 separate experiments. C. GCN2+/+ and -/- BMDM were subjected to AAR or not for 16 hrs prior to stimulation with LPS for 6 hrs. CXCL1 mRNA levels were determined by northern hybridization and quantified as in Fig 1A. Results are presented as the mean of duplicate determinations +/- ½ range. CHOP, XBP1s, and BIP mRNA levels were determined by real time PCR as in A.
Tm-induced stress enhances TLR4 signaling via RIPK1 kinase
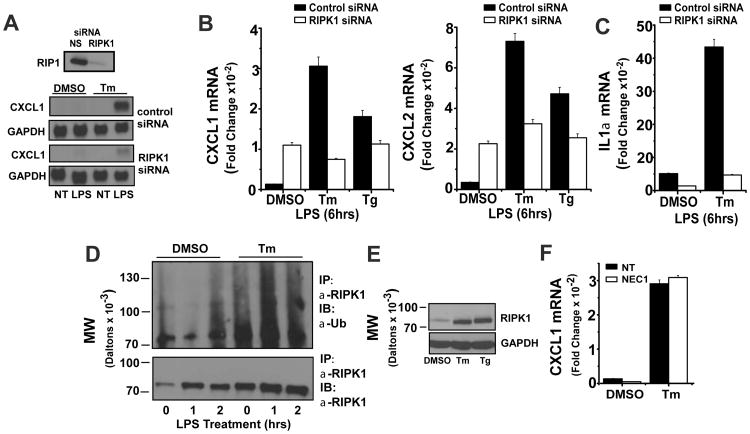
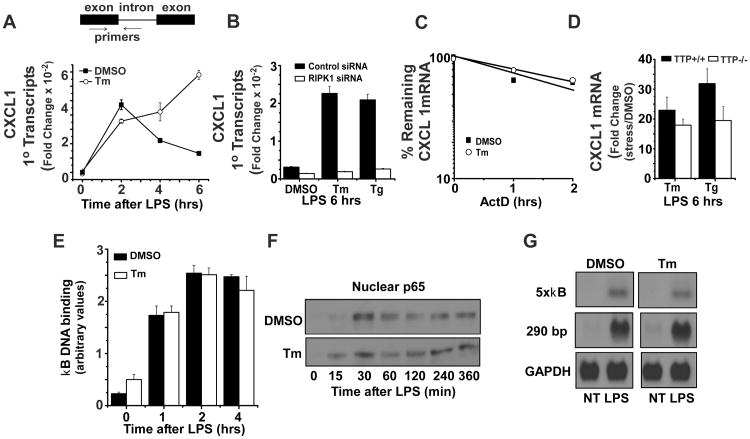
Our finding that Tm-driven amplification of chemokine expression is independent of CHOP and XBP1 prompted us to consider other stress sensitive mediators. Because of the requirement for TRIF but not IRF3 in stress mediated amplification of LPS-induced chemokine expression, we examined RIPK1, a kinase that participates in TNFR, TLR3 and TLR4 signaling to NFκB (28, 29, 56, 57). Importantly, RIPK1 is also required for NFκB activation during the DNA damage response and is an essential component of the necroptosis pathway (58-60). siRNA-mediated depletion of RIPK1 almost fully abrogated the amplification of LPS-induced CXCL1 levels in Tm-treated BMDM as measured by northern hybridization (Fig 4A). Interestingly, the depletion of RIPK1 did not compromise the basal TLR4-induced CXCL1 and CXCL2 mRNA levels and indeed resulted in a modest elevation that cannot be appreciably increased by either Tm or Tg (Fig 4B). It is noteworthy however, that the effects of RIPK1 depletion vary with the specific target gene as levels of IL1α in response to LPS were markedly reduced in both resting and stressed BMDM (Fig 4C). It has been shown that the kinase activity of RIPK1 is not essential for NFκB activation (57, 61, 62). Rather, NFκB activation depends upon ubiquitin modification of RIPK1 on lys377 (56, 57, 62, 63). Tm treatment of BMDM for 6 hrs markedly enhanced the abundance of cellular RIPK1 protein measured by western blot. Importantly, stress treatment resulted in ubiquitination of RIPK1 as well (Fig 4D). Treatment with Tg also induced increased levels of RIPK1 protein and ubiquitination (Fig 4E and not shown). Stress-mediated enhancement of TLR4-stimulated CXCL1 expression is not compromised in BMDM treated with the specific RIPK1 kinase inhibitor NEC1 suggesting that the kinase activity is not essential for the role of RIPK1 in mediating the effects of cell stress on CXCL1 expression (Fig 4F).
Figure 4. Tm-induced stress enhances TLR4 signaling via RIPK1 kinase.
A. BMDM were transfected with control or RIPK1 siRNAs and 72 hrs later aliquots of cells were lysed to determine residual levels of RIPK1 protein by western blot. Cultures were treated with DMSO or Tm for 6 hrs followed by LPS for 6 hrs and levels of CXCL1 and GAPDH mRNAs were determined by northern hybridization. Results are representative of more than 3 separate experiments. B. BMDM were transfected with control or RIPK1 siRNAs and treated with DMSO, Tg, or Tm for 6 hrs followed by LPS for 6 hrs. Levels of CXCL1 and CXCL2 mRNAs were normalized to GAPDH as determined by real-time PCR. Values are presented as the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. C. BMDM were transfected with control or RIPK1 siRNAs and treated with DMSO or Tm for 6 hrs followed by LPS for 6 hrs. The level of IL1α mRNA was normalized to GAPDH as determined by real-time PCR. Values are presented as the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. D. BMDM were treated with DMSO or Tm for 6 hrs followed by LPS for the indicated times. Cell lysates were used to determine levels of RIPK1 protein and RIPK1 ubiquitination as described in Materials and Methods. Results are representative of 3 separate experiments. E. BMDM were treated with DMSO, Tm, or Tg for 6 hrs. RIPK1 and GAPDH protein levels were determined by western blot and are representative of 3 separate experiments. F. BMDM were pre-treated with NEC1 (10 μM) or DMSO for 30 min, then treated with or without Tm for 6 hrs followed by LPS for 6 hrs. CXCL1 mRNA levels were determined by real-time PCR as in B.
A recent report demonstrated that apoptosis induced via FAS/CD95 interaction can promote RIPK1-dependent chemokine production as part of a program to help clear apoptotic cells by soliciting phagocytes (64). In our experiments, we did not find evidence for significant increases in apoptosis during treatment of stressed macrophages with LPS (not shown), suggesting that apoptosis was not the basis for enhanced chemokine expression. Cullen et al also identified a requirement for cellular inhibitors of apoptosis (cIAPs) in the FAS-mediated chemokine response (64). To determine if cIAPs were required for stress-enhanced chemokine expression, we treated BMDM with the cIAP inhibitor AT 406, a smac mimetic that results in the rapid degradation of cIAP1/2 (65) along with Tg for 6 hrs prior to stimulation with LPS. While AT 406 resulted in significant reduction in levels of cIAP, there was no effect on the pattern of CXCL1 mRNA expression in either resting or stressed macrophages (supplemental Fig 1A). Furthermore, macrophages treated with a monoclonal anti-CD95 antibody that can trigger apoptosis did not induce CXCL1 mRNA expression in either resting or stressed BMDM (supplemental Fig 1B). Finally we considered the possibility that LPS-induced TNFα, the expression of which is also elevated in stressed cells, might be responsible for the enhanced chemokine production. The addition of TNFα (20 ng/mL) to BMDM alone or along with LPS and/or stress inducers did not alter the pattern of CXCL1 mRNA expression nor did the inclusion of a neutralizing antibody against TNFα (supplemental Fig 1C). Hence we concluded that the mechanism described by Cullen et al (64) was not responsible for the elevated chemokine expression observed in LPS-stimulated stressed macrophages.
Stress enhances TLR4-induced CXCL1 gene transcription
The magnitude and duration of chemokine gene expression is known to be controlled both by transcription and mRNA half-life (66, 67). To assess CXCL1 transcription in TLR4-stimulated macrophages, a set of primers amplifying a fragment that contains both intronic and exonic sequence was employed as a measure of primary transcript abundance (see schematic in Fig 5A). Using total RNA prepared from stressed and unstressed macrophages treated for various times with LPS, the levels of primary transcript predicted the temporal pattern of mature mRNA accumulation (compare Fig 5A and Fig 1B) indicating that gene transcription is a target for the action of cell stress. Furthermore, depletion of RIPK1 protein also compromised the stress-mediated increase in CXCL1 primary transcript abundance (Fig 5B). While mRNA half-life is well documented to control CXCL1 expression (67), the half-life of CXCL1 mRNA in BMDM does not appear to be regulated by cell stress. CXCL1 mRNA decay was not significantly changed by cell stress (Fig 5C) and the abundance of CXCL1 mRNA was comparably amplified by Tm in BMDM prepared from wild type mice or mice deficient in Tristetraprolin, a protein known to be the predominant regulator of CXCL1 half-life (47) (Fig 5D). The transcription factor NFκB is an important determinant of LPS-induced CXCL1 gene transcription and is a major downstream mediator in RIPK1-dependent signaling (36, 57, 62). The activation and function of this factor does not, however, appear to be the major target for the action of cell stress in enhancing CXCL1 expression. This is based upon several observations. First, the magnitude and kinetics of nuclear localization and DNA binding activity of p65 are similar in untreated or Tm-pretreated BMDM following TLR4 stimulation (Fig 5E,F). Secondly, we evaluated RAW264.7 cells transiently transfected with reporter plasmids in which luciferase expression is driven by either 5 tandemly arranged κB sites or a 290 bp fragment from the CXCL1 promoter (36) that contains 2 κB sites and observed that κB-driven transcripts exhibit sensitivity to TLR4 but not cell stress (Fig. 5G). Hence the enhanced transcription apparently depends upon sequences not contained within the 290 nucleotide fragment from the CXCL1 promoter used in this experiment. Taken together, these data suggest that cell stress amplifies both the magnitude and duration of CXCL1 mRNA expression via modulation of gene transcription, though the effect seems not to be achieved by enhancing the activity of NFκB.
Figure 5. Stress enhances TLR4-induced CXCL1 gene transcription.
A. BMDM were treated with DMSO or Tm for 6 hrs followed by LPS for the indicated times and total RNA was used to determine levels of CXCL1 primary transcripts by real-time PCR as described in Materials and Methods. Values presented are the fold induction relative to cultures treated with DMSO alone for 12 hrs and are the mean of duplicate determinations +/- ½ range. The schematic above the graph shows the position of primers amplifying CXCL1 transcripts containing both intronic and exonic sequence. B. BMDM transfected with control or RIPK1 siRNAs were treated with DMSO, Tm or Tg for 6 hrs followed by 6 hrs of LPS treatment. CXCL1 primary transcripts were determined as in A. C. BMDM were treated with DMSO or Tm for 6 hrs and LPS for 5 hrs prior to addition of actinomycin D. RNA was prepared at the indicated time and used to determine the remaining CXCL1 mRNA by northern hybridization. Autoradiographs were quantified as in Fig 1A and results are representative of 2 separate experiments. D. BMDM from TTP+/+ or TTP-/- mice were subjected to DMSO or Tm treatment for 6 hrs followed by LPS treatment for 6 hrs. CXCL1 mRNA was measured by real-time PCR using primers to detect mature mRNA and quantified as in A. E. BMDM were treated with DMSO or Tm for 6 hrs followed by LPS for the indicated times. Nuclear extracts were prepared and used to analyze NFκB p65 DNA binding activity as described in Materials and Methods. Results are presented as the mean of duplicate determinations +/- ½ range. F. BMDM were treated with DMSO or Tm for 6 hrs, then stimulated with LPS for indicated times. Nuclear extracts were prepared and used to determine levels of NFκB p65 by western blot. Results are representative of 3 separate experiments. G. RAW 264.7 cells transiently transfected with the indicated luciferase reporter plasmids were treated with DMSO or Tm for 6 hrs followed by LPS for 6 hrs. Total RNA was prepared and used to determine luciferase mRNA by northern hybridization as in Fig 1. Results are representative of 3 separate experiments.
Sensitivity to cell stress-mediated amplification of chemokine expression varies with myeloid cell origin
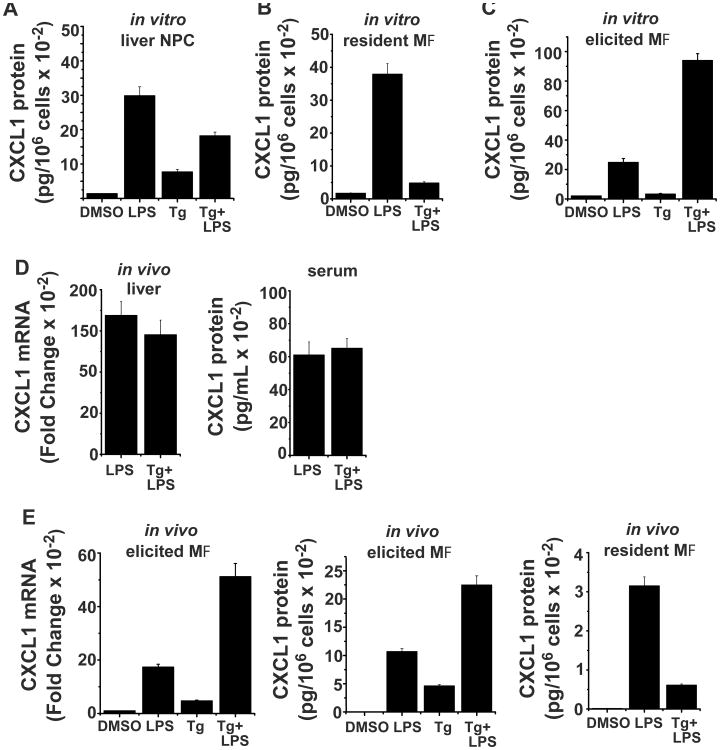
Pro-inflammatory cytokine expression is commonly observed at sites of injury or infection within populations of infiltrating myeloid cells. Hence we reasoned that the ability of cell stress to enhance chemokine expression might be limited to certain myeloid cell populations, particularly those infiltrating damaged tissues. To test this hypothesis we initially determined the sensitivity to stress in resident myeloid cell populations prepared from the liver and the peritoneal cavity (Fig 6A,B). Though chemokine expression can be induced in both cell populations by treatment with LPS, pretreatment with Tg does not alter the magnitude of this response. In contrast, inflammatory macrophages obtained from the peritoneal cavity following injection of thioglycollate broth show substantial amplification of CXCL1 mRNA and protein levels when treated with Tg followed by LPS as compared to cells treated with LPS alone (Fig 6C). Similar outcomes were obtained in vivo when mice were injected ip with the stress inducer Tg followed 6 hr later by LPS. Stress did not alter LPS-induced CXCL1 mRNA expression in the liver and did not appreciably alter levels of circulating chemokine in the serum (Fig 6D). The sequential exposure of macrophages to Tg followed by LPS within the peritoneal cavity resulted in a dramatic increase in levels of CXCL1 mRNA in the elicited cell population and in secreted protein in the peritoneal wash as compared to those receiving LPS alone (Fig 6E). Similar treatment of unmanipulated mice did not show stress-mediated enhancement of chemokine secretion from resident macrophages, consistent with the results of in vitro treatments (Fig 6E). These findings support the hypothesis that stress-dependent modulation of myeloid cell cytokine production occurs selectively within the population of macrophages infiltrating tissues in response to inflammatory conditions. In contrast, resident myeloid cell populations do not exhibit this sensitivity.
Figure 6. Sensitivity to cell stress-mediated amplification of chemokine expression is cell type restricted.
A. Non-parenchymal cells (NPC) were prepared from the liver as described in Materials and Methods and were treated with DMSO or Tg for 6 hrs followed by LPS for 6 hrs. The supernatants were used to measure CXCL1 protein levels by ELISA and normalized to cell number. 35-40% of the cells were CD11b+ by flow cytometry. Values presented are the mean of duplicate determinations +/- ½ range. B. Resident peritoneal cells were obtained by peritoneal lavage and CXCL1 protein production was determined with and without Tg pretreatment as described in A. C. Elicited peritoneal macrophages were obtained 48 hrs following i.p. injection of thioglycollate broth and used to determine CXCL1 protein as in A and B. D. Mice were injected i.p. with DMSO or Tg (1 μg/g body weight) and after 6 hrs were given LPS (25 μg/mouse). 6 hrs after the last injection mice were sacrificed and total RNA was prepared from liver for determination of CXCL1 mRNA levels by real-time PCR. Serum was used to determine CXCL1 protein levels by ELISA. Values are the mean of duplicate determinations +/- ½ range. E. Untreated mice (resident macrophages) or mice injected with thioglycollate broth for 48 hrs (elicited macrophages) were injected i.p. with DMSO or Tg (1 μg/g body weight) for 2 hrs followed LPS (25 μg/mouse) for an additional 4 hrs. The peritoneal cavity was lavaged with 2 mL HBSS and the wash fluid recovered was used for determination of CXCL1 protein by ELISA. Elicited cells were used to extract RNA and CXCL1 mRNA levels were determined by real-time PCR. Values are the mean of duplicate determinations +/- ½ range.
Discussion
Several recent studies have demonstrated that expression of specific inflammatory cytokine genes can be markedly amplified in TLR-stimulated macrophages undergoing cellular stress responses but the spectrum of cytokine loci that are sensitive and the mechanistic appreciation of how cellular stress inducers impact on TLR signaling remains poorly defined (23, 24, 26, 27). In particular, the effects of stress on expression of chemokines that recruit neutrophils, amongst the earliest events following injury or infectious challenge, have not been explored. In the present study we examined how different environmental stress conditions impact on TLR-induced chemokine expression. The findings support the following conclusions. 1) Macrophages engaged in cell stress exhibit a dramatic increase in the transcription of a subset of chemokine and cytokine genes that include CXCL1 and CXCL2 but not CXCL10 or CCL5. 2) Though multiple stress inducers can amplify chemokine and cytokine gene expression in macrophages following stimulation through TLRs, responses from the TLR3/TLR4/TRIF pathway exhibit the greatest sensitivity. 3) While XBP1 and CHOP, components of the UPR, are implicated in stress-mediated increases in TLR-induced IFNβ, IL-23p19, and IL-6 expression, the effects of stress on CXCL1 expression are independent of both factors. 4) RIPK1, a stress sensitive signaling protein in the TLR3 and TLR4 pathways, is necessary for the amplified magnitude and duration of chemokine gene transcription in stressed macrophages. 5) Surprisingly, while the effects involve amplification of transcription, neither IRF3 nor NFκB appear to be the primary determinants of stress-mediated amplification of chemokine production. 6) Finally, the sensitivity of myeloid cells for stress enhancement of chemokine expression appear to be restricted to inflammatory macrophages while resident myeloid cells from the liver or peritoneal cavity do not exhibit sensitivity. It is worth noting that the three mechanistically distinct treatments used to induce cellular stress responses all had similar effects on chemokine and cytokine expression induced in response to TLR4. We did, however, observe TLR selectivity in responses obtained using different stress mechanisms (e.g., TLR3/4 but not TLR2 are sensitive to Tm-induced stress). Hence there appears to be substantial mechanistic complexity and diversity in the interface between cellular stress and inflammatory TLR signaling pathways. This mechanistic diversity makes it particularly challenging to identify common molecular events and this represents an area for further exploration.
Because multiple laboratories have identified a relationship between responses to environmental stress and the magnitude and duration of inflammatory cytokine expression, particularly in myeloid cell populations, it is important to consider the present findings in the context of these earlier studies (23-27, 52, 68). Stress responses that enhance expression of IFNβ, IL-6 and IL23p19 are well documented (23-26) but the mechanism(s) involved in amplification of chemokine transcription observed in the present study is readily distinguished from these based upon differential dependency on IRF3 and CHOP; while IFNβ requires IRF3 and both IL-6 and IL-23p19 require CHOP, CXCL1 expression is comparably amplified in macrophages deficient in either gene. Effects on a broader spectrum of inflammatory cytokine genes have been linked to ER stress responses involving the IRE1/XBP1 pathway (24, 27). Our findings however, demonstrate that the modulation of chemokine gene expression is not dependent upon XBP1 based upon siRNA-mediated depletion. Furthermore, under conditions of AAR in GCN2-deficient macrophages, the XBP1 pathway is not activated and yet the amplification of gene expression remains intact (see Fig 3C). While the UPR-mediated activation of ATF6 has been reported to contribute to acute phase gene expression (69), we have not explicitly examined this pathway in the effect of cell stress on chemokine expression. Thus a mechanistic connection with UPR stress remains a possibility. Nevertheless, three mechanistically distinct routes of inducing cell stress in mouse macrophages result in markedly elevated and prolonged transcription of CXCL1 that does not appear to be linked with the specific UPR pathways reported for other stress-sensitive inflammatory cytokine genes.
The absence of a role for the UPR prompted a search for additional routes/mechanisms through which stress and inflammatory signaling might be linked. RIPK1 is known to contribute to inflammatory cytokine expression downstream of TNFR, TLR3, and TLR4, and also as a sensor of cellular stress response to DNA damage (57, 62, 70, 71). Moreover, the requirement of TRIF for stress-mediated amplification of both TLR3- and TLR4-driven chemokine responses strongly suggested that RIPK1 might be involved. The known roles for RIPK1 in cytokine gene expression require its polyubiquitination and associated capacity to promote activation of NFκB but are independent of its kinase activity (56, 57, 61, 62). The depletion of RIPK1 markedly compromises the stress-amplified transcriptional response to stimulation through TLR4 but does not reduce the basal response and a similar finding was made in TRIF-/- macrophages (Figs 2E and 4B). These observations suggest that TRIF/RIPK1 are not requisite for basal CXCL1 transcription in LPS-stimulated BMDM. Moreover, stress engagement leads to an increase in cellular RIPK1 protein as well as ubiquitination status. The TRIF signaling pathway is believed to be responsible for prolonged NFκB-mediated transcription in LPS-stimulated macrophages (72) and the effects of cell stress are most evident in this context (See Fig 1B). It is noteworthy that RIPK1 depletion does not compromise the effects of stress on responses to TLR2 or TLR7 (data not shown).
Though mRNA half-life is well recognized as an important regulatory mechanism governing the expression of many cytokine and chemokine genes (67, 73), it does not appear to be the mechanistic route through which environmental stresses amplify chemokine expression levels. Rather changes in target gene transcription, measured as an increase in the abundance of non-spliced primary transcripts, appear to be responsible. Furthermore, though prior reports have shown that both IRF3 and NFκB exhibit sensitivity to stress pathways (74-77), the stress enhanced transcription of CXCL1 mRNA is observed in IRF3-deficient macrophages and does not appear to be a consequence of altered NFκB activation. Multiple criteria support the latter conclusion. 1) While cell stress has been linked to the activation of NFκB through several mechanistic pathways (indeed, there is both modest activation of NFκB and associated cytokine gene expression in macrophages treated with stress inducers) there is no significant amplification of κB binding activity or nuclear localization of p65 in stressed macrophages stimulated with LPS. 2) Importantly, cell stress does not enhance LPS-stimulated κB-dependent reporter gene expression in transiently transfected RAW264.7 cells. These findings support the conclusion that stress operates via alternate mechanisms.
Myeloid cells are among the sentinels that detect and provide first response to signals resulting from disruption of the local tissue microenvironment. The link between cell stress and inflammatory cytokine expression provides a mechanism for adjusting the magnitude and duration of pro-inflammatory gene expression and operates in vivo as evidenced by the dramatic change in chemokine expression when stress signaling is engaged at a site of inflammatory response (see Fig 6). Sensitivity to such environmental cues does not, however, appear to be a property shared by all myeloid cell populations. Indeed, resident myeloid cells within the liver or peritoneal cavity do not appear to show altered LPS-induced CXCL1 expression following stress either in vitro or in vivo. This selectivity may reflect the phenotypic heterogeneity within myeloid cell populations (78-81). Importantly, recent findings suggest the existence of at least two populations of circulating monocytes, of which one is a precursor to inflammatory infiltrating macrophages. Moreover, there is substantial evidence supporting the idea that inflammatory cytokine expression occurs selectively within infiltrating myeloid cell populations (82, 83). Our findings suggest that the sensitivity for stress-mediated amplification of cytokine and chemokine expression may be restricted to such inflammatory cell populations. Interestingly, a recent report identifies RIPK1 as a critical determinant of systemic inflammatory disease associated with the tyr208asn mutation of the SHP1 protein tyrosine phosphatase (PTPN6spin) (84). Deficiency of RIPK1 in mice with this mutation protects against neutrophilia, tissue inflammation, and elevated systemic cytokine production. Hence the pattern of chemokine and cytokine expression described in the present report may reflect a mechanism for controlling the intensity of inflammation that operates on infiltrating inflammatory macrophages in specific pathophysiologic contexts and depends upon, at least in some circumstances, the stress sensitive kinase RIPK1.
Supplementary Material
Acknowledgments
This work was supported by USPHS grants CA039621 and CA062220.
Abbreviations used
- UPR
Unfolded Protein Response
- TRIF
TIR domain containing adaptor inducing IFNβ
- P3C
Pam3CSK4 (TLR2 ligand)
- Tm
Tunicamycin
- Tg
Thapsigargin
- RIPK1
Receptor interacting protein kinase 1
- NEC1
Necrostatin 1
- BMDM
Bone marrow derived macrophages
- AAR
Amino acid restriction
- CHOP
c/EBP homologous protein
- XBP1
X box binding protein 1
- IRE1
Inositol requiring kinase and endonuclease 1
References
- 1.Thorp E, Iwawaki T, Miura M, Tabas I. A reporter for tracking the UPR in vivo reveals patterns of temporal and cellular stress during atherosclerotic progression. J Lipid Res. 2011;52:1033–1038. doi: 10.1194/jlr.D012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitamura M. Endoplasmic reticulum stress and unfolded protein response in renal pathophysiology: Janus faces. Am J Physiol Renal Physiol. 2008;295:F323–334. doi: 10.1152/ajprenal.00050.2008. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Glimcher LH. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr Opin Immunol. 2011;23:35–40. doi: 10.1016/j.coi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Bauer S, Muller T, Hamm S. Pattern recognition by Toll-like receptors. Adv Exp Med Biol. 2009;653:15–34. doi: 10.1007/978-1-4419-0901-5_2. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 12.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 13.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 15.Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2011;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rath E, Haller D. Inflammation and cellular stress: a mechanistic link between immune-mediated and metabolically driven pathologies. Eur J Nutr. 2011;50:219–233. doi: 10.1007/s00394-011-0197-0. [DOI] [PubMed] [Google Scholar]
- 21.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol Sci. 33:53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Martinon F. The endoplasmic reticulum: a sensor of cellular stress that modulates immune responses. Microbes Infect. 14:1293–1300. doi: 10.1016/j.micinf.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Schwabe RF, DeVries-Seimon T, Yao PM, Gerbod-Giannone MC, Tall AR, Davis RJ, Flavell R, Brenner DA, Tabas I. Free cholesterol-loaded macrophages are an abundant source of tumor necrosis factor-alpha and interleukin-6: model of NF-kappaB- and map kinase-dependent inflammation in advanced atherosclerosis. J Biol Chem. 2005;280:21763–21772. doi: 10.1074/jbc.M501759200. [DOI] [PubMed] [Google Scholar]
- 24.Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, Gaston JS. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci U S A. 2010;107:17698–17703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60:2633–2643. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-{kappa}B activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 29.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 30.He S, Liang Y, Shao F, Wang X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci U S A. 108:20054–20059. doi: 10.1073/pnas.1116302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandal P, Hamilton T. Signaling in lipopolysaccharide-induced stabilization of formyl peptide receptor 1 mRNA in mouse peritoneal macrophages. J Immunol. 2007;178:2542–2548. doi: 10.4049/jimmunol.178.4.2542. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori Y, Hamilton TA. The interferon stimulated response element and a kB site mediate synergistic induction of murine IP-10 gene transcription by IFN gamma and TNFa. J Immunol. 1995;154:5235–5244. [PubMed] [Google Scholar]
- 34.Ohmori Y, Tebo J, Nedospasov S, Hamilton TA. kB binding activity in a murine macrophage-like cell line. Sequence-specific differences in kB binding and transcriptional activation functions. J Biol Chem. 1994;269:17684–17690. [PubMed] [Google Scholar]
- 35.Dai Y, Datta S, Novotny M, Hamilton TA. TGFbeta inhibits LPS-induced chemokine mRNA stabilization. Blood. 2003;102:1178–1185. doi: 10.1182/blood-2002-12-3771. [DOI] [PubMed] [Google Scholar]
- 36.Ohmori Y, Fukumoto S, Hamilton TA. Two structurally distinct kappaB sequence motifs cooperatively control LPS-induced KC gene transcription in mouse macrophages. J Immunol. 1995;155:3593–3600. [PubMed] [Google Scholar]
- 37.Yu MC, Chen CH, Liang X, Wang L, Gandhi CR, Fung JJ, Lu L, Qian S. Inhibition of T-cell responses by hepatic stellate cells via B7-H1-mediated T-cell apoptosis in mice. Hepatology. 2004;40:1312–1321. doi: 10.1002/hep.20488. [DOI] [PubMed] [Google Scholar]
- 38.Novotny M, Datta S, Biswas R, Hamilton T. Functionally independent AU-rich sequence motifs regulate KC (CXCL1) mRNA. J Biol Chem. 2005;280:30166–30174. doi: 10.1074/jbc.M502280200. [DOI] [PubMed] [Google Scholar]
- 39.Zhao C, Datta S, Mandal P, Xu S, Hamilton T. Stress-sensitive regulation of IFRD1 mRNA decay is mediated by an upstream open reading frame. J Biol Chem. 2010;285:8552–8562. doi: 10.1074/jbc.M109.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arima Y, Harada M, Kamimura D, Park JH, Kawano F, Yull FE, Kawamoto T, Iwakura Y, Betz UA, Marquez G, Blackwell TS, Ohira Y, Hirano T, Murakami M. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell. 148:447–457. doi: 10.1016/j.cell.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Pagano MB, Bartoli MA, Ennis TL, Mao D, Simmons PM, Thompson RW, Pham CT. Critical role of dipeptidyl peptidase I in neutrophil recruitment during the development of experimental abdominal aortic aneurysms. Proc Natl Acad Sci U S A. 2007;104:2855–2860. doi: 10.1073/pnas.0606091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu YP, Zeng L, Tian A, Bomkamp A, Rivera D, Gutman D, Barber GN, Olson JK, Smith JA. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 189:4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 474:96–99. doi: 10.1038/nature09982. [DOI] [PubMed] [Google Scholar]
- 45.Zak DE, Schmitz F, Gold ES, Diercks AH, Peschon JJ, Valvo JS, Niemisto A, Podolsky I, Fallen SG, Suen R, Stolyar T, Johnson CD, Kennedy KA, Hamilton MK, Siggs OM, Beutler B, Aderem A. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc Natl Acad Sci U S A. 108:11536–11541. doi: 10.1073/pnas.1107577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang K, Kaufman RJ. Identification and characterization of endoplasmic reticulum stress-induced apoptosis in vivo. Methods Enzymol. 2008;442:395–419. doi: 10.1016/S0076-6879(08)01420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta S, Biswas R, Novotny M, Pavicic P, Herjan T, Mandal P, Hamilton TA. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 48.Lennarz WJ. Lipid linked sugars in glycoprotein synthesis. Science. 1975;188:986–991. doi: 10.1126/science.167438. [DOI] [PubMed] [Google Scholar]
- 49.Lytton J, Westlin M, Hanley MR. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J Biol Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 50.Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ, Vogel SN. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 51.Ohmori Y, Hamilton TA. Requirement for STAT1 in LPS-induced gene expression in macrophages. J Leukoc Biol. 2001;69:598–604. [PubMed] [Google Scholar]
- 52.Chen L, Jarujaron S, Wu X, Sun L, Zha W, Liang G, Wang X, Gurley EC, Studer EJ, Hylemon PB, Pandak WM, Jr, Zhang L, Wang G, Li X, Dent P, Zhou H. HIV protease inhibitor lopinavir-induced TNF-alpha and IL-6 expression is coupled to the unfolded protein response and ERK signaling pathways in macrophages. Biochem Pharmacol. 2009;78:70–77. doi: 10.1016/j.bcp.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng L, Liu YP, Sha H, Chen H, Qi L, Smith JA. XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages. J Immunol. 2010;185:2324–2330. doi: 10.4049/jimmunol.0903052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, Cavener DR. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menu P, Mayor A, Zhou R, Tardivel A, Ichijo H, Mori K, Tschopp J. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis. 2012;3:e261. doi: 10.1038/cddis.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 57.Christofferson DE, Li Y, Yuan J. Control of Life-or-Death Decisions by RIP1 Kinase. Annu Rev Physiol. 2013 doi: 10.1146/annurev-physiol-021113-170259. in press. [DOI] [PubMed] [Google Scholar]
- 58.Vandenabeele P, Declercq W, Van Herreweghe F, Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal. 2012;3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- 59.Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- 60.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 62.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14:727–736. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 63.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 64.Cullen SP, Henry CM, Kearney CJ, Logue SE, Feoktistova M, Tynan GA, Lavelle EC, Leverkus M, Martin SJ. Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic cells. Mol Cell. 2013;49:1034–1048. doi: 10.1016/j.molcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Cai Q, Sun H, Peng Y, Lu J, Nikolovska-Coleska Z, McEachern D, Liu L, Qiu S, Yang CY, Miller R, Yi H, Zhang T, Sun D, Kang S, Guo M, Leopold L, Yang D, Wang S. A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J Med Chem. 54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamilton TA. Molecular Basis of Macrophage Activation: From Gene Expression to Phenotypic Diversity. In: Lewis C, Burke B, editors. The Macrophage. 2. Oxford University Press; Oxford: 2002. pp. 73–102. [Google Scholar]
- 67.Hamilton T, Li X, Novotny M, Pavicic PG, Jr, Datta S, Zhao C, Hartupee J, Sun D. Cell type- and stimulus-specific mechanisms for post-transcriptional control of neutrophil chemokine gene expression. J Leukoc Biol. 2012;91:377–383. doi: 10.1189/jlb.0811404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahadevan NR, Rodvold J, Sepulveda H, Rossi S, Drew AF, Zanetti M. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc Natl Acad Sci U S A. 2011;108:6561–6566. doi: 10.1073/pnas.1008942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 70.Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 71.Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, Look AT, Wang J, Bouchier-Hayes L, Sidi S. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol Cell. 47:681–693. doi: 10.1016/j.molcel.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 73.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 74.Liu YP, Zeng L, Tian A, Bomkamp A, Rivera D, Gutman D, Barber GN, Olson JK, Smith JA. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 2012;189:4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kitamura M. Control of NF-kappaB and inflammation by the unfolded protein response. Int Rev Immunol. 2011;30:4–15. doi: 10.3109/08830185.2010.522281. [DOI] [PubMed] [Google Scholar]
- 76.Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, Ron D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol. 2004;24:10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, Wek RC. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 79.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 80.Sunderkotter C, Nikolic T, Dillon MJ, Van Rooijen N, Stehling M, Drevets DA, Leenen PJ. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 81.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 82.Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J Immunol. 2010;184:6843–6854. doi: 10.4049/jimmunol.0903987. [DOI] [PubMed] [Google Scholar]
- 83.Tsuji H, Mukaida N, Harada A, Kaneko S, Matsushita E, Nakanuma Y, Tsutsui H, Okamura H, Nakanishi K, Tagawa Y, Iwakura Y, Kobayashi K, Matsushima K. Alleviation of lipopolysaccharide-induced acute liver injury in Propionibacterium acnes-primed IFN-gamma-deficient mice by a concomitant reduction of TNF-alpha, IL-12, and IL-18 production. J Immunol. 1999;162:1049–1055. [PubMed] [Google Scholar]
- 84.Lukens JR, Vogel P, Johnson GR, Kelliher MA, Iwakura Y, Lamkanfi M, Kanneganti TD. RIP1-driven autoinflammation targets IL-1alpha independently of inflammasomes and RIP3. Nature. 2013;498:224–227. doi: 10.1038/nature12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.