Abstract
Carcinogenesis is a mechanistically complex and variable process with a plethora of underlying genetic causes. Cancer development consists of a multitude of steps that occur progressively starting with initial driver mutation(s), to tumorigenesis, and ultimately metastasis. During these transitions, cancer cells accumulate a series of genetic alterations that confer upon the cells an unwarranted survival and proliferative advantage. During the course of development, however, cancer cells also encounter a physiologically ubiquitous cellular program that aims to eliminate damaged or abnormal cells: Apoptosis. Thus, it is essential that cancer cells acquire instruments to circumvent programmed cell death. Here we discuss emerging evidence indicating how cancer cells adopt various strategies to override apoptosis including amplifying the anti-apoptotic machinery, downregulating the pro-apoptotic program, or both.
Keywords: BH3, Caspase, MOMP, Phosphorylation, Ubiquitination
Regulation of apoptosis in normal and cancer cells
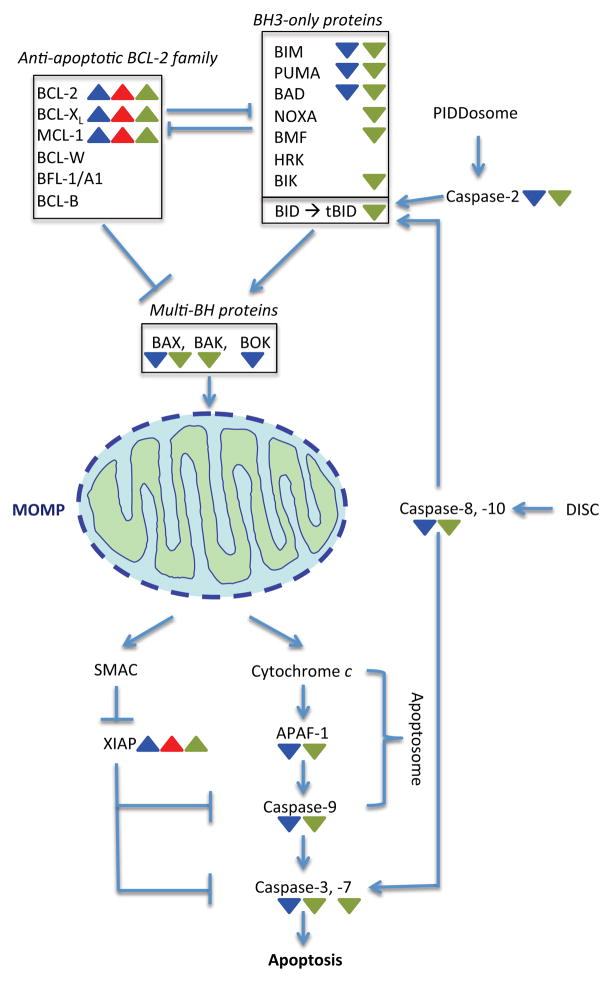
Apoptosis is a cellular suicide program that organisms have evolved to eliminate unnecessary or unhealthy cells from the body in the course of development or following cellular stress. It involves a series of cellular events that ultimately leads to activation of a family of cysteine proteases called caspases. In response to various apoptotic stimuli, “initiator” caspases (caspase-2, -8, -9, or -10) are activated. Initiator caspases, in turn, cleave and activate the zymogenic forms of “executioner” caspases (e.g. caspase-3 or -7), resulting in the proteolytic cleavage of specific cellular substrates and, consequently, cell death (Figure 1). In this regard, the cleavage (activation) of executioner caspases is a hallmark of apoptosis. There are two routes to apoptosis: extrinsic and intrinsic. In the extrinsic pathway, initiator caspase-8 and -10 are activated through the formation of a death-inducing signal complex (DISC) in response to engagement of extracellular ligands (e.g. Fas or TNF [Tumor necrosis factor]) by cell surface receptors (Figure 1). In the intrinsic pathway of apoptosis, mitochondrial outer membrane permeabilization (MOMP) is involved; MOMP triggers the release of a group of pro-apoptotic proteins, including cytochrome c and second mitochondria-derived activator of caspases (SMAC), from the mitochondrial intermembrane space to the cytoplasm (Figure 1). In the cytoplasm, cytochrome c binds to the adaptor protein APAF-1 (apoptotic protease activating factor 1) to form a caspase-9-activating complex, called the apoptosome (Figure 1). SMAC augments cytochrome c-induced caspase activation by binding and neutralizing XIAP (X-linked inhibitor of apoptosis protein), an inhibitor of caspase-3, -7, and -9 (Figure 1).
Figure 1. Apoptosis is triggered in response to internal or external stimuli.
The intrinsic pathway to apoptosis is initiated by activation of BH3-only protein. Among the BH3-only proteins, whose expression is often transcriptionally induced following an apoptotic stimulus, BID is unique in that it is activated by caspase cleavage (by caspases-2 and -8/10); cleaved BID, termed tBID. BH3 proteins trigger the activation of multi-BH domain proteins by direct binding or by inhibition of anti-apoptotic BCL-2 (B-cell lymphoma 2) family proteins. Once activated, multi-BH domain proteins form oligomers that induce MOMP. MOMP prompts pro-apoptotic proteins SMAC and cytochrome c to be released to the cytoplasm, where cytochrome c induces the formation of the Apoptosome. The initiator caspase-9 activated within the Apoptosome initiates the activation of executioner caspase-3 and -7, resulting in apoptosis. Of note, caspase-2 is activated via formation of the PIDDosome, a complex which consists of the adaptor proteins PIDD (p53-induced death domain protein) and RAIDD (RIP-associated protein with a death domain). However, despite identification of this complex, the precise mechanism of caspase-2 activation remains elusive. In addition, only a few cellular substrates of caspase-2, including BID, have been identified. Colored triangles denote that the indicated gene or gene product may be transcriptionally (blue), translationally (red), and post-translationally (green) up- or down-regulated in cancer cells (see text and Table 1).
The balance between pro- and anti-apoptotic BCL-2 family proteins is a fundamental determinant for the initiation of MOMP. The BCL-2 family proteins are classified based on the presence of shared blocks of sequence homology, termed BCL-2 homology (BH). The pro-apoptotic BCL-2 family members, which promote MOMP, include BH3-only proteins (e.g. BIM [Bcl2-interacting mediator of cell death], BID [BH3-interacting domain death agonist], and BAD [Bcl2-associated agonist of cell death] that share only a single block, BH3 domain, of BCL-2 homology) and multi-BH domain proteins (e.g. BAX [Bcl2-associated protein X] and BAK [Bcl2 antagonist/killer], which share BH1-BH3 domains). Following cytotoxic or genotoxic stress, BH3-only proteins are activated and promote oligomerization of BAX (or BAK), resulting in MOMP, whereas pro-survival members (e.g. BCL-2, BCL-XL [B-cell lymphoma extra large], and MCL-1 [Induced myeloid leukemia cell differentiation protein], which contain all four BH domains) counteract this process by sequestering pro-apoptotic family members (Figure 1). Importantly, the interactions between pro- and anti-apoptotic BCL-2 family proteins are determined with specific binding specificities [1, 2]; for instance, BIM interacts with all six anti-apoptotic BCL-2 family proteins as well as BAX and BAK [3], whereas NOXA (Phorbol-12-myristate-13-acetate-induced protein 1) only binds to MCL-1 and BFL-1/A1.
Unlike normal cells, cancer cells are under constant stress, such as oncogenic stress, genomic instability, and cellular hypoxia. In response to such internal apoptotic stimuli, the intrinsic pathway of apoptosis would normally be activated. Yet, cancer cells can often avoid this cellular response by disabling the apoptotic pathways. Notably, mouse genetic models have shown that genetic inactivation of a BH3-only protein or a caspase can not only lead to resistance to certain pro-apoptotic stimuli but also accelerate tumor formation in mice [4–6]. Moreover, forced expression of anti-apoptotic BCL-2 family proteins can significantly augment tumor development induced by an oncogene, such as MYC, although overexpression of an anti-apoptotic BCL-2 family protein alone may not result in similar tumor formation as seen with an oncogene alone [7–10]. Thus, it is strongly suggested that inhibition of apoptosis plays a critical role in cancer cell survival and tumor development.
Cancer cells can modulate apoptotic pathways transcriptionally, translationally, and post-translationally. In some cases, cancer cells may escape apoptosis by increasing or decreasing expression of anti- or pro-apoptotic genes, respectively. Alternatively, they may inhibit apoptosis by stabilizing or de-stabilizing anti- or pro-apoptotic proteins, respectively. Moreover, cancer cells may also prevent apoptosis by changing the functions of anti- or pro-apoptotic proteins through post-translational modifications, such as phosphorylation. Importantly, these mechanisms are not mutually exclusive and cancer cells may employ one or multiple mechanisms to evade apoptosis. In this review, we highlight the core anti-apoptotic machinery that cancer cells may employ for survival, focusing our discussion on the transcriptional, translational, and post-translational modifications to evade apoptosis (note that recent studies suggest that cancer metabolism can modulate this machinery to protect cells from apoptosis [7, 8]; however, this is outside the current scope of this discussion).
Transcriptional/translational regulation
Genomic and epigenomic abnormalities are a characteristic of cancer cells. This includes gene copy number amplification, gene deletion, gene silencing by DNA methylation, and activation (or inactivation) of transcription factors, which impact expression of apoptotic regulators [13]. In addition, microRNAs negatively control gene expression by targeting 3′UTR of mRNAs, although they could function as tumor suppressors or oncogenes, depending on the target messengers [14]. Thus, transcriptional and translational alternations can be a means for cancer cells to directly raise a threshold for apoptotic induction at gene expression levels. In this section, we will discuss how cancer cells may increase or block expression of anti- or pro-apoptotic gene products, respectively (see Table 1 for loss or amplification of apoptotic genes identified in cancer tissues).
Table 1.
Cancer and Apoptotic Regulators
| Loss or Amplification# | Kinases (Phosphatases) | Sites* | Outcomes of Phosphorylation | Cancer Connections## | |
|---|---|---|---|---|---|
| Caspase-2 | CDK1/CYCLIN B1 | Ser308 | Suppression of caspase activity [122] | Suppressing mitotic cell death | |
| CaMKII (PP1) | Ser164 | Suppression of caspase activity [123] | Metabolically regulated | ||
| CK2 | Ser157 | Suppression of caspase activity [124] | TRAIL resistance Esophageal cancer cells Colon cancer cells Glioma cells |
||
| Caspase-3 | PKCδ | ND | Enhancement of caspase activity [125] | ||
| p38 MAPK (PP2A) | Ser150 | Suppression of caspase activity [126] | |||
| Caspase-7 | PAK2 | Ser30 Thr173 Ser239 |
Suppression of caspase activity [127] | Breast cancer cells | |
| Caspase-8 | ND | Tyr310 (Tyr 293) | SHP1 binding required for dephosphorylation of Tyr397 and Tyr465 [128] | ||
| p38 MAPK | Ser364 | Suppression of caspase activity [126] | |||
| SRC FYN LYN (SHP1) |
Tyr397 (Tyr380) | Suppression of caspase activity [128, 129] | Colon cancer | ||
| Lyn (SHP1) | Tyr465 | Suppression of caspase activity [128] | |||
| Caspase-9 | PKA | Ser99 Ser183 Ser195 |
No effects [130] | ||
| CDK1, DYRK1A, ERK1/2, p38 MAPK (PP1α) | Thr125 | Suppression of caspase activity [131–136] | Suppressing mitotic cell death | ||
| PKCζ | Ser144 | Suppression of caspase activity [137] | |||
| c-ABL | Tyr153 | Promoting caspase activation [138] | Promoting DNA damage-induced apoptosis | ||
| AKT | Ser196 (Human specific)** | Suppression of caspase activity [139] | Prostate cancer cells Colon cancer cells |
||
| BID | CK1 CK2 |
Thr58 Ser61 Ser64 |
Prevents cleavage by caspase 8 [121] | ||
| ATM | Ser61 Ser78 |
S phase arrest in response to DNA damage [140] | |||
| BIM*** | <Loss> Chronic myeloid leukemia and non-small cell lung cancer [60] Cutaneous T-cell lymphoma [142] | ERK | Ser55 Ser65 Ser100 (mouse) |
Inactivation [116] | Under the control of cytokine IL3 |
| JNK p38 MAPK |
Ser65 (mouse) | Activation [114, 115, 117] | |||
| ERK JNK |
Ser69 | Ubiquitination and degradation by promoting phosphorylation at Ser93/Ser94/Ser9 8 by RSK [99,101,141] | Non-small cell lung cancer (NSCLC) cells Chronic Myeloid Leukemia(CML) cells T-cell acute lymphoblastic leukemia (T-ALL) cells |
||
| PKA | Ser86 | Prevents degradation [143] | |||
| RSK | Ser93 Ser94 Ser98 |
SCFβ-TrCP-mediated ubiquitination and degradation Following phosphorylation at Ser69 [101] | |||
| JNK | Thr112 (mouse) | Activation [4] | |||
| PUMA | <Loss> Breast, Hepatocellular, Lung NSC, Medulloblastoma, Ovarian, Renal [23] | IKK | Ser10 | Ubiquitination and degradation [144, 145] | Under the control of cytokine IL3 |
| BAD | IKK | Ser26 | Inactivation [146] | TNFα resistance | |
| PAK1 | Ser111 | Inactivation [147] | Malignant peripheral nerve sheath tumor (MPNST) cells Lung cancer cells |
||
| PAK1 PAK4 PAK5 PKA PKCι RAF1 RSK |
Ser112 | Inactivation [147–155] | Malignant peripheral nerve sheath tumor (MPNST) cells Lung cancer cells Under the control of cytokine IL3 |
||
| PAK1 AKT PKCι RSK |
Ser136 | Inactivation [112, 155, 156] | Lung cancer cells Under the control of IGF-1 |
||
| PKA PKCι RSK |
Ser155 | Inactivation [155, 157, 158] | Lung cancer cells | ||
| NOXA | CDK5 | Ser13 | Inactivation [159] | Metabolically regulated T-cell leukemia cells | |
| BMF | JNK ERK2 |
Ser74 | Activation [160, 161] | ||
| ERK2 | Ser77 | Inactivation [161] | Melanoma cells | ||
| BIK | ERK1/2 | Thr124 | Ubiquitination and degradation [162] | Lung cancer cells Colon cancer cells |
|
| BAX | <Loss> Colon Cancer [45] | PKA | Ser60 | Activation Following phosphorylation at Thr167 and dephosphorylation at Ser184 [163] | |
| ERK1/2 | Thr167 | Inactivation via interaction with Pin1 [164] | |||
| GSK3 JNK p38 MAPK |
Activation [165, 166] | Hepatoma cells | |||
| AKT PKCζ (PP2A) |
Ser184 | Inactivation [167–169] | Small cell lung cancer (SCLC) cells Non-small cell lung cancer (NSCLC) cells |
||
| p38 MAPK | ND | Activation [170] | Required during anoikis | ||
| BAK | Kinase unidentified (PTPN2) (PTPN5) (PTPN23) | Tyr108 | Inactivation [171] | Fibrosarcoma cells Colon cancer cells |
|
| Kinase unidentified (PP2A) | Ser117 | Activation [172] | Colon cancer cells | ||
| BCL-XL | <Amplification> Non-small cell lung cancer (NSCLC) Pancreas cancer [21] giant-cell tumor of bone [22] Breast cancer Ovarian [23] | PLK3 | Ser49 | Cell cycle (G2) arrest [173] | Lymphoma cells |
| CDK1/CYCLIN B1 PLK1 JNK2 |
Ser62 | Cell cycle (G2) arrest Inactivation [174, 175] | Cervical carcinoma cells | ||
| BCL-2 | <Amplification> B-cell follicular lymphoma [18–20] | ERK1/2 PKC |
Ser70 | Activation [176, 177] | Under the control of cytokine IL3 |
| ASK1 JNK1 |
Thr69 Ser70 Ser87 |
Inactivation [178] | |||
| ERK1/2 (MKP-3) | Thr 56 Thr 74 Ser 87 |
Prevents degradation [179,180] | |||
| CDK1/CYCLIN B1 | ND | Inactivation [174] | |||
| MCL-1 | <Amplification> Lung cancer [23, 24], Breast cancer Dedifferentiated liposarcoma, Esophageal adenocarcinoma, Hepatocellular, Melanoma, Ovarian, Prostate, Renal [23] | CDK1 CDK2 JNK1 |
Ser64 | Enhances binding to BIM, NOXA, BAK [181] | TRAIL resistance Cholangiocarcinoma cells |
| CDK1/CYCLIN B | Thr92 | APC/CCDC20-mediated ubiquitination and degradation [89] | |||
| JNK p38 MAPK CKII |
Ser121 | SCFFBW7 – mediated ubiquitination and degradation [91] | Ovarian cancer Non-small-cell lung cancer (NSCLC) |
||
| GSK3 JNK p38 MAPK CKII |
Ser155 Ser159 Ser163 |
Ubiquitination and degradation mediated by TRIM17, SCFβ-TrCP and SCFFBW7 Inhibits USP9X binding [81–85, 90, 91, 97] | Breast cancer cells Ovarian cancer Non-small-cell lung cancer (NSCLC) T-cell acute lymphoblastic leukaemia (T-ALL) Follicular lymphomas Diffuse large B-cell lymphomas Multiple myeloma Under the control of cytokine IL3 |
||
| ERK1/2 | Ser163 | Prevents degradation [86,87] | Hepatoma cells Burkitt lymphoma cells |
||
| APAF-1 | <Loss> Melanoma [68–70], Colorectal cancer [71,72], Gastric cancer [73], Bladder cancer [74,75] <Amplification> Glioblastoma/Me dulloblastoma/As trocytoma [182] |
RSK | Ser268 | Inhibition [118] | Prostate cancer cells |
| XIAP | <Amplification> Cervical cancer [38] Acute myeloid leukemia [183] Squamous carcinomas [184] Rectal [185] | AKT | Ser87 | Prevents ubiquitination and degradation [186] | Ovarian cancer cells |
| TBK1 IKKε |
Ser430 | Autoubiquitination and degradation [187] | Colon cancer cells |
confirmed at copy number, mRNA, or protein levels in cancer tissues
demonstrated or suggested in cancer cell lines or cancer tissues
Human sites unless specified,
not conserved in other species,
BIMEL
Inducing Anti-apoptotic Protein Expression
To overcome stress signals, cancer cells commonly overexpress anti-apoptotic proteins, especially anti-apoptotic BCL-2 family proteins. Cancer cells prevent MOMP by amplification of anti-apoptotic BCL-2 family proteins and inhibition of one or multiple anti-apoptotic BCL-2 family proteins cause apoptosis in cancer cells, but not in healthy normal cells – it is often said that cancer cells are primed to death or addicted to anti-apoptotic BCL-2 family proteins. Overexpression of anti-apoptotic BCL-2 family proteins is often associated with poor prognosis, recurrence, and resistance to cancer therapeutics [15–17]. The BCL-2 gene was originally found in B-cell follicular lymphoma where genetic translocation resulted in constitutive expression of BCL-2 [18–20] (Table 1). Amplification of the BCL-XL gene has been reported in various cancers, including lung and giant-cell tumor of bone [21–23] (Table 1). Likewise, frequent amplification of the MCL-1 gene has also been reported in breast and lung cancer samples [23,24] (Table 1).
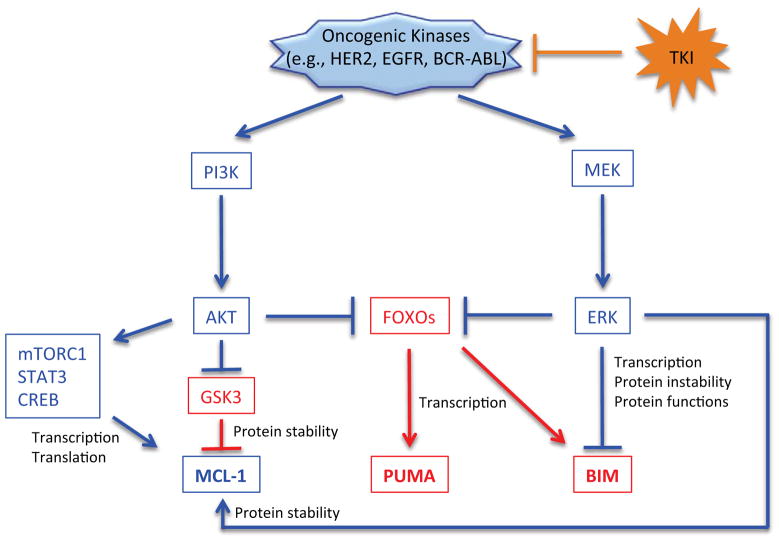
In normal cells, MCL-1 transcription is induced upon cytokine and growth factor stimulation [25, 26], while its synthesis is inhibited upon various cellular stresses, including DNA damage [27–29]. In particular, it was shown that the transcription factor E2F1, known to control proliferation and apoptosis, could repress MCL-1 gene expression by directly binding to the MCL-1 promoter [30], resulting in increased apoptosis. Alternatively, the transcription factors CREB (cAMP response element-binding protein) and STAT3 (signal transducer and activator of transcription 3), known to activate genes responding to pro-survival stimuli, can transactivate the MCL-1 gene [25,27]. Importantly, whereas these signaling pathways operate to maintain cellular homeostasis in normal cells, they are often deregulated in cancer cells. Especially, since the phosphatidylinositol 3-kinase (PI3K)/AKT pathway is frequently hyperactivated in cancer cells by mutations and other mechanisms, the transcriptional regulation of this pathway (e.g., activation of CREB or STAT3) could promote MCL-1 expression in certain cancer cells (Figure 2). In this regard, it was also shown that MCL-1 mRNA translation could be facilitated by mTORC1 (mammalian target of rapamycin complex 1), a downstream target of PI3K/AKT signaling, in a mouse lymphoma model (Figure 2) [31].
Figure 2. Oncogenic kinase signaling regulates MCL-1, BIM, and PUMA.
Oncogenic kinases regulate BIM, PUMA, and MCL-1 by changing gene expression, protein stability, and protein functions by activating two major pro-survival pathways: the AKT and ERK (Extracellular signal-regulated kinases) pathways. Pro-survival and pro-apoptotic pathways are denoted in blue and red, respectively.
microRNAs also play a role in the regulation of anti-apoptotic proteins. miR-15 and miR-16 inhibit BCL-2 mRNA translation [32]. Likewise, miR-29b, miR-101, and miR-193a-3p suppress MCL-1 mRNA translation, making cells susceptible to apoptosis [33–35] – note that the role of miR-29b in cell death may be complex as it also targets several BH3-only proteins, inhibiting apoptosis in neurons [36]. Expression of miR-15 and miR-16 is reduced in chronic lymphocytic leukemia [32], whereas expression of miR-101 is diminished in hepatocellular carcinoma [35] suggesting a possible mechanism to evade apoptosis by these cancers. Last, two recent studies have shown that expression of miR-7 and miR-24, which inhibit XIAP mRNA, is suppressed in various cancer cells including cervical cancer and lung cancer cells, resulting in elevated XIAP protein levels and blocked caspase activation [37, 38]. It would be interesting to fully investigate whether expression of these microRNAs is altered in various cancer tissues and whether their expression patterns are associated with prognostic outcomes.
Suppressing Pro-apoptotic Protein Expression
In normal cells, genotoxic and cytotoxic stress can induce expression of pro-apoptotic genes, particularly some of the BH3-only proteins that trigger apoptosis. In cancer cells, this mechanism is often nullified by mutation and silencing of a key apoptotic gene(s).
In normal cells, p53 is among the most critical transcription factor that modulates the expression of an array of genes known to trigger apoptosis following various cellular stresses. Given the fact that TP53 encoding p53 is the most frequently mutated, or inactivated, tumor suppressor gene in cancer (~50%) [39, 40], apoptotic pathways induced by this gene, including BAX, PUMA (p53 upregulated modulator of apoptosis), NOXA, APAF-1, may be widely affected in cancer cells [41–44] – note that frameshift mutations in the BAX gene have also been reported in colon cancer [45] (Table 1). Interestingly, p53 could also activate BAX (and BAK) in a transcription-independent manner in response to DNA damage and metabolic changes [46–48]. Thus, loss of p53 in cancer cells will elevate a threshold for MOMP by limiting BAX expression and reducing a chance of BAX oligomerization.
The transcription of the BH3-only protein BIM is induced upon growth factor withdrawal and other apoptotic stimuli [5, 49]. Various transcription factors have been implicated in BIM expression, including FOXOs [49–51], RUNX3 [52], AP-1 [53], and E2F1 [54]. Among the transcription factors, FOXOs are of particular interest since its activity can be suppressed by two major pro-survival kinases, AKT [55] and ERK [56–59]. Thus, in cancer cells with high levels of AKT or ERK, BIM expression is suppressed; conversely, agents that directly or indirectly inhibit AKT or ERK could trigger expression of BIM (Figure 2).
BIM plays an important role in cancer cells when apoptosis is induced by tyrosine kinase inhibitors (TKIs) [5, 59]. Since AKT and ERK are key effectors downstream of oncogenic kinases (e.g., EGFR [epidermal growth factor receptor], HER2 [Human epidermal growth factor receptor 2], BCR-ABL), inhibition of the tyrosine kinase could result in suppression of AKT and ERK activity, resulting in the expression of BIM (Figure 2). Notably, a recent study showed that BIM RNA levels can predict clinical responsiveness to targeted cancer therapy, such as EGFR, HER2, and PI3K inhibitors; higher BIM RNA levels detected prior to the inhibitor treatments are associated with better clinical outcomes [59]. Additionally, a BIM deletion polymorphism has been reported in certain human populations, and is significantly associated with innate resistance to TKI therapies in chronic myeloid leukemia and non-small cell lung cancer [60] (Table 1). Therefore, BIM induction is critical for the efficacy of TKI therapies and BIM RNA levels may be used as a biomarker to predict cancer patients’ prognoses and susceptibility to TKI therapies.
A large somatic copy number analysis identified another BH3-only family member PUMA as one of the pro-apoptotic BCL-2 family genes that are most frequently deleted in cancer (the study identified two genes and the other deleted gene was BOK) [23] (Table 1). PUMA was originally discovered as a p53- and p73 target gene [42, 61]. However, FOXOs can also transactivate PUMA, independently of p53, in response to growth factor withdrawal or TKI treatments [5, 62] (Figure 2). In this regard, a recent study has shown that PUMA may be implicated in apoptosis triggered by FOXOs following inhibition of the PI3K-AKT pathway, while BIM may be primarily under control of the ERK pathway in HER2-positive breast cancer and EGFR mutant lung cancer (as described below, BIM is also regulated at the protein levels by ERK); for full execution of apoptosis, inhibition of both pathways by TKIs, i.e., expression of both PUMA and BIM, is required [5]. This finding sheds light on an underappreciated role of PUMA in TKI-induced apoptosis. Since BIM is also known to be regulated by FOXOs, how PUMA and BIM are independently regulated remains to be fully elucidated (Figure 2).
In addition to the signaling pathways that impede activation of BAX and BAK, a defect in the pathway following cytochrome c release could further provide cancer cells an additional survival advantage, independent of BAX and BAK inhibition (Figure 1). Expression of some caspase genes (caspases-2, 7, -8, and -9) is transcriptionally regulated in response to E2F1 or p53 [63–65]; in cancer cell where these pathways are inactivated, expression of these caspases could be compromised resulting in apoptotic protection. APAF-1 gene is transactivated by p53 [43, 44] and E2F1 [44, 66] and its mRNA translation is initiated through an internal ribosome entry site (IRES) [67]. Loss of APAF-1 expression has been reported in melanoma [68–70], colorectal cancer [71, 72], gastric cancer [73] and bladder cancer [74, 75], and its loss is often associated with poor clinical outcomes. Studies show that loss of APAF-1 expression in cancer cells can be secondary to promoter methylation [68, 72, 73, 75] and loss of heterogeneity at the APAF-1 locus [68–70, 73] (Table 1).
Collectively, it is evident that the transcriptional network is deregulated in cancer cells, which significantly impacts the expression of apoptotic regulators and, consequently, contributes to tumor development and resistance to therapeutic agents.
Post-translational regulation
Various apoptotic regulators are modulated not only by transcriptional/translational machinery but also by post-translational modifications, including ubiquitination and phosphorylation. Protein ubiquitination is implicated in protein stability and is dynamically regulated through the ubiquitin/proteasome pathway [76], whereas phosphorylation can change protein functions [77, 78]. It should be noted, however, that phosphorylation can also influence ubiquitination of the target protein by providing a docking site for an E3 ubiquitin ligase (phospho-degron).
The ubiquitin/proteasome pathway involves a series of biochemical events, which are mediated by E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases [79]. In normal cells, expression levels of an anti-apoptotic protein with a short half-life (e.g., MCL-1) may readily decline by inhibition of transcription or translation following an apoptotic stimulus. In contrast, an apoptotic signal may prevent constant degradation of a pro-apoptotic protein (e.g., BIM), resulting in the accumulation of that protein. Importantly, cancer cells may deregulate this process by enhancing the degradability of specific pro-apoptotic proteins or by promoting the stability of anti-apoptotic proteins. This may also be the case when cancer cells acquire resistance to a therapeutic agent. It was recently shown that HER2-amplified breast cancer cells modulate stabilities of pro- and anti-apoptotic proteins, both upstream and downstream of MOMP, when acquiring resistance to the HER2 inhibitor lapatinib; consequently, the resistant cancer cells exhibit strong protection against cell death induced by HER2 inhibition [80].
Here we discuss how the stability of some apoptotic regulators (MCL-1, BIM, and BAX) is controlled in normal cells and how this stability may be altered in cancer cells, contributing to apoptotic protection. Then, we will highlight some examples where phosphorylation plays a role to alter functions of apoptotic regulators.
Stabilizing MCL-1
Whereas BCL-2 and BCL-XL proteins are relatively stable, MCL-1 protein is characterized by its rapid turnover [28, 81]. As described above, MCL-1 transcription is stimulated, at least in part, by pro-survival factors, such as cytokines. Hence, once its transcription is suppressed following apoptotic stimuli, MCL-1 protein levels quickly drop, potentially facilitating MOMP. Importantly, emerging evidence strongly suggest that the rapid turnover of MCL-1 protein may be abrogated in some cancer cells. Thus far, five E3 ligases have been discovered to ubiquitinate MCL-1 for degradation, including the most recently identified TRIM17 (tripartite motif containing17), which facilitates MCL-1 degradation during neuronal apoptosis [82]. It should be noted, however, that MCL-1 protein could also be degraded in a ubiquitination independent manner [83] although the precise mechanism remains elusive.
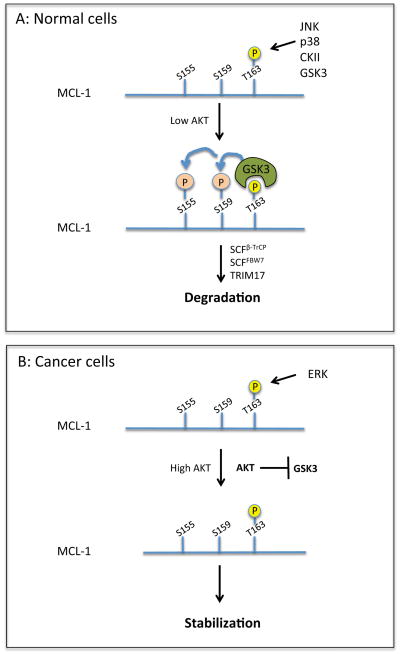
Phosphorylation also plays a key role in the stability of MCL-1 protein. Phosphorylation of MCL-1 at T163 followed by phosphorylation at S155/S159 targets the protein for proteasomal degradation, which is mediated by the E3 ligase SCFβ-TrCP (Skp, Cullin, F-box containing complex with β-transducin repeat-containing protein) [81, 84]. The priming phosphorylation site T163 is a target of multiple kinases (Figure 3). An initial study identified glycogen synthase kinase 3 (GSK3) as the major kinase that phosphorylates this residue; however, a more recent study demonstrated that JNK (c-Jun N terminal kinase) primarily phosphorylates T163; in both cases, T163 phosphorylation will trigger subsequent phosphorylation of S159 (and then S155) by GSK3 [81, 85]. Collectively, phosphorylation at S155/S159/T163 serves as a degron sequence for SCFβ-TrCP (Table 1 and Figure 3).
Figure 3. MCL-1 protein stability is controlled by multiple kinases.
(A) MCL-1 can be phosphorylated at T163 by several kinases including GSK3. AKT inhibits GSK3. Thus, in low levels of AKT, phosphorylation by GSK3 sequentially occurs at S155/S159, targeting MCL-1 for degradation by E3 ligases, SCFβ-TrCP, SCFFBW7 (F-box/WD repeat-containing protein 7), and TRIM17. (B) In cancer cells with high levels of AKT and ERK, ERK is likely to be the main kinase that phosphorylates MCL-1. Phosphorylation does not continue to S155/S159 and MCL-1 is stabilized.
Paradoxically, it has also been reported that phosphorylation at T163 by ERK can lead to marked MCL-1 stabilization [86, 87] (Table 1). How can we reconcile these conclusions? Is the mechanism of MCL-1 degradation simply cell type specific? We speculate that it may be phosphorylation of S159 that determines the fate of MCL-1 protein following T163 phosphorylation. It is known that GSK3 activity is negatively regulated by AKT, which is commonly upregulated in cancer cells; therefore, in cancer cells with high AKT and ERK activity, MCL-1 may be only phosphorylated at T163 but not at S155 and S159, which promotes stabilization of MCL-1 (Figure 3). Supporting this, it is reported that the abundance of MCL-1 protein is associated with low GSK3 activity in breast cancer tissues [88]. In contrast, in normal cells, cellular stress signals, such as growth factor deprivation, would lead to inactivation of AKT (and activation of JNK), which elicits serial phosphorylation events at S155/S159/T163; this would result in degradation of the MCL-1 protein (Figure 3). It should be noted, however, that there is at least one additional E3 ligase SCFFBW7, that also regulates MCL-1 stability depending on the phosphorylation status of T163 (see below). Thus, the regulation of MCL-1 protein stability mediated by this phosphorylation motif is apparently complex and the mechanism remains to be fully elucidated.
A failure in the cell cycle can trigger apoptosis in normal cells. Furthermore, prolonged mitotic arrest can lead to the activation of the intrinsic apoptotic pathway. For this reason, some of microtubule inhibitors that induce mitotic arrest by inhibiting mitotic spindle assembly (e.g., paclitaxel) are clinically used to treat various cancers. However, the emergence of cancer cells with intrinsic or acquired resistance to this type of chemotherapeutic agents is an unresolved clinical problem. Three independent studies described the mechanism of MCL-1 degradation during prolonged mitotic arrest, suggesting a possible role for MCL-1 stabilization in the resistance to mitotic apoptosis. These studies have shown that in addition to the mechanisms described above, two E3 ligases, APC/CCDC20 (Anaphase-promoting complex/cyclosome with cell-division cycle protein 20) and SCFFBW7, target MCL-1 for destruction during mitosis [89–91]. APC/CCDC20 ubiquitination of MCL-1 is mediated by phosphorylation at T92 by CDK1 (cyclin-dependent kinase 1)/CYCLIN B and the D-box motif during mitotic arrest [89]. Ubiquitination of MCL-1 by SCFFBW7 is mediated by two phospho-degrons on MCL-1 (S121/E125 and S159/T163), of which S159/T163 appears to play a major role [90, 91]. Interestingly, JNK, p38, and CKII (Casein kinase II) phosphorylate both degrons during mitotic arrest [91] although GSK3 phosphorylation of MCL-1 at S159/T163 may be sufficient to target MCL-1 for SCFFBW7-mediated degradation [90] (Table 1). Since, unlike CDK1/CYCLIN B, these kinases are not directly controlled by the cell cycle, the mechanism of mitotic phosphorylation of MCL-1 remains undetermined. In addition, it is not known whether APC/CCDC20 and SCFFBW7 compete for the mitotic degradation of MCL-1 or work independently. From the clinical viewpoint, it is interesting to note that mutations or loss of FBW7 (a substrate-binding component of the multi-subunit E3 complex SCFFBW7) has been reported in various cancer types [92–94]. Moreover, cancer cells with low FBW7 levels have elevated MCL-1 protein and exhibit intrinsic resistance to ABT-737, a small molecule inhibitor of BCL-2, BCL-XL, and BCL-W [90] (BOX 1).
BOX 1. BH3 mimetics and cancer therapy.
The binding between pro- and anti-apoptotic BCL-2 family proteins is mediated through a hydrophobic groove on the anti-apoptotic proteins and a BH3 domain. Several small molecule inhibitors, so-called BH3 mimetics (e.g., ABT-737, ATB-263, ATB-199), have been engineered to specifically bind to the hydrophobic pocket, thereby antagonizing the function of anti-apoptotic BCL-2 family proteins. The initial inhibitor of this class, ABT-737, and its orally bioavailable analog ABT-263, both of which inhibit both BCL-2 and BCL-XL, were a pre-clinical success [188, 189]. Nonetheless, the clinical success was hampered by thrombocytopenia caused by BCL-XL inhibition. To circumvent this problem, a BCL-2 specific inhibitor ABT-199 was recently developed and is now being tested in clinical trials [190]. Another challenge in BCL-2/BCL-XL inhibitors is that cancer cells can acquire resistance to this class of inhibitors by overexpressing MCL-1 [191]. Recently, two groups independently identified MCL-1 inhibitors through genomic or biochemical screenings: Golub and colleagues identified compounds that suppress MCL-1 transcription [192], whereas Walensky and colleagues discovered a small molecule, MIM1, which binds to the BH3-domain binding pocket of MCL-1 [193] – these inhibitors may overcome resistance to BCL-2/BCL-XL inhibitors. Of note, the short protein half-life of MCL-1 makes MCL-1-addicted cancer cells more susceptible to chemotherapy than cancer cells depending on BCL-2 or BCL-XL for survival [8]. However, given the multiple mechanisms of MCL-1 stabilization in cancer cells (see text), it is assumed that this approach may not work in all of MCL-1-addicted cancer cells. For cancer cells with stabilized MCL-1, agents that directly inhibit MCL-1 protein (e.g., MIM1) or promote MCL-1 protein degradation may be an option.
A HECT-domain containing E3 ligase, HUWE1 (MULE, Mcl-1 ubiquitin ligase E3), also targets MCL-1 for proteasomal degradation through a pathway that does not require MCL-1 phosphorylation by GSK3; the binding between MCL-1 and HUWE1 is mediated through the BH3 domains on each protein [95]. Interestingly, it has been reported that ERK activity, directly or indirectly, reduces the interaction between MCL-1 and HUWE1 although the detailed molecular mechanism remains to be elucidated [96]. It would be interesting to test whether the aforementioned T163 phosphorylation by ERK is implicated in the MCL-1-HUWE1 interaction.
Deubiquitinating enzymes (DUBs) are negative regulators of the ubiquitination-mediated protein degradation as DUBs can stabilize their substrate proteins by removing polyubiquitin chains. Ubiquitin specific peptidase 9 X-linked, USP9X, reverses ubiquitination of MCL-1 [97]. As opposed to SCFβ-TrCP and SCFFBW7, the binding of USP9X to MCL-1 is negatively regulated by GSK3 phosphorylation of MCL-1 at S155/S159/T163, suggesting that USP9X may directly counteract MCL-1 degradation controlled by SCFβ-TrCP and SCFFBW7. USP9X and MCL-1 protein expression levels were positively correlated in several cancer types, including follicular lymphomas [97]. Elevated USP9X transcription was also associated with poor prognosis in multiple myelomas [97].
Destroying Pro-apoptotic Proteins
As cancer cells can attenuate MCL-1 degradation, they may also suppress accumulation of pro-apoptotic proteins, such as BIM. As with MCL-1, BIM is also characterized by its short protein half-life. BIM stability is regulated by ERK phosphorylation at S69; phosphorylation of this site promotes BIM degradation through the ubiquitin-proteasome pathway [98, 99]. A transgenic mouse study demonstrated that replacement of the ERK phosphorylation sites with Ala prolongs BIM protein half-life and increases apoptotic cell death in response to serum starvation [4]. SCFβ-TrCP, c-CBL, and TRIM2 are the E3 ligases responsible for ERK-dependent BIM degradation [100–102]. Notably, SCFβ-TrCP-mediated BIM ubiquitination requires phosphorylation of the degron sequence containing S93/S94/S98 by RSK (ribosomal S6 kinase), which is promoted by ERK phosphorylation at S69 [101] (Table 1). Whether BIM ubiquitination by c-CBL and TRIM2 may also require RSK phosphorylation remains elusive. Last, the ubiquitin E3 ligase complex ElonginB/C-Cullin2-CIS is also implicated in BIM ubiquitination and degradation [103]. BIM ubiquitination mediated by ElonginB/C-Cullin2-CIS is enhanced following paclitaxel treatment and a defect in this ubiquitination pathway may confer resistance to this agent in cancer cells [103].
BAX is a central component of apoptosis as its activation is directly linked to MOMP (Figure 1). Thus, causing degradation of BAX protein may be a “last minute” safeguard for cancer cells to prevent apoptosis. BAX protein mainly localizes in the cytoplasm. Upon genotoxic or cytotoxic stress, BAX translocates to the mitochondria outer membrane, forms oligomeric structures, and triggers MOMP [104]. A recent study has shown, however, that the subcellular localization of BAX protein may be in a more dynamic equilibrium between the cytoplasm and the mitochondrial outer membrane [105]. Although the detailed mechanism of how the equilibrium may change following apoptotic stimuli remains elusive, the BAX equilibrium hypothesis suggests there is a mechanism that prevents activation of BAX on the mitochondrial outer membrane. Interestingly, it has been known that BCL-XL can block the mitochondrial translocation of BAX, by re-directing BAX back to the cytoplasm [106]. Moreover, BAX degradation takes place on mitochondrial outer membrane following apoptotic stimuli [107, 108]. BAX is a protein with a short half-life [109, 110] and the rapid degradation of BAX protein has been associated with poor clinical outcomes and high tumor grades in chronic lymphocytic leukemia and prostate cancer [109]. The E3 ligase IBRDC2 (IBR domain containing 2; also known as p53RFP or RNF144B) was recently identified and shown to ubiquitinate BAX specifically on the mitochondrial outer membrane [108]. Overexpression of IBRDC2, however, did not block apoptosis triggered by actinomycin D or staurosporine treatment [108], suggesting that there may be additional factors (e.g., co-factors or other ligases) involved in this process.
Altering protein functions
In addition to modulating a docking site for an E3 ligase and subsequent ubiquitin-mediated protein degradation, phosphorylation can also alter the function of apoptotic regulators. In particular, phosphorylation by pro-survival kinases (e.g., AKT and ERK) often results in suppression of the pro-apoptotic function of the target protein, whereas phosphorylation by pro-apoptotic kinases (e.g., GSK3 and JNK) can lead to activation of pro-apoptotic proteins or inhibition of anti-apoptotic proteins (see Table 1 for a comprehensive list of known phosphorylation sites of apoptotic regulators). For instance, the pro-apoptotic activity of the BH3-only protein BAD is inhibited by a number of pro-survival kinases, including PAK1 and AKT [111–113]. Likewise, ERK phosphorylation of BIM prevents the interaction between BIM and BAX, thereby inhibiting apoptosis, whereas phosphorylation of BIM by JNK or p38 MAP kinase promotes the pro-apoptotic activity of BIM by enhancing binding to anti-apoptotic Bcl-2 family proteins [4,114–117]. As described above, ERK also promotes suppression of BIM expression and degradation of BIM protein [4, 98, 99] (Figure 2), further highlighting a critical role for ERK in BIM inhibition.
Pro-survival kinases also negatively regulate apoptotic pathways downstream of mitochondrial cytochrome c release (Figure 1). Activation of APAF-1 in response to cytochrome c is inhibited via phosphorylation of APAF-1 by RSK [118], a kinase often amplified in prostate cancer [119] (Table 1). Leukemic tyrosine kinases, such as BCR-ABL, also inhibit APAF-1 oligomerization by modulating the phosphorylation status of the Apaf-1 inhibitor HSP90β [120]. In addition to APAF-1, caspases are direct targets of various kinases; activation of caspases can be suppressed by direct phosphorylation (Table 1). Furthermore, it is known that phosphorylation can protect some caspase substrates from proteolytic cleavage [78]. For example, phosphorylation by CK1 or CK2 protects BID from caspase-8, thereby blocking the activation of this BH3-only protein and subsequent cytochrome c release from mitochondria [121] (Table 1).
In summary, post-translational modifications play a critical role in the regulation of apoptosis by modulating protein stabilities and functions.
Concluding remarks
In order to survive and proliferate, cancer cells need to override a number of barriers that otherwise would cause apoptosis. It is conceivable that cancer cells employ multiple mechanisms, instead of a single pathway. Given the multiplicity of anti-apoptotic mechanisms utilized by cancer cells, it is speculated that agents that can globally alter cancer’s apoptotic resistance by targeting multiple pathways, may provide us with the most valuable therapeutic benefits in the future of cancer treatment. To target cancer cells most efficiently and selectively, we must further elucidate the mechanisms of how they evade the apoptotic program (see Outstanding Questions box).
Outstanding Questions.
Do cancer cells simply depend on the net abundance of pro-survival BCL-2 family proteins relative to the total BH3-only proteins? Alternatively, may the regulation of a particular BCL-2 family member selectively play a role for survival in certain cancer types? How do the functional redundancy and specificity of pro-survival BCL-2 family impact cancer cell survival?
Cancer cells employ multiple mechanisms to silence the apoptotic machinery. Letai and colleagues have developed a functional assay, called BH3 profiling, to gauge the dependency of cancer cells on anti-apoptotic BCL-2 family proteins. Although the BH3 profiling made a successful breakthrough [194], is there any other way to predict which pathways are altered in particular cancer cells?
Abundance of MCL-1 is one of the mechanisms for the resistance to a BCL-2/BCL-XL inhibitor in cancer (BOX 1). If a BCL-2/BCL-XL inhibitor is used concomitantly with an MCL-1 inhibitor, can we cure all of cancer? Alternatively, will cancer cells eventually acquire the resistance to the combination of a BCL-2/BCL-XL inhibitor and an MCL-1 inhibitor? If so, how?
Why is the pathway downstream of mitochondrial cytochrome c often inhibited in cancer cells in addition to the inhibition of MOMP [195]?
The failure to activate caspases in response to an apoptotic stimulus can result in caspase-independent cell death [196]. Does caspase-independent cell death play a role in cancer cell survival?
Acknowledgments
We are grateful to Leta Nutt, Joseph Opferman, Marisa Buchakjian, and Matthew Ung for critical reading and feedback on the manuscript. This work was supported by an NCI Career Development Award R00 CA140948 (to MK).
Glossary Box
- APAF-1
Apoptotic protease activating factor 1
- APC/C
Anaphase-promoting complex/cyclosome
- BAD
Bcl2-associated agonist of cell death
- BAK
Bcl2 antagonist/killer
- BAX
Bcl2-associated protein X
- β-TrCP
β-transducin repeat-containing protein
- BCL-2
B-cell lymphoma 2
- BCL-XL
B-cell lymphoma extra large
- BH3
Bcl-2 homology domain 3
- BID (tBid)
(truncated) BH3-interacting domain death agonist
- BIM
Bcl2-interacting mediator of cell death (Bcl2-like 11)
- CDC20
Cell-division cycle protein 20
- CDK1
Cyclin-dependent kinase 1
- CKII
Casein kinase II
- CREB
cAMP response element-binding protein
- DISC
Death inducing signaling complexes
- EGFR
Epidermal growth factor receptor
- ERK
Extracellular signal-regulated kinases
- FBW7
F-box/WD repeat-containing protein 7
- GSK3
Glycogen synthase kinase 3
- HER2
Human epidermal growth factor receptor 2
- HUWE1
HECT, UBA and WWE domain-containing protein 1
- IBRDC2
IBR domain containing 2 (E3 ubiquitin-protein ligase RNF144B)
- JNK-c
Jun N terminal kinase
- MAPK
Mitogen-activated protein kinases
- MCL-1
Induced myeloid leukemia cell differentiation protein
- MOMP
Mitochondrial outer membrane permeabilization
- MULE
Mcl-1 ubiquitin ligase E3
- mTORC1
Mammalian target of rapamycin complex 1
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NOXA
Phorbol-12-myristate-13-acetate-induced protein 1
- PI3K
phosphatidylinositol 3-kinase
- PIDD
p53-induced death domain protein
- PUMA
p53 upregulated modulator of apoptosis
- RAIDD
RIP-associated protein with a death domain
- RSK
Ribosomal S6 kinase
- SCF
Skp, Cullin, F-box containing complex
- SMAC
Second mitochondria-derived activator of caspases
- STAT3
Signal transducer and activator of transcription 3
- TNF
Tumor necrosis factor
- TRIM
Tripartite motif containing
- XIAP
X-linked inhibitor of apoptosis protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strasser A, et al. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMJO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rautureau GJ, et al. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012;13:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hübner A, et al. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell. 2008;30:415–425. doi: 10.1016/j.molcel.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bean GR, et al. PUMA and BIM Are Required for Oncogene Inactivation-Induced Apoptosis. Sci Sinal. 2013:6,ra20. doi: 10.1126/scisignal.2003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons MJ, et al. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.38. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strasser A, et al. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 8.Brunelle JK, et al. MCL-1-dependent leukemia cells are more sensitive to chemotherapy than BCL-2-dependent counterparts. J Cell Biol. 2009;187:429–442. doi: 10.1083/jcb.200904049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaux DL, et al. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1998;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P, et al. Mcl-1 in transgenic mice promotes survival in a spectrum of hematopoietic cell types and immortalization in the myeloid lineage. Blood. 1998;92:3226–3239. [PubMed] [Google Scholar]
- 11.Vander-Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11:715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Cakouros D. Transcriptional control of the core cell-death machinery. Trends Biochem Sci. 2004;29:193–199. doi: 10.1016/j.tibs.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann SH, et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- 16.Taniai M, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–3524. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- 17.Wuilleme-Toumi S, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–1252. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- 18.Bakhshi A, et al. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41:899–906. doi: 10.1016/s0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- 19.Cleary ML, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci USA. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsujimoto Y, et al. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 21.Tonon G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith LT, et al. 20q11.1 amplification in giant-cell tumor of bone: Array CGH, FISH, and association with outcome. Genes Chromosomes & Cancer. 2006;45:957–966. doi: 10.1002/gcc.20354. [DOI] [PubMed] [Google Scholar]
- 23.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JM, et al. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;9:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang HM, et al. Mcl-1 is a common target of stem cell factor and interleukin-5 for apoptosis prevention activity via MEK/MAPK and PI-3K/Akt pathways. Blood. 2000;96:1764–1771. [PubMed] [Google Scholar]
- 27.Liu H, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- 28.Nijhawan D, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fritsch RM, et al. Translational repression of MCL-1 couples stress-induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. J Biol Chem. 2007;282:22551–22562. doi: 10.1074/jbc.M702673200. [DOI] [PubMed] [Google Scholar]
- 30.Croxton R, et al. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–1369. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- 31.Mills JR, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mott JL, et al. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon JE, et al. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis. 2013;18:896–909. doi: 10.1007/s10495-013-0841-7. [DOI] [PubMed] [Google Scholar]
- 35.Su H, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 36.Kole AJ, et al. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, et al. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2012;32:2442–2451. doi: 10.1038/onc.2012.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, et al. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.05.054. In press. [DOI] [PubMed] [Google Scholar]
- 39.Hollstein D, et al. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 40.Feki I, Irminger-Finger I. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol. 2004;52:103–116. doi: 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Oda E, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 42.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 43.Fortin A, et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol. 2001;155:207–216. doi: 10.1083/jcb.200105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moroni MC, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 45.Rampino N, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 46.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 47.Chipuk JE, et al. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2004;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 48.Nieminen A, et al. Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1208530110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahl M, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 50.Dijkers PF, et al. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 51.Sunters A, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 52.Yano T, et al. The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol Cell Biol. 2006;26:4474–4488. doi: 10.1128/MCB.01926-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin HO, et al. Up-regulation of Bak and Bim via JNK downstream pathway in the response to nitric oxide in human glioblastoma cells. J Cell Physiol. 2006;206:477–486. doi: 10.1002/jcp.20488. [DOI] [PubMed] [Google Scholar]
- 54.Gogada R, et al. Bim, a proapoptotic protein, up-regulated via transcription factor E2F1-dependent mechanism, functions as a prosurvival molecule in cancer. J Biol Chem. 2013;288:368–381. doi: 10.1074/jbc.M112.386102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilley J, et al. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa DB, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cragg MS, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–1689. doi: 10.1371/journal.pmed.0040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong Y, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4(10):e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faber AC, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng KP, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 61.Melino G, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 62.You H, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liedtke C, et al. The human caspase-8 promoter sustains basal activity through SP1 and ETS-like transcription factors and can be up-regulated by a p53-dependent mechanism. J Biol Chem. 2003;278:27593–27604. doi: 10.1074/jbc.M304077200. [DOI] [PubMed] [Google Scholar]
- 64.Nahle Z, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2003;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 65.Baptiste-Okoh N, et al. Caspase 2 is both required for p53-mediated apoptosis and downregulated by p53 in a p21-dependent manner. Cell Cycle. 2008;7:1133–1138. doi: 10.4161/cc.7.9.5805. [DOI] [PubMed] [Google Scholar]
- 66.Furukawa Y, et al. Apaf-1 is a mediator of E2F-1-induced apoptosis. J Biol Chem. 2002;277:39760–39768. doi: 10.1074/jbc.M200805200. [DOI] [PubMed] [Google Scholar]
- 67.Coldwell MJ, et al. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 68.Soengas MS, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 69.Dai DL, et al. Reduced Apaf-1 expression in human cutaneous melanomas. Br J Cancer. 2004;91:1089–1095. doi: 10.1038/sj.bjc.6602092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujimoto A, et al. Allelic imbalance of 12q22-23 associated with APAF-1 locus correlates with poor disease outcome in cutaneous melanoma. Cancer Res. 2004;64:2245–2250. doi: 10.1158/0008-5472.can-03-2932. [DOI] [PubMed] [Google Scholar]
- 71.Umetani N, et al. Allelic imbalance of APAF-1 locus at 12q23 is related to progression of colorectal carcinoma. Oncogene. 2004;23:8292–8300. doi: 10.1038/sj.onc.1208022. [DOI] [PubMed] [Google Scholar]
- 72.Zlobec I, et al. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur J Cancer. 2007;43:1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 73.Wang HL, et al. Rationales for expression and altered expression of apoptotic protease activating factor-1 gene in gastric cancer. World J Gastroenterol. 2007;13:5060–5064. doi: 10.3748/wjg.v13.i38.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christoph F, et al. A gene expression profile of tumor suppressor genes commonly methylated in bladder cancer. J Cancer Res Clin Oncol. 2007;133:343–9. doi: 10.1007/s00432-006-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hinz S, et al. EZH2 polycomb transcriptional repressor expression correlates with methylation of the APAF-1 gene in superficial transitional cell carcinoma of the bladder. Tumour Biol. 2007;28:151–157. doi: 10.1159/000103380. [DOI] [PubMed] [Google Scholar]
- 76.Vucic D, et al. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 77.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weissman AM, et al. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurokawa M, et al. A Network of Substrates of the E3 Ubiquitin Ligases MDM2 and HUWE1 Control Apoptosis Independently of p53. Sci Signal. 2013:6 ra32. doi: 10.1126/scisignal.2003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maurer U, et al. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2005;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 82.Magiera MM, et al. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 2013;20:281–292. doi: 10.1038/cdd.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stewart DP, et al. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol Cell Biol. 2010;30:3099–3110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding Q, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morel C, et al. Mcl–1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–3852. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao M, et al. Role of bile salt in regulating Mcl-1 phosphorylation and chemoresistance in hepatocellular carcinoma cells. Mol Cancer. 2011;10:44. doi: 10.1186/1476-4598-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nifoussi SK, et al. Thr 163 phosphorylation causes Mcl-1 stabilization when degradation is independent of the adjacent GSK3-targeted phosphodegron, promoting drug resistance in cancer. PloS ONE. 2012;7:e47060. doi: 10.1371/journal.pone.0047060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding Q, et al. Myeloid cell leukemia-1 inversely correlates with glycogen synthase kinase-3beta activity and associates with poor prognosis in human breast cancer. Cancer Res. 2007;67:4564–4571. doi: 10.1158/0008-5472.CAN-06-1788. [DOI] [PubMed] [Google Scholar]
- 89.Harley ME, et al. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inuzuka H, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 92.Akhoondi S, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 93.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;47:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 95.Zhong Q, et al. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 96.Pervin S, et al. Reduced association of anti-apoptotic protein Mcl-1 with E3 ligase Mule increases the stability of Mcl-1 in breast cancer cells. Br J Cancer. 2011;105:428–437. doi: 10.1038/bjc.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 98.Ley R, et al. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 99.Luciano F, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- 100.Akiyama T, et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dehan E, et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thompson S, et al. Identification of a novel Bcl-2-interacting mediator of cell death (Bim) E3 ligase, tripartite motif-containing protein 2 (TRIM2), and its role in rapid ischemic tolerance-induced neuroprotection. J Biol Chem. 2011;286:19331–19339. doi: 10.1074/jbc.M110.197707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W, et al. RACK1 and CIS mediate the degradation of BimEL in cancer cells. J Biol Chem. 2008;283:16416–16426. doi: 10.1074/jbc.M802360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walter KG, et al. Movement of Bax from the Cytosol to Mitochondria during Apoptosis. J Cell Biol. 1998;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schellenberg B, et al. Bax exists in a dynamic equilibrium between the cytosol and mitochondria to control apoptotic priming. Mol Cell. 2013;49:959–971. doi: 10.1016/j.molcel.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edlich F, et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu FT, et al. Bortezomib blocks Bax degradation in malignant B cells during treatment with TRAIL. Blood. 2008;111:2797–2805. doi: 10.1182/blood-2007-08-110445. [DOI] [PubMed] [Google Scholar]
- 108.Benard G, et al. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 2010;29:1458–1471. doi: 10.1038/emboj.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agrawal SG, et al. Increased proteasomal degradation of Bax is a common feature of poor prognosis chronic lymphocytic leukemia. Blood. 2008;111:2790–2796. doi: 10.1182/blood-2007-10-110460. [DOI] [PubMed] [Google Scholar]
- 110.Fu NY, et al. Baxβ: a constitutively active human Bax isoform that is under tight regulatory control by the proteasomal degradation mechanism. Mol Cell. 2009;33:15–29. doi: 10.1016/j.molcel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 111.Zha J, et al. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 112.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 113.del Peso L, et al. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 114.Putcha GV, et al. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 115.Becker EB, et al. Characterization of the c-Jun N-terminal kinase-BimEL signaling pathway in neuronal apoptosis. J Neurosci. 2004;24:8762–8770. doi: 10.1523/JNEUROSCI.2953-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harada H, et al. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci USA. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai B, et al. p38 MAP kinase mediates apoptosis through phosphorylation of BimEL at Ser-65. J Biol Chem. 2006;281:25215–25222. doi: 10.1074/jbc.M512627200. [DOI] [PubMed] [Google Scholar]
- 118.Kim J, et al. Rsk-mediated phosphorylation and 14-3-3_ binding of Apaf-1 suppresses cytochrome c-induced apoptosis. EMBO J. 2012;31:1279–1292. doi: 10.1038/emboj.2011.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clark DE, et al. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res. 2005;65:3108–3116. doi: 10.1158/0008-5472.CAN-04-3151. [DOI] [PubMed] [Google Scholar]
- 120.Kurokawa M, et al. Inhibition of apoptosome formation by suppression of Hsp90β phosphorylation in tyrosine kinase-induced leukemias. Mol Cell Biol. 2007;28:5494–5506. doi: 10.1128/MCB.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Desagher S, et al. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 122.Andersen JL, et al. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 2009;28:3216–3227. doi: 10.1038/emboj.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nutt KL, et al. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shin S, et al. Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J. 2005;24:3532–3542. doi: 10.1038/sj.emboj.7600827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Voss OH, et al. Regulation of monocyte apoptosis by the protein kinase Cdelta-dependent phosphorylation of caspase-3. J Biol Chem. 2005;280:17371–17379. doi: 10.1074/jbc.M412449200. [DOI] [PubMed] [Google Scholar]
- 126.Alvarado-Kristensson M, et al. p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J Exp Med. 2004;199:449–458. doi: 10.1084/jem.20031771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li X, et al. Phosphorylation of caspase-7 by p21-activated protein kinase (PAK) 2 inhibits chemotherapeutic drug-induced apoptosis of breast cancer cell lines. J Biol Chem. 2011;286:22291–22299. doi: 10.1074/jbc.M111.236596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jia SH, et al. Dynamic regulation of neutrophil survival through tyrosine phosphorylation or dephosphorylation of caspase-8. J Biol Chem. 2008;283:5402–5413. doi: 10.1074/jbc.M706462200. [DOI] [PubMed] [Google Scholar]
- 129.Cursi S, et al. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J. 2006;25:1895–1905. doi: 10.1038/sj.emboj.7601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin MC, et al. Protein kinase A regulates caspase-9 activation by Apaf-1 downstream of cytochrome c. J Biol Chem. 2005;280:15449–15455. doi: 10.1074/jbc.M414325200. [DOI] [PubMed] [Google Scholar]
- 131.Allan LA, et al. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol. 2003;5:647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 132.Dessauge F, et al. Identification of PP1alpha as a caspase-9 regulator in IL-2 deprivation-induced apoptosis. J Immunol. 2006;177:2441–2451. doi: 10.4049/jimmunol.177.4.2441. [DOI] [PubMed] [Google Scholar]
- 133.Allan AL, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell. 2007;26:301–310. doi: 10.1016/j.molcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 134.Laguna A, et al. The protein kinase DYRK1A regulates caspase-9-mediated apoptosis during retina development. Dev Cell. 2008;15:841–853. doi: 10.1016/j.devcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 135.Seifert A, et al. DYRK1A phosphorylates caspase 9 at an inhibitory site and is potently inhibited in human cells by harmine. FEBS J. 2008;275:6268–6280. doi: 10.1111/j.1742-4658.2008.06751.x. [DOI] [PubMed] [Google Scholar]
- 136.Seifert A, Clarke PR. p38alpha- and DYRK1A-dependent phosphorylation of caspase-9 at an inhibitory site in response to hyperosmotic stress. Cell Signal. 2009;21:1626–1633. doi: 10.1016/j.cellsig.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 137.Brady SC, et al. Regulation of caspase 9 through phosphorylation by protein kinase C zeta in response to hyperosmotic stress. Mol Cell Biol. 2005;25:10543–10555. doi: 10.1128/MCB.25.23.10543-10555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Raina D, et al. c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J Biol Chem. 2005;280:11147–11151. doi: 10.1074/jbc.M413787200. [DOI] [PubMed] [Google Scholar]
- 139.Cardone MH, et al. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 140.Zinkel SS, et al. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 141.Leung KT, et al. Activation of the JNK pathway promotes phosphorylation and degradation of BimEL--a novel mechanism of chemoresistance in T-cell acute lymphoblastic leukemia. Carcinogenesis. 2008;29:544–551. doi: 10.1093/carcin/bgm294. [DOI] [PubMed] [Google Scholar]
- 142.Chakraborty AR, et al. MAPK pathway activation leads to Bim loss and histone deacetylase inhibitor resistance: rationale to combine romidepsin with an MEK inhibitor. Blood. 2013;121:4115–4125. doi: 10.1182/blood-2012-08-449140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moujalled D, et al. Cyclic-AMP-dependent protein kinase A regulates apoptosis by stabilizing the BH3-only protein Bim. EMBO Rep. 2011;12:77–83. doi: 10.1038/embor.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fricker M, et al. Phosphorylation of Puma modulates its apoptotic function by regulating protein stability. Cell Death Dis. 2010;1:e59. doi: 10.1038/cddis.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sandow JJ, et al. Cytokine receptor signaling activates an IKK-dependent phosphorylation of PUMA to prevent cell death. Cell Death Differ. 2012;19:633–641. doi: 10.1038/cdd.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yan J, et al. Inactivation of BAD by IKK inhibits TNFα-induced apoptosis independently of NF-κB activation. Cell. 2013;152:304–315. doi: 10.1016/j.cell.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ye DZ, et al. p21-Activated kinase 1 (Pak1) phosphorylates BAD directly at serine 111 in vitro and indirectly through Raf-1 at serine 112. PloS one. 6:e27637. doi: 10.1371/journal.pone.0027637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harada H, et al. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 149.Bonni A, et al. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 150.Fang X, et al. Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene. 1999;18:6635–6640. doi: 10.1038/sj.onc.1203076. [DOI] [PubMed] [Google Scholar]
- 151.Tan Y, et al. p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J Biol Chem. 1999;274:34859–34867. doi: 10.1074/jbc.274.49.34859. [DOI] [PubMed] [Google Scholar]
- 152.Schürmann A, et al. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Moll Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gnesutta N, et al. The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J Biol Chem. 2001;276:14414–14419. doi: 10.1074/jbc.M011046200. [DOI] [PubMed] [Google Scholar]
- 154.Cotteret S, et al. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jin Z, et al. Survival function of protein kinase Cι as a novel nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-activated bad kinase. J Biol Chem. 2005;280:16045–16052. doi: 10.1074/jbc.M413488200. [DOI] [PubMed] [Google Scholar]
- 156.Harada H, et al. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc Natl Acad Sci U S A. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tan Y, et al. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 158.Lizcano JM, et al. Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J. 2000;349:547–557. doi: 10.1042/0264-6021:3490547. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 159.Lowman XH, et al. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell. 2010;40:823–833. doi: 10.1016/j.molcel.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 160.Lei K, Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100:2432–2437. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shao Y, Aplin AE. ERK2 phosphorylation of serine 77 regulates Bmf pro-apoptotic activity. Cell Death Dis. 2012;3:e253. doi: 10.1038/cddis.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lopez J, et al. Src tyrosine kinase inhibits apoptosis through the Erk1/2- dependent degradation of the death accelerator Bik. Cell Death Dis. 2012;19:1459–1469. doi: 10.1038/cdd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arokium H, et al. Substitutions of potentially phosphorylatable serine residues of Bax reveal how they may regulate its interaction with mitochondria. J Biol Chem. 2007;282:35104–35112. doi: 10.1074/jbc.M704891200. [DOI] [PubMed] [Google Scholar]
- 164.Shen ZJ, et al. The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat Immunol. 2009;10:257–265. doi: 10.1038/ni.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim BJ, et al. JNK- and p38 kinase-mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J Biol Chem. 2006;281:21256–21265. doi: 10.1074/jbc.M510644200. [DOI] [PubMed] [Google Scholar]
- 166.Linseman DA, et al. Glycogen synthase kinase-3beta phosphorylates Bax and promotes its mitochondrial localization during neuronal apoptosis. J Neurosci. 2004;24:9993–10002. doi: 10.1523/JNEUROSCI.2057-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gardai SJ, et al. Phosphorylation of Bax Ser184 by Akt regulates its activity and apoptosis in neutrophils. J Biol Chem. 2004;279:21085–21095. doi: 10.1074/jbc.M400063200. [DOI] [PubMed] [Google Scholar]
- 168.Xin M, Deng X. Protein phosphatase 2A enhances the proapoptotic function of Bax through dephosphorylation. J Biol Chem. 2006;281:18859–18867. doi: 10.1074/jbc.M512543200. [DOI] [PubMed] [Google Scholar]
- 169.Xin M, et al. Protein kinase Cζ abrogates the proapoptotic function of Bax through phosphorylation. J Biol Chem. 2007;282:21268–21277. doi: 10.1074/jbc.M701613200. [DOI] [PubMed] [Google Scholar]
- 170.Owens TW, et al. Apoptosis commitment and activation of mitochondrial Bax during anoikis is regulated by p38MAPK. Cell Death Differ. 2009;16:1551–1562. doi: 10.1038/cdd.2009.102. [DOI] [PubMed] [Google Scholar]
- 171.Fox JL, et al. Tyrosine dephosphorylation is required for Bak activation in apoptosis. EMBO J. 2010;29:3852–3868. doi: 10.1038/emboj.2010.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Azad A, et al. Blockade of the BAK hydrophobic groove by inhibitory phosphorylation regulates commitment to apoptosis. PLoS ONE. 2012;7:e49601. doi: 10.1371/journal.pone.0049601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang J, et al. Bcl-xL phosphorylation at Ser49 by polo kinase 3 during cell cycle progression and checkpoints. Cell Signal. 2011;23:2030–2038. doi: 10.1016/j.cellsig.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Terrano D, et al. Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol. 2010;30:640–656. doi: 10.1128/MCB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang J, et al. Phospho-Bcl-x(L)(Ser62) plays a key role at DNA damage-induced G(2) checkpoint. Cell Cycle. 2012;11:2159–2169. doi: 10.4161/cc.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.May WS, et al. Bcl-2 phosphorylation required for anti-apoptosis function. J Biol Chem. 1997;272:11671–11673. doi: 10.1074/jbc.272.18.11671. [DOI] [PubMed] [Google Scholar]
- 177.Deng X, et al. Survival function of ERK1/2 as IL-3-activated, staurosporine-resistant Bcl2 kinases. Proc Natl Acad Sci U S A. 2000;97:1578–1583. doi: 10.1073/pnas.97.4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yamamoto K, et al. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dimmeler S, et al. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med. 1999;189:1815–1822. doi: 10.1084/jem.189.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Breitschopf K, et al. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Mol Cell Biol. 2000;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kobayashi S, et al. Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem. 2007;282:18407–18417. doi: 10.1074/jbc.M610010200. [DOI] [PubMed] [Google Scholar]
- 182.Johnson CE, et al. Differential Apaf-1 levels allow cytochrome c to induce apoptosis in brain tumors but not in normal neural tissues. Proc Natl Acad Sci U S A. 2007;104:20820–20825. doi: 10.1073/pnas.0709101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tamm I, et al. XIAP expression correlates with monocytic differentiation in adult de novo AML: impact on prognosis. Hematol J. 2004;5:489–495. doi: 10.1038/sj.thj.6200549. [DOI] [PubMed] [Google Scholar]
- 184.Burstein DE, et al. Immunohistochemical detection of the X-linked inhibitor of apoptosis protein (XIAP) in cervical squamous intraepithelial neoplasia and squamous carcinoma. Ann Diagn Pathol. 2008;12:85–89. doi: 10.1016/j.anndiagpath.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 185.Moussata D, et al. XIAP as a radioresistance factor and prognostic marker for radiotherapy in human rectal adenocarcinoma. Am J Pathol. 2012;181:1271–1278. doi: 10.1016/j.ajpath.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 186.Dan HC, et al. Akt phosphorylation and stabilization of X-linked inhibitor of apoptosis protein (XIAP) J Biol Chem. 2004;279:5405–5412. doi: 10.1074/jbc.M312044200. [DOI] [PubMed] [Google Scholar]
- 187.Nakhaei P, et al. IκB kinase ε-dependent phosphorylation and degradation of X-linked inhibitor of apoptosis sensitizes cells to virus-induced apoptosis. J Virol. 2012;86:726–737. doi: 10.1128/JVI.05989-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2012;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 189.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 190.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 191.Yecies D, et al. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–3313. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wei G, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell. 2012;21:547–562. doi: 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cohen NA, et al. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol. 2012;19:1175–1186. doi: 10.1016/j.chembiol.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vo TT, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Parrish AB, et al. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol. 2013 doi: 10.1101/cshperspect.a008672. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tait SW, et al. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]