Summary
Background
In Drosophila embryos, the mid-blastula transition (MBT) dramatically remodels the cell cycle during the fourteenth interphase. Before the MBT, each cycle is composed of only a short S phase and mitosis. At the MBT, S phase is dramatically lengthened by the onset of late replication, and a G2 phase is introduced. Both changes set the stage for gastrulation and require downregulation of Cdc25 phosphatase, which was previously attributed to the elimination of its transcripts at the MBT.
Results
Premature removal of Cdc25 transcripts by RNAi did not affect progression to the MBT. Instead, an antibody against the Cdc25 isoform, Twine, showed that Twine protein was abundant and stable until the MBT, when it was destabilized and rapidly eliminated. Persistence of pre-MBT levels of Twine was sufficient to prevent cell cycle slowing. Twine protein destruction was timed by the nucleo-cytoplasmic ratio and depended on the activation of zygotic transcription at the MBT, including expression of the gene tribbles, whose activity was sufficient to trigger Twine destruction and was required for prompt Twine disappearance.
Conclusions
We propose that the developmentally regulated destruction of Twine protein is a critical switch that contributes to the cell cycle change at the MBT, including the addition of a G2 phase and onset of late replication. Moreover, we show that this destruction is triggered by the nucleo-cytoplasmic ratio-dependent onset of zygotic transcription of tribbles and other unknown genes.
Keywords: Drosophila, mid-blastula transition, Cdc25, destruction, Twine
Introduction
When the Drosophila embryo is first fertilized, its development is entirely controlled by a program orchestrated by its mother. Its nuclei sit transcriptionally silent in a vast syncytial cytoplasm packed with transcripts and proteins funneled in during oogenesis. Nuclei divide rapidly, on a dedicated mission to fill the expansive cytoplasm with enough DNA so that zygotic transcription can, when it is later activated, direct development. The nuclei progress directly from DNA replication to mitosis and then back again without pausing in gap phases. Even replication is fast during these cycles. Though some later S phases will take up to 8 hours, these earliest S phases range from only 3.4–12 minutes. Then, at the mid-blastula transition (MBT)—the first major developmental transition after fertilization—the embryo takes control of its own development. Cellularization envelops each nucleus in a cell membrane, so it no longer mingles with a massive cytoplasm. Many maternally-loaded messages are degraded, while zygotic transcription fills each cell with embryonically-derived messages. Cells scramble to organize themselves as they begin to gastrulate and differentiate into precursors of their eventual tissues. Finally, the cell cycle slows drastically, in concert with these other developmental processes, through two mechanisms. First, the length of DNA replication is dramatically extended by nearly four-fold at the MBT, to approximately 50 minutes, largely because sequences that replicated simultaneously before the MBT now replicate in a distinct and structured order[1]. Second, cells no longer enter mitosis immediately after completing replication—instead, they pause in a G2 phase of variable length, determined by each now-distinct cell[2].
Both changes have been linked to MBT-associated decline in Cdc25 activity. Cdc25 phosphatase activates the major mitotic kinase, Cdk1, by removing inhibitory phosphates from its ATP binding pocket[3]. In Drosophila, Cdc25 exists as two homologs—String and Twine—whose transcripts are abundant before the MBT[4]. Both homologs are functionally similar, though Twine is specifically required for the meiotic divisions[5, 6]. In pre-MBT embryos, both String and Twine are present, and maternal string is dispensable[4]. Following the MBT, the soma expresses only string, thus it is required for subsequent mitoses. String protein levels decline gradually over the pre-MBT blastoderm cycles, eventually vanishing shortly before the MBT[7]. Moreover, maternally-loaded transcripts encoding both String and Twine turn over at the MBT, during cycle 14[4].
Cdc25 downregulation allows Cdk1 kinase to accumulate inhibitory phosphorylation in cycle 14, leading to its inactivation[7]. One consequence is the addition of the G2 phase in cycle 14. When cells finish replication, they cannot enter mitosis until Cdk1 activity is restored by zygotic expression of cdc25, specifically string, which removes inhibitory phosphates from Cdk1 and activates it. Premature heat-shock expression of string is sufficient to cause cells to exit G2 and enter mitosis[2]. Our lab recently showed that S phase lengthening at the MBT also requires decline in Cdc25 and Cdk1 activity at the MBT[8]. Increased String or Twine in cycle 14 shortened post-MBT S phase by stimulating late-replicating sequences to replicate early. Premature downregulation of Cdk1 before the MBT extended the duration of replication in normally short S phases. Thus, downregulation of Cdc25 and consequent inhibition of Cdk1 seem to regulate the length of embryonic S phase. Given the importance of Cdc25 downregulation, we wanted to explore its mechanism and timing.
Results
Premature destruction of Cdc25 transcripts does not slow the cell cycle
Previous reports had noted destruction of cdc25 mRNA (both string and twine) in interphase 14 and attributed the decline in Cdc25 activity and slowing of the cell cycle to this[4]. To test whether decline in Cdc25 transcripts was sufficient to lengthen the cell cycle, we used dsRNA against both string and twine to eliminate these transcripts prematurely. The dsRNAs elicited strong knockdown within 25 minutes (Fig. 1A). Histone-GFP embryos[9] were injected with the dsRNAs 45 minutes before cycle 12, but imaging revealed no change in the length of either interphase 12 or 13 (Fig. 1B). This indicates that the elimination of string and twine transcripts is insufficient to trigger the cell cycle change that defines the MBT. Given the evidence that Cdc25 downregulation is important at the MBT, this suggested its regulation was at another level.
Figure 1. RNAi against string and twine does not lengthen the cell cycle.
(A) RT-PCR of string and twine in embryos injected with combined dsRNA against string and twine shows that the dsRNA triggers near complete destruction of these transcripts by 25 minutes after injection. (B) The length of interphases 12 and 13 was measured by H2AvD-GFP in uninjected embryos and embryos injected with dsRNA against string and twine 45 minutes before cycle 12.
Twine protein is degraded at the MBT
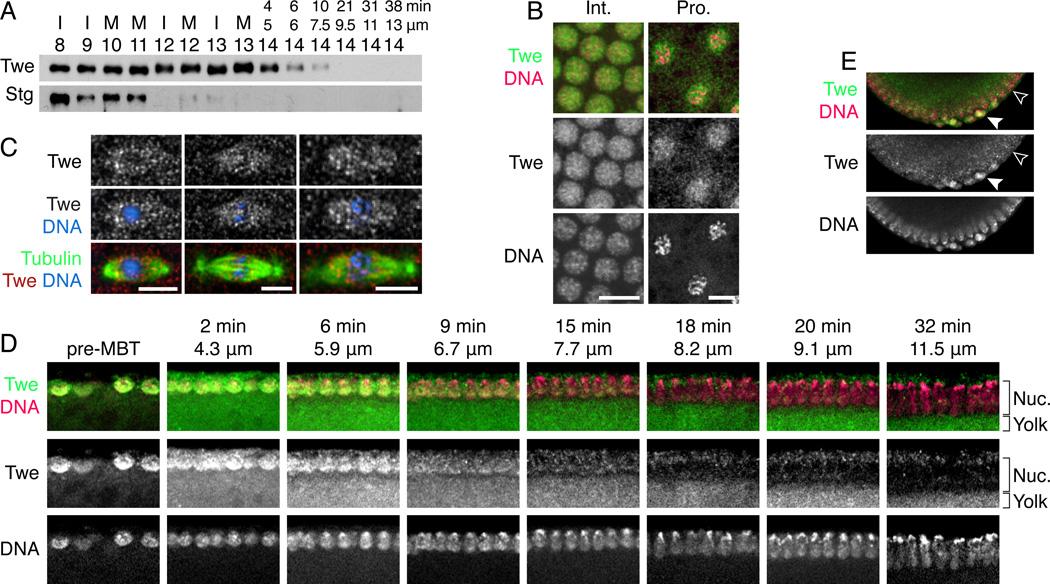
String protein declines during the blastoderm cycles, well before the MBT, making it an unlikely candidate for triggering the cell cycle change [7]. However, the behavior of Twine protein at the MBT had not been studied, so to test it, we raised an antibody against full-length Twine (Fig. S1). We probed the behavior of String and Twine proteins in the early embryo by performing a Western blot on precisely-staged single embryos [1]. String protein behaved as previously described—its level was highest in cycle 8 and declined gradually (Fig. 2A), becoming very low by cycle 12, though not completely eliminated until cycle 14. Twine protein, however, behaved quite differently. Its levels remained consistently high throughout the pre-MBT cycles, and it began to decline only upon entry to cycle 14. At that point, however, Twine levels dropped precipitously; a slight decrease was evident 4 minutes into cycle 14, and Twine became almost undetectable after 20 minutes.
Figure 2. Twine is destroyed in cycle 14, later than String.
(A) Western blot of single embryos showing the destruction of String (Stg) and Twine (Twe). Embryos are labeled with their cycle number (determined by nuclear spacing) and either cell cycle stage (I = interphase, M = mitosis) or nuclear length (µm) and calculated time (min) if they are in cycle 14. (B) Twine localization during interphase and prophase. DNA (Pico Green dye, magenta) and Twine (anti-Twe, green). (C) Twine enrichment on the spindle during metaphase and anaphase. Tubulin (anti-Tubulin, green), Twine (anti-Twine, red), and DNA (Hoecsht, blue). (D) Twine destruction over the course of cycle 14. Twine (anti-Twe, green) and DNA (Pico Green dye, magenta). Micrographs are labeled with nuclear length (µm) and calculated time in cycle 14 (min). Staining of Twine is the signal that colocalizes with nuclei, whereas signal below the nuclei is fluorescence of the yolk. (E) Twine is destroyed later in the pole cells (solid arrowhead) than somatic cells (open arrowhead). Twine (anti-Twe, shown in green), DNA (Pico Green, shown in magenta).
By immunofluorescence, Twine exhibited strong nuclear localization during interphase and prophase (Fig. 2B) and weak spindle enrichment during metaphase and anaphase (Fig. 2C). Spindle localization may be relevant to Twine’s requirement in meiosis, as its mutant phenotypes include defects in meiotic spindles[5]. Immunofluorescence also confirmed Twine’s destruction during cycle 14. During interphase 14, cellularization of the syncytial embryo results in highly stereotyped elongation of the nuclei. Thus, in sagittal sections of embryos, age can be determined by nuclear length [1]. Twine antibody stained nuclei at the beginning of cycle 14 about as intensely as in pre-MBT cycles (Fig. 2D). Note that the yolk, pictured directly below the nuclei, fluoresces, but this does not represent staining of Twine protein. As cycle 14 progressed, evidenced by the lengthened nuclei, Twine staining diminished from 2–21 minutes, in accord with our analysis by Western blot. The stainings revealed an interesting exception to the cycle 14 destruction of Twine; Twine persisted in the pole cells (Fig. 2E, closed arrowhead) after its disappearance from somatic cells (open arrowhead). We conclude that Twine protein level declines abruptly in the soma early in interphase 14.
Twine protein is highly stable before the MBT
The decline in Twine protein levels could represent either a change in protein stability at the MBT, or a decrease in translation that results in elimination of already unstable protein. To differentiate between these possibilities, we knocked down twine transcript several cycles early by dsRNA injection (Fig. 1A). Embryos injected with twine dsRNA retained at least half their protein for as much as an hour after injection (Fig. 3A–C, 64 min vs. 1/2 pre). Since twine mRNA was effectively removed by 25 minutes after injection, this means that, before the MBT, half-destruction of Twine requires approximately 30 minutes or more.
Figure 3. Twine protein is relatively stable until the MBT, and pre-MBT Twine level causes a penetrant phenotype.
(A–C) Western blots of embryos treated with twine RNAi, showing that Twine protein level does not depend on continued translation of its mRNA. Controls: a pre-MBT embryo (“pre”) and a 1:2 dilution (“1/2 pre”), an embryo in cycle 14 after Twine destruction (“late”). Embryos were injected at different times, aged for the time marked above each lane, and picked in cycle 11 (A), cycle 12 (B), or cycle 13 (C), as gauged by nuclear distance. (D–E) Blots showing stability of Twine protein after injection with cycloheximide (CHX), an inhibitor of translation. Dilutions of an uninjected embryo are shown (“1:1”, “1:2”, and “1:4”). Embryos were injected with CHX in cycle 12 (D) or cycle 13 (E) (and thereby arrested in that cycle) and aged for the number of minutes indicated above the lane, before being picked. (F) H2AvD-GFP embryos were injected in one end with twine mRNA at varying concentrations, and imaged to determine how many embryos underwent a shortened S phase or G2 as a result. (G) Western blot of Twine in embryos either before the MBT (“pre-MBT”) or 15 minutes into cycle 14, injected with twine mRNA at varying concentrations.
We separately investigated Twine protein stability before the MBT by assaying its persistence following the inhibition of translation. We injected cycle 12 and 13 embryos with cycloheximide, an inhibitor of translation, aged them, and then blotted single embryos to measure remaining Twine protein. Embryos treated with cycloheximide in cycle 12 showed almost no change in protein level after treatment—even 50 minutes later, embryos did not seem to have significant degradation of the protein (Fig. 3D). Embryos treated in cycle 13 exhibited slightly different behavior—there was gradual destruction of Twine (Fig. 3E). We conclude that Twine stability changes as the embryo approaches the MBT. It is stable in early cycles and half-destruction of Twine takes about 30 min in cycle 13 (Fig 3: compare 30/31 min vs. 50 or 60 min). The dramatic decline in its levels in cycle 14 suggests a much shorter time for half-destruction after the MBT, around 5 min (Fig. 2A: compare 4 min vs. 10 min). Thus, Twine protein is destabilized at the MBT, likely due to the activation of a protein destruction mechanism. Moreover, Twine’s stability before the MBT explains why dsRNA against string and twine did not affect the cell cycle—removal of Cdc25 transcripts does not eliminate Twine protein.
We previously established that expression of Twine in cycle 14 prevents the MBT changes to the cell cycle—both shortening S phase and eliminating the G2 [8]. To estimate how essential activation of Twine destruction is for development, we wanted to compare the level of Twine required to shorten the cell cycle to the amount present before the MBT. Thus, we injected different concentrations of twine mRNA into histone-GFP embryos and assayed the percentage with a shortened cell cycle. Increased twine mRNA resulted in increased phenotype penetrance (Fig. 3F). We then injected embryos with twine mRNA and blotted them to assay the amount of Twine protein produced by each treatment, compared to pre-MBT embryos. Increased twine transcript resulted in increased production of Twine protein (Fig. 3G). Injection of 600 ng/µl twine mRNA produced slightly less Twine protein in cycle 14 than was present before the MBT, but was sufficient to cause a cell cycle phenotype in nearly half of treated embryos. This underscores that the timed destruction of Twine protein is critical for executing the MBT, as the amount of Twine protein present before the MBT prevents proper slowing of the cell cycle.
Twine is destroyed according to the nucleo-cytoplasmic ratio
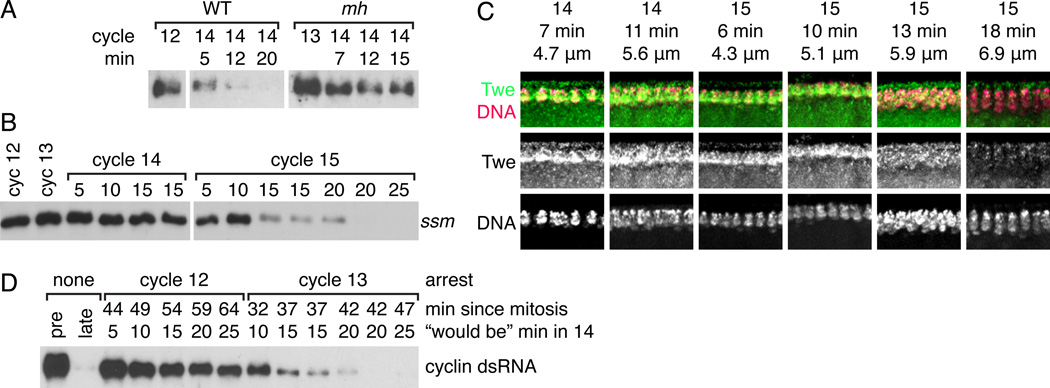
Since the MBT is exquisitely timed, and Twine destruction is important for triggering it, how then is the destruction of Twine scheduled? Slowing of the cell cycle is timed in Drosophila embryos by the nucleo-cytoplasmic ratio (NC ratio)—that is, its activation occurs when the embryo, through DNA replication and mitosis, amasses a particular amount of DNA. Normally, this threshold is reached in cycle 14, but this can be altered by changing the DNA content of each nucleus (e.g. with compound chromosomes or translocations), which changes the number of cell cycles required to amass the threshold amount of DNA and trigger the cell cycle change[10, 11]. Since the NC ratio times the cell cycle slowing, we tested whether it also times Twine destruction by looking in two maternal-effect mutants, mh and ssm, that produce haploid embryos (here called mh embryos and ssm embryos)[10–14]. These embryos do not incorporate the paternally provided chromosomes into the initial mitotic spindle, so they are lost and each nucleus has half the normal amount of DNA[14, 15]. These embryos lengthen interphase and S phase 15 instead of 14[10].
Western blots showed that Twine remained stable during cycle 14 in mh and ssm embryos (Fig. 4A–B). An extended time course in ssm embryos showed Twine persistence in cycle 14 with a decline in cycle 15 similar to that which usually occurs in cycle 14 in wild-type embryos (Fig. 4B). Twine antibody staining of ssm embryos (Fig. 4C) showed that levels remained high throughout cycle 14 and into early 15, and then declined over the course of cycle 15 (5.9µm and 6.7µm), all as seen by Western blot. These results show that the timing of Twine destruction is determined by the NC ratio, as it is delayed a cycle when the DNA content of the embryo is reduced.
Figure 4. The onset of Twine destruction responds to the nucleo-cytoplasmic ratio.
(A) Western blot comparing destruction of Twine in Sevelen (WT) embryos and maternal haploid embryos (mh). The cycle and time in cycle 14 (determined visually on the microscope) is marked above each lane. (B) Western blot showing destruction of Twine in haploid ssm embryos. Time in cycle 14 or 15 is marked above each lane. (C) Immunofluoresence showing Twine destruction in cycle 15 in ssm embryos. Twine (anti-Twe, green) and DNA (Pico Green dye, magenta). Micrographs are labeled with cycle, nuclear length (µm) and calculated time in cycle 14 or 15 (min) (see Fig. S2). Twine staining is signal that colocalizes with nuclei, whereas signal below the nuclei is fluorescence from the yolk. (D) Western blot showing Twine stability in embryos arrested with dsRNA against the mitotic cyclins. Lanes are labeled from top to bottom with: the cycle in which the embryo arrested, the number of minutes since it entered that cycle, and finally a calculated time, describing how far into the cycle 14 the embryo would be if it had not been arrested. Controls: a pre-MBT embryo (“pre”) and an embryo in cycle 14 after Twine destruction (“late”).
We also performed an experiment to see whether embryos had to reach a particular NC ratio to activate destruction of Twine protein. To do this, we arrested the cell cycle using injection of dsRNA against the mitotic cyclins, which prevented embryos from entering mitosis (and thus subsequent S phases)[16]. We arrested embryos in both cycles 12 and 13, meaning that those embryos never amassed cycle 14 amounts of DNA. Following their last mitosis, the embryos were aged for a length of time equivalent to that required to enter cycle 14, when Twine is normally destroyed in wild-type embryos. We assayed levels of Twine in these embryos and found that embryos arrested in cycle 13 eventually destroyed Twine rapidly, much as would have been seen in cycle 14 (Fig. 4D). Embryos arrested in cycle 12, however, only showed gradual destruction of Twine, rather than the rapid destruction characteristic of the MBT. Thus, we conclude that cycle 13 nuclear density in a diploid embryo triggers the events that, after a short lag, result in the destruction of Twine in cycle 14.
Twine destruction does not require Chk1 phosphorylation
But, how does the NC ratio trigger the destruction of Twine? Data from several systems suggested that gradual activation of Chk1 DNA damage kinase as DNA content increased could be responsible. For instance, in Xenopus, Cdc25 must also be inactivated to slow the cell cycle at the MBT, including the Cdc25A isoform, which seems to be targeted for degradation by Chk1 phosphorylation[17]. Chk1-targeting of Cdc25A for destruction is also conserved to mammals[18]. In Drosophila, maternal grapes (Chk1 homolog) is required for cell cycle slowing the MBT [19, 20], and String protein does not decline on time in grapes mutant embryos[21]. Thus, it seemed that Twine’s destruction may also depend on phosphorylation by Chk1.
To test this, we assayed Twine destruction in grapes mutants by Western blot. We confirmed that String protein was stabilized in a grapes mutant (Fig. 5A), as reported previously[21]. Twine protein was also stabilized, persisting at high levels for more than 40 minutes, far longer than in a wild-type embryo. These findings suggested that degradation of Twine may require phosphorylation by grapes. However, the grapes mutant is highly disturbed, and the embryo arguably never truly starts an interphase 14—for instance, grapes embryos do not activate zygotic transcription, do not cellularize, and do not attempt to gastrulate [22].
Figure 5. Transcription, but not Chk1 phosphorylation, is required for Twine destruction at the MBT.
(A) Western blot of String (Stg) and Twine (Twe) in embryos from mothers mutant for grapes (grp) (the Drosophila homolog of Chk1). Lanes are labelled with cycle and time after mitosis 13 (measured visually on scope) (B) Western showing normal destruction of String and Twine in embryos mutant for grapes and mnk (also known as loki, the Drosophila homolog of Chk2). Embryos are labeled with cycle and nuclear length (µm). Embryos labeled 14/15* are either cycle 14 or 15, which cannot be differentiated by nuclear spacing in this strain. (C) Western of embryos injected with either PBS or alpha-amanitin, an inhibitor of RNA polymerase II. Lanes are labeled with the cycle of the embryo and the time in cycle 14. (D) Embryos injected in late cycle 13 with either vehicle (1% DMSO) or alpha-amanitin (an inhibitor of RNA polymerase II) and cycloheximide (an inhibitor of translation), dissolved in 1% DMSO.
Mutation of maternal mnk (the Drosophila homolog of Chk2, also known as loki) rescues some defects of the grapes mutant[22]. Embryos from grapes mnk mutant mothers do not hatch, but they cellularize, exhibit gastrulation movements, and activate zygotic transcription. Using this less pleiotropic double mutant that still cannot confer Chk1 phosphorylation on substrates, we tested whether String and Twine proteins were degraded normally in maternal mutant grapes mnk embryos. String protein declined between cycles 12 and 13, delayed one cycle compared to wild-type. Destruction of Twine protein also occurred in grapes mnk mutants (Fig. 5B), though we are unsure of the timing of this destruction, because we conducted this analysis with fixed embryos. grapes mnk embryos go through an extra short cycle without segregating their chromosomes, thus cycle 14 and 15 embryos cannot be distinguished by nuclear spacing[22]. Because the embryos are cellularizing at the time Twine protein is destroyed, we suspect the destruction occurs in cycle 15, which would be a delay of one cycle, or about 12 minutes later than normal. Though the coordination of MBT events with cycle 14, including Twine destruction, appears disturbed in the double mutant, the activation of an abrupt destruction program for Twine is still observed, leading us to conclude that direct phosphorylation by Grapes (or Chk1) is not required for the MBT-associated destruction of Twine.
Transcription is required for Twine destruction
What other factors might promote the onset of Twine destruction? One of the most dramatic changes in the embryo at the MBT, aside from the remodeling of the cell cycle, is the onset of zygotic transcription. The transcripts produced enable many of the other changes that happen at the MBT, including cellularization and gastrulation. Moreover, the transcription of many genes responds to the NC ratio and is delayed one cycle in haploid mutants[11]. Inhibition of zygotic transcription prevents the onset of late replication, which requires the destruction of Twine[1]. Finally, zygotic transcription marks a difference between grapes and grapes mnk double mutants—grapes mnk embryos activate transcription, whereas grapes embryos do not[22]. This suggested that new gene products expressed upon reaching a critical NC ratio could be an important trigger for the destruction of Twine. Thus, we assayed Twine levels in embryos injected with alpha-amanitin, a toxin that inhibits RNA polymerase II, or injected with both alpha-amanitin and cycloheximide, inhibitors of transcription and translation. In vehicle-injected embryos, Twine declined rapidly after the MBT (Fig. 5C, “PBS”), but in embryos injected with alpha-amanitin, Twine protein was not eliminated. It persisted more than 35 minutes into cycle 14 with no apparent change in protein level (Fig. 5C, “alpha-amanitin”). Embryos injected with both alpha-amanitin and cycloheximide (Fig. 5D) showed a gradual decline in Twine (half-destruction took longer than 28 min, Fig. S3), similar to the destruction rate we measured for Twine in cycle 13. Thus, the addition of drugs that block gene expression prevent the MBT associated increase in Twine turnover in cycle 14. Since the slow turnover of Twine in the absence of protein synthesis (Fig. 5D) is similar to the turnover in pre-MBT cycle 13 embryos, we conclude that new zygotic gene expression is needed to drive the acceleration of Twine turnover at the MBT. The absence of any apparent decay of Twine in alpha-amanitin alone seems to occur because, in the absence of cycloheximide, translation of unusually persistent Twine message[4] replaces the Twine lost to slow decay.
The tribbles gene is a key, but not exclusive, contributor to Twine destruction
Of course, the next major question was to determine the identity of the zygotically expressed genes involved in Twine destruction. Published results highlighted the kinase-like tribbles as a good candidate for a zygotically expressed gene required for Twine destruction. Tribbles reduced levels of transfected HA-Twine in cell culture[23], and injection of tribbles mRNA arrested embryos in cycle 13[24], which could be consistent with premature destruction of Twine.
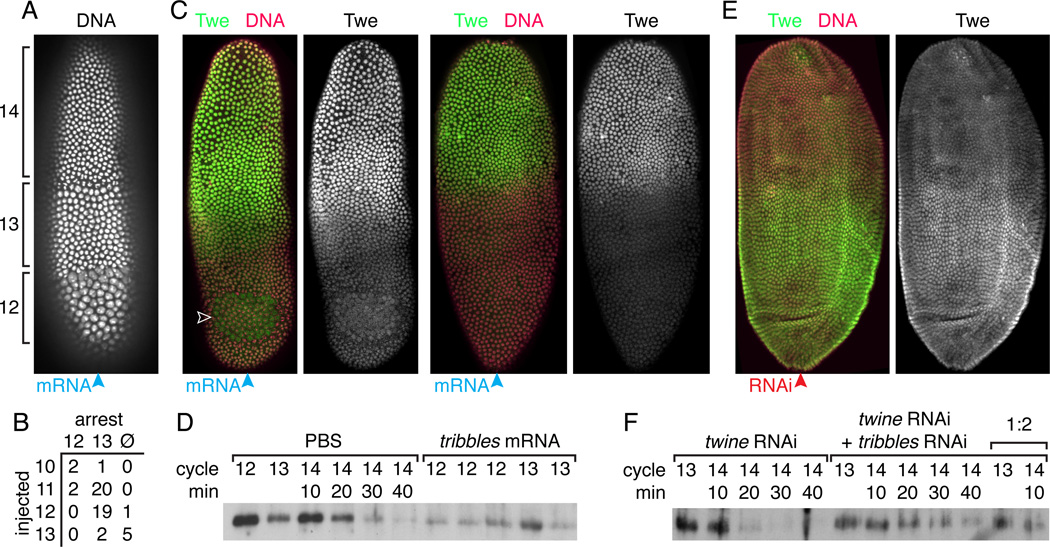
We first verified that Tribbles could arrest embryos. Injection of tribbles mRNA did arrest embryos in interphase near the site of injection (Fig. 6A), but contrary to what was previously shown, the arrest was not limited to cycle 13; embryos also arrested in cycle 12, dependent on the time of their injection (Fig. 6A–B). To see whether this arrest was associated with the destruction of Twine, we injected embryos with tribbles mRNA in one pole, fixed them after 20 minutes, and stained for Twine. We consistently saw reduced Twine staining near the site of injection (Fig. 6C). First shown is an embryo where the tribbles mRNA caused a small zone of cycle 13 arrest (white arrowhead) and significantly reduced Twine staining in the half of the embryo near the injected pole. Next shown is an embryo where the tribbles mRNA did not cause an arrest, but the injected end of the embryo showed reduced staining, indicating that tribbles accelerated Twine destruction. Moreover, this shows that tribbles acceleration of Twine destruction does not require cell cycle arrest. We also injected embryos three times along their length with tribbles mRNA to express it throughout the embryo and analyzed these embryos by Western blot. We saw significant reduction in Twine levels in cycle 12 and cycle 13 embryos (Fig. 6D), so that they resembled those midway through the Twine destruction program that normally occurs in cycle 14. These results show that tribbles expression is sufficient to trigger destruction of Twine protein. Moreover, consistent with the result that tribbles expression can induce arrest in cycles 12 or 13, it can also trigger Twine destruction in cycles 12 and 13.
Figure 6. Tribbles induces the destruction of Twine protein at the MBT.
(A) H2AvD-GFP showing the cycle 12 and 13 arrests induced by injection of tribbles mRNA into one end (blue arrowhead) of an embryo. (B) Table comparing numbers of embryos that arrested in cycle 12, cycle 13, or did not arrest (Ø), based on the cycle in which they were injected. (C) Images of two embryos injected at one pole with tribbles mRNA (blue arrowhead), demonstrating the local destruction of Twine protein near the site of injection and induced cell cycle arrest (open arrowhead). Twine (anti-Twe, green) and DNA (Pico Green dye, magenta). (D) Western blot, probed with anti-Twe, showing decreased Twine in embryos injected with tribbles mRNA relative to control. (E) An embryo injected at one pole with tribbles RNAi (red arrowhead) demonstrating local stabilization of Twine protein near the site of injection. Twine (anti-Twe, green) and DNA (Pico Green dye, magenta). (F) Western blot, probed with anti-Twe, showing that in embryos treated with twine RNAi to prevent continued Twine translation, Twine persists longer after the MBT in embryos also treated with tribbles RNAi. Lanes are labelled with cycle and time into cycle 14, determined visually on the scope. 1:2 indicates a 1/2 dilution of a sample.
In addition to being capable of triggering Twine destruction, tribbles seems to be transcribed at the proper time to cause the observed program of Twine destruction. RNAseq experiments with single, staged embryos have shown that tribbles mRNA is upregulated at the MBT[25]. There is a small amount of maternal transcript loaded, but this begins to increase, first slightly in cycle 13, then dramatically in the first half of cycle 14. This is consistent with the very slight destabilization of Twine protein in cycle 13 and its dramatic destabilization in cycle 14. Moreover, a microarray study suggests that the transcription of tribbles responds to the NC ratio[11]. Thus, the transcriptional profile mirrors what would be expected of a zygotic transcript that regulated Twine destruction.
However, while the phenotype of zygotic tribbles mutants suggested that it plays a role in triggering the MBT, it did not support the hypothesis that tribbles alone is responsible for the destruction of Twine. A small percentage of embryos that are zygotically mutant for tribbles have a short interphase 14[23], but far fewer embryos than would be expected if there were no turnover of Twine. To determine the degree to which tribbles mutants stabilized Twine, we injected wild-type embryos with dsRNA against tribbles to prevent its expression and stained them with Twine antibody (Fig. 6E). These embryos showed continued presence of Twine protein near the site of injection after it was already degraded in the rest of the embryo. This suggested that tribbles expression is involved in the MBT destruction of Twine. We then injected embryos along their length with either twine dsRNA or a combination of twine and tribbles dsRNAs. Knockdown by twine dsRNA prevented further translation of Twine, allowing comparison of the perdurance of Twine protein at the MBT with and without expression of tribbles at the MBT, depending on whether it was knocked down with dsRNA. Twine protein persisted longer (half-destruction took approximately 20 minutes) in embryos injected with tribbles dsRNA than those that were not (Fig. 6F), but it was not as stable as in pre-MBT embryos (Fig. 3C). These results indicate that zygotic transcription of tribbles contributes to the destruction of Twine at the MBT. Since it is sufficient to destroy Twine, and its transcription is strongly upregulated at the MBT in an NC-dependent manner, we suggest tribbles is one of the transcripts that regulates Twine destruction and thus the cell cycle change at the MBT. However, the low-penetrance cell cycle phenotype and only partial stabilization of Twine in the absence of tribbles suggests that there may be another gene that participates as well.
Discussion
Our findings outline a linear regulatory cassette that contributes to MBT timing and slowing of the cell cycle—increasing NC ratio triggers zygotic expression, including tribbles, which activates Twine protein destruction. This promotes the downregulation of Cdk1, which underlies both extension of S phase and introduction of a G2. We propose that the primary mode of Cdc25 downregulation at the MBT is via destruction of Twine protein, rather than the elimination of its mRNA as has been previously proposed. Moreover, we propose that the NC ratio times cell cycle slowing by triggering transcription of multiple genes that act in parallel to downregulate Cdk1 in cycle 14, including by activation of Twine destruction.
Destruction of Twine protein is critical for the MBT
Slowing of the cell cycle at the MBT—both onset of late replication and addition of a G2—correlates with the decline in Cdk1 activity [2, 7, 8]. Premature downregulation of Cdk1 advanced the slowing of the cell cycle and increased Cdk1 activity in cycle 14 prevented cell cycle slowing by causing early replication of late replicating DNA sequences and early entry into mitosis[8]. Together, these results argued that the downregulation of Cdk1 regulates the slowing of the cell cycle.
Cdk1 is downregulated in cycle 14 by accumulation of inhibitory phosphorylation that is normally counteracted by Cdc25, showing the importance of Cdc25 downregulation[7]. Increased dose of maternal twine, but not maternal string, generates a low-penetrance extra syncytial division (ESD) phenotype where the cell cycle fails to slow in cycle 14[4]. Twine protein is destroyed right at the beginning of the MBT, and sustained Twine at its pre-MBT level causes a high-penetrance ESD phenotype. Since the elimination of twine transcript does not remove Twine protein, this shows that the destruction of Twine protein is a key regulator of the downregulation of Cdk1 and thus slowing of the cell cycle at the MBT.
Twine destruction cooperates with other inputs into Cdk1 downregulation
Previous publications identified other conditions that generate partially penetrant ESD phenotypes at the MBT, and thereby identified other inputs into MBT cell cycle slowing. One such input is frühstart, an inhibitor of cyclin-Cdk that is first expressed at the MBT[26]. Like the early induction of Twine destruction, early expression of frühstart arrested the cell cycle by opposing cyclin-Cdk1, though likely through a mechanism independent of inhibitory phosphorylation. Mutants in frühstart have a low-penetrance ESD phenotype. However, frühstart acts in concert with the destruction of Twine, as increased maternal dose of twine in frühstart mutants, strongly intensifies the penetrance of the ESD phenotype. Another experiment showed that high levels of cyclin B injected during the first few minutes of cycle 14 (but not after) can cause an ESD phenotype[27]. This suggests that cyclin degradation during mitosis 13 provides a buffer by keeping Cdk1 inactive early in cycle 14, allowing time for these other inputs into Cdk1 downregulation—Twine destruction and frühstart expression—to take effect. However, in the end, all of these inputs seem to be a set of partially redundant pathways that feed into the ultimate downregulation of Cdk1 at the MBT to accomplish the slowing of the cell cycle.
Zygotic genome activation triggers Cdk1 downregulation
Several models that involve the titration or rundown of replicative or mitotic factors have been proposed to explain the MBT slowing of the cell cycle, and such models could generally be compatible with the downregulation of Cdk. However, the inhibition of zygotic transcription by alpha-amanitin invariably shortened S phase and eliminated G2, which argues against a direct titration model and instead suggests that zygotic transcription is actually upstream of cell cycle slowing[1]. This is in accord with findings that the onset of transcription of numerous genes is dependent on the NC ratio[11] and with the findings we present here. It also suggests that if detection of the NC ratio depends on titration of some component, it is likely some form of transcriptional repressor. Despite zygotic transcription’s importance for triggering the MBT, thorough screening and genome-wide analysis by chromosomal aneuploids has not revealed any single gene that is absolutely required for the MBT, suggesting that multiple, partially redundant transcripts work together[24, 28, 29]. This is consistent with the multiple inputs into the downregulation of Cdk1 described above.
The novel expression of frühstart at the MBT obviously requires the activation of the zygotic genome. However, in this study, we show that the destruction of Twine is also downstream of the activation of zygotic transcription. We uncover one gene, tribbles, that is sufficient to destroy Twine, and is expressed at the MBT, but it does not seem to be fully required. Removal of tribbles results in only partial stabilization of Twine and a much lower penetrance ESD phenotype than would be expected from complete stabilization of Twine. The simplest, but not the only, explanation for this is that there are one or more redundant, as yet undiscovered genes also expressed at the MBT, which compensate for tribbles in its absence. Regardless, these results show that both major inputs into Cdk1 downregulation and slowing of the cell cycle at the MBT are triggered by the activation of zygotic transcription at the MBT.
NC as a trigger of cell cycle slowing at the MBT
Recent research has shown zygotic genome activation is not a single event—some genes respond to the NC ratio and delay transcription in haploid embryos, whereas other genes are transcribed in cycle 14 regardless of embryo ploidy[11]. Both the slowing of the cell cycle[10], generally, and the destruction of Twine, specifically, are regulated by the NC ratio. Both of the identified transcriptional inputs into this process—frühstart and tribbles—are genes regulated by the NC ratio[26]. Furthermore, by arresting the cell cycle with cyclin RNAi, we show that the capacity to begin accumulating Twine degradation capability first appears in cycle 13. This suggests that, upon reaching the nuclear density corresponding to a diploid cycle 13 embryo, transcription of genes that promote Twine destruction commences, as has been observed for tribbles. Thus, we propose that the primary input of the NC ratio into timing the MBT is to activate the transcription of a group of genes that collectively act to downregulate Cdk1.
The mechanism by which the NC ratio can determine the time of transcription remains a mystery. However, we do think our experiments eliminate some previously popular theories. The interphases of the pre-MBT cycles are impressively short, but each syncytial blastoderm interphase is slightly longer than the one preceding it[30]. Since the entry into mitosis aborts nascent transcripts in progress[31], one theoretical mechanism for limiting transcription until cycle 13 would be to have a gene that was too long to be fully transcribed in the slightly shorter preceding interphases. However, if this were the mechanism limiting the transcription of tribbles, then extending an earlier, shorter cycle should enable transcription of tribbles to be completed and trigger premature destruction of Twine, but we do not observe this when cycle 12 is extended by cyclin RNAi. Another popular model is that Cdk1 phosphorylation of RNA polymerase II’s C-terminal domain limits its activity and prevents zygotic transcription[32]. Then, as Cdk1 activity declines in the early embryo, for instance through progressive destruction of cyclins, this would enable transcription to begin. If this limited tribbles transcription, however, premature downregulation of Cdk1 should enable premature transcription of tribbles and destruction of Twine, but we do not observe this after cyclin RNAi. Thus, some other mechanism must respond to the NC ratio and govern the onset of transcription of tribbles and presumably frühstart, and thus cell cycle slowing.
Not all aspects of the MBT, however, are regulated by the NC ratio. Morphogenetic events, such as cellularization and gastrulation are not delayed in haploid embryos or embryos prevented from reaching cycle 14 nuclear density by cyclin RNAi [10, 16]. However, these events are sensitive to the cell cycle. Cellularization and early gastrulation movements, such as ventral furrow formation, are disrupted by entry into mitosis [10, 24, 26, 33]. This means that a properly slowed cell cycle is a required permissive condition even for many events that are not directly timed by the NC ratio. Thus, we suggest that the study of the regulation and activation of zygotic transcription determined by the NC ratio, especially tribbles and frühstart, is the next frontier in understanding the MBT.
Experimental Procedures
Considerably more detailed procedures included in the Supplement, including strains, constructs, primers, and detailed protocols.
Twine antibody
Full-length Twine protein was cloned into pET28, expressed in BL21(DE3) cells, and then purified using nickel agarose beads (Qiagen) under denaturing conditions. 2 rabbits were immunized by Pacific Immunology, and the sera was then purified against a column loaded with purified Twine protein.
Western blotting
Single embryos were either fixed and staged by nuclear length (Figs. 2A and 5B) using Pico Green (Molecular Probes) or by time after mitosis 13 and fixed in 1:1 methanol:heptane. Individual embryos were lysed in 2x SDS sample buffer and blotted using 1:1000 Twine antibody overnight and 1:10000 Donkey anti-HRP (Jackson Labs).
Immunofluorescence
Fixed embryos (3.7% formaldehyde for 20 minutes, or 1:1 methanol heptane for spindle localization) were stained with 1:500 anti-Twine or a mixture of 1:50 each anti-Tubulin AA12.1, AA4.3, and E7 mouse monoclonals (DSHB) overnight and counter-stained for 15 minutes with 1:500 Pico Green (Invitrogen, Molecular Probes).
Embryo Injection
dsRNA was prepared as described [16] and injected at concentrations of approximately 1–2mg/ml in PBS. mRNA was produced as previously described [8] and injected at 800ug/µl, unless otherwise noted. Templates for all dsRNA and mRNA are detailed in the Supplement. Cycloheximide (Sigma-Aldrich) was injected at 1mg/ml in 1% DMSO. Alphaamanitin (Calbiochem) was injected at 1mg/ml.
Supplementary Material
Highlights.
-
–
Premature knockdown of cdc25 mRNA does not slow the cell cycle
-
–
Twine (Cdc25) protein is stable until the MBT, when it is destabilized and degraded
-
–
Twine destruction is timed by the nucleo-cytoplasmic ratio
-
–
Twine destruction requires zygotic transcription, including transcription of tribbles
Acknowledgments
We thank the Wieschaus lab, Theurkauf lab, and Bloomington Stock Center for strains. We thank members of the O’Farrell laboratory, Stefano di Talia, Eric Wieschaus, Barbara Panning, David Morgan, and Lauren Booth for helpful comments. This research was supported by a National Science Foundation Graduate Research Fellowship (to J.A.F.) and National Institutes of Health grant GM037193 (to P.H.O.).
References
- 1.Shermoen AW, McCleland ML, O'Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20:2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar BA, O'Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 4.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 5.White-Cooper H, Alphey L, Glover DM. The cdc25 homologue twine is required for only some aspects of the entry into meiosis in Drosophila. J Cell Sci. 1993;106(Pt 4):1035–1044. doi: 10.1242/jcs.106.4.1035. [DOI] [PubMed] [Google Scholar]
- 6.Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- 7.Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell JA, Shermoen AW, Yuan K, O'Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 2012;26:714–725. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- 10.Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleocytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zalokar M, Audit C, Erk I. Developmental defects of female-sterile mutants of Drosophila melanogaster. Dev Biol. 1975;47:419–432. doi: 10.1016/0012-1606(75)90295-x. [DOI] [PubMed] [Google Scholar]
- 14.Loppin B, Docquier M, Bonneton F, Couble P. The maternal effect mutation sésame affects the formation of the male pronucleus in Drosophila melanogaster. Dev Biol. 2000;222:392–404. doi: 10.1006/dbio.2000.9718. [DOI] [PubMed] [Google Scholar]
- 15.Loppin B, Berger F, Couble P. Paternal chromosome incorporation into the zygote nucleus is controlled by maternal haploid in Drosophila. Dev Biol. 2001;231:383–396. doi: 10.1006/dbio.2000.0152. [DOI] [PubMed] [Google Scholar]
- 16.McCleland ML, O'Farrell PH. RNAi of Mitotic Cyclins in Drosophila Uncouples the Nuclear and Centrosome Cycle. Current Biology. 2008;18:245–254. doi: 10.1016/j.cub.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21:3694–3703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 19.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 20.Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 21.Su TT, Campbell SD, O'Farrell PH. Drosophila grapes/CHK1 mutants are defective in cyclin proteolysis and coordination of mitotic events. Curr Biol. 1999;9:919–922. doi: 10.1016/s0960-9822(99)80399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada S, Kwak S, Koppetsch BS, Theurkauf WE. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development. 2007;134:1737–1744. doi: 10.1242/dev.02831. [DOI] [PubMed] [Google Scholar]
- 23.Mata J, Curado S, Ephrussi A, Rørth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 24.Grosshans J, Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell. 2000;101:523–531. doi: 10.1016/s0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- 25.Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9:e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosshans J, Müller HAJ, Wieschaus E. Control of cleavage cycles in Drosophila embryos by frühstart. Dev Cell. 2003;5:285–294. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- 27.Royou A, McCusker D, Kellogg DR, Sullivan W. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J Cell Biol. 2008;183:63–75. doi: 10.1083/jcb.200801153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wieschaus E, Sweeton D. Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development. 1988;104:483–493. doi: 10.1242/dev.104.3.483. [DOI] [PubMed] [Google Scholar]
- 29.Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- 30.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 31.Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebara MM, Sayre MH, Corden JL. Phosphorylation of the carboxyterminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J. Cell. Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- 33.Seher TC, Leptin M. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr Biol. 2000;10:623–629. doi: 10.1016/s0960-9822(00)00502-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.