Abstract
Design of materials to aid intracellular delivery of agents can greatly improve medical treatments. While DNA is a molecule difficult to introduce into cells, DNA can be engineered into devices capable of intracellular delivery. Yet, transport mediated by DNA-devices void of other structural material, with size greater than that associated with non-specific penetration, and with targeting capacity enough to overcome non-specific pathways has not been achived. This study demonstrates that this is possible. Submicrometer (200-nm) dendrimers built of DNA (nucleodendrimers (NDs)) are coupled to antibodies against selected cell-surface receptors and compared to polymer nanoparticles (NPs). NDs and NPs bind specifically to cells expressing these targets and efficiently enter cells via the pathway associated with the selected receptor. While NPs traffic to perinuclear endo-lysosomes, NDs remain scattered throughout the cell, suggesting endosomal escape. This is confirmed in vitro, where NDs disrupt membranous vesicles at endosomal-like pH and in cell culture, where they: provide endosomal escape of model drugs, sugars, proteins, and nucleic acids; allow access to other intracellular compartments; result in measurable effects of cargoes; and do not cause cytotoxicity. Therefore, these DNA-nanodevices can be used to selectively overcome intracellular barriers, underscoring the growing range of applications of DNA materials.
Keywords: DNA-devices, targeted dendrimers, intracellular drug delivery, endocytosis, endosomal escape
1. Introduction
Safe and efficient delivery of research tools, diagnostic probes, and therapeutic agents to the interior of cells remains a major challenge.[1] This is due to the fact that most of these molecules cannot transverse cellular membranes, including the plasmalemma and membranes of endocytic vesicles.[2] This prevents access to the cell cytosol and other intracellular compartments, hindering our ability to exploit intracellular delivery of compounds for basic research and, primarily, translational applications.[3] Numerous strategies aim to overcome this limitation, including those that enhance permeation across the plasmalemma or rely on endocytosis and transport through the endo-lysosomal route to then permeate acidic endosomal compartments.[4] Most of these strategies employ cell-penetrating or fusogenic peptides, peptidomimetics, toxins, synthetic or natural polycationic polymers or lipids, capitalize on the proton sponge effect or volume changes within endosomes, etc.[5-12] Yet, despite this considerable advancement, intracellular delivery remains elusive. Aspects requiring optimization relate to the safety and mechanism of action of these approaches since often they: (a) are inherently toxic,[5,6,13] (b) disturb the plasma membrane (instead of or concomitantly to the endosomal membrane),[8-10] (c) involve materials that cannot be fully degraded and/or their products cannot be completely eliminated,[6,9,11] (d) recognize broadly displayed signatures (e.g. cell-surface charge, etc.), suffering from lack of specificity,[5,8,12] (e) are limited in their loading or transport potential to certain types of cargo molecules,[9,12] and/or (f) have poor applicability beyond local administration.[6,11]
DNA represents an alternative material toward this goal. While in nature DNA exists as a linear or circular polymer, it can also be engineered into branched, dendritic, or networked forms, such as the case of Y-, T-, and X-shaped DNA, tiles, origami, nanocages, etc.[14-16] DNA synthesis and assembly, as well as DNA disassembly and degradation can be controlled by chemical, enzymatic, and physical methods.[14-16] Indeed, DNA-devices have been proven valuable in multiple applications, including molecular detection, diagnosis, biotechnology, and others.[14-16] Although DNA represents the most classical example of a molecule difficult to introduce into cells, it also has properties amenable for drug delivery. For instance, DNA is water soluble, biodegradable, can interact with positively-charged carriers and cargoes, can solubilize hydrophobic drugs between its bases, has 5’-phosphate and 3’-hydroxyl ends for conjugation, and can be modified with other functional elements.[15,17,18] While delivery of compounds by DNA-based devices has been less investigated, recent publications have shown considerable promise also on this regard. As examples of this potential, DNA spherical particles, origami, cages, tubes, hydrogels, and liposomes-like DNAsomes can carry and release hydrophobic drugs, oligonucleotides, proteins, and imaging particles.[19-27]
However, most previous studies on this application have employed DNA-devices which: (a) contained an additional material as a part of their structural scaffold (lipidic, polymeric, inorganic, etc.),[17,23,24,27] (b) had sizes small enough so that they may not be able to avoid non-specific penetration through certain tissues (≤ 20-nm)[19,24] or were too large to enter non-immune cells by endocytosis (macro-assemblies),[15, 17] and/or (c) were shown to bind and gain access into cells largely by non-specific pathways (scavenger receptors, macropinocytosis, etc.)[20, 23, 24] even when targeted to selected cell-surface receptors.[24] The study presented here has explored intracellular delivery of diverse compounds (model drugs and biologicals) mediated by DNA-devices void of other structural materials, large enough to avoid broad penetration through tissues yet still small enough to allow endocytosis, and with enough selectivity toward molecular targets as to overcome non-specific receptors and endocytic pathways.
2. Results
2.1. Cell Binding and Endocytosis
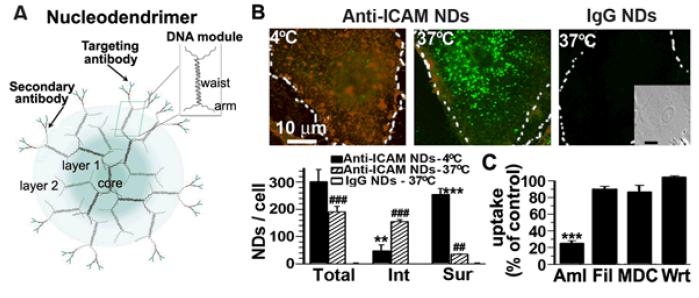
To determine if DNA-devices can provide cell targeting and endocytosis via selected cell-surface receptors and pathways, commercial DNA dendrimers[28] were tested. These dendrimers consist of DNA modules (Figure 1A), where each module has two DNA strands that form a central dsDNA region (waist) with four ssDNA ends (arms). Driven by complementarity between the arms of individual DNA modules, these dendrimers self-assemble to contain a core, a selected number of layers (generations), and a tunable number of outer arms for functionalization.[28] These devices are available as scaffolds to build signal amplifiers by conjugation to molecular probes,[29, 30] but had not been tested for drug delivery. Generation 4 DNA dendrimers bearing ~20 anti-mouse IgG molecules (a secondary antibody to which targeting antibodies could be coupled by affinity) were selected as a model. These dendrimers are referred to as nucleodendrimers (NDs) thereafter. The size of these commercial NDs was 175.4±9.2-nm in diameter and 0.3±0.1 polydispersity index (PDI), measured by dynamic light scattering (DLS). Binding of mouse IgG, used as a model antibody, increased ND size (p = 0.04) to 211.3±13.9-nm and 0.4±0.01 PDI. This result indicates that a coat of antibodies (which could be chosen to bind to selected targets) could be similarly incorporated.
Figure 1.
Effect of model cargoes delivered intracellularly by targeted nucleodendrimers. Fluorescence microscopy (top) and quantification (bottom) of HUVECs viability after 16 h exposure to 50 μM H2O2. Cells had been pre-incubated with catalase and anti-ICAM NDs or anti-ICAM NPs 3 h prior to H2O2 oxidative challenge. Green = calcein-positive live cells. Red = ethidium homodimer-1-positive dead cells. Scale bar = 30-μm. *Compares NDs or NPs in control condition vs. H2O2 insult. #Compares NDs vs. NPs. Two or three symbols represent p < 0.01 or p < 0.001, respectively.
To then evaluate if specific targeting and endocytosis is possible, NDs were coupled to an antibody recognizing intercellular adhesion molecule-1 (ICAM-1).[31] This is a glycoprotein overexpressed on several cell types in many pathologies,[31] which has been well-studied in the context of targeting and transport of drug delivery systems.[32] Anti-ICAM NDs were incubated for 1 h at 37°C with human umbilical vein endothelial cells (HUVECs) activated with TNFα, which induces ICAM-1 expression mimicking a pathological state.[31] Using an established protocol (see Experimental Section and [33]) that allows fluorescence microscopy quantification of total cell-associated, surface-bound, and internalized carriers (Figure 1B), anti-ICAM NDs were observed to bind to cells with high specificity: ~190 NDs/cell (~190-fold over IgG NDs). Binding was blocked by co-incubation with anti-ICAM (89% inhibition; p < 0.001), supporting further targeting specificity. This level and specificity of binding is comparable to previously described anti-ICAM nanoparticles (anti-ICAM NPs) built of polymers (e.g. non-degradable polystyrene or degradable poly(lactic-co-glycolic acid),[34,35] implying that the DNA scaffold of NDs does not impact this parameter.
With regard to transport into cells, anti-ICAM NDs were efficiently internalized after 1 h incubation at 37°C (Figure 1B): ~82% of total cell-associated NDs, while this parameter was undetectable for control IgG NDs. Internalization was inhibited at 4°C (~20% uptake; Figure 1B), suggesting that this is an energy dependent event, likely and endocytic process. In accord, uptake by cells was impaired upon incubation at 37°C in the presence of inhibitors of endocytosis mediated by ICAM-1[33] (amiloride; ~20% uptake), but not inhibitors of different pathways such as caveoli (filipin; ~71% uptake), clathrin (monodansylcadaverine; ~70% uptake), or macropinocytosis (wortmannin; ~85% uptake) (Figure 1C is notmalized to control). This set of data suggests that the rate and mechanism of uptake of ICAM-1-targeted NDs is similar to other ICAM-1-targeted systems, such as anti-ICAM NPs, previously reported.[33,34] Therefore, the DNA scaffold of NDs does not seem to impact this parameter. Indeed, comparison of anti-ICAM NDs to anti-ICAM NPs of similar size (220±18-nm; 0.2±0.02 PDI) showed similar uptake by HUVECs (~80-90%; Figure 2).
Figure 2.
Intracellular distribution of targeted nucleodendrimers. Fluorescence microscopy (top) and quantification (bottom) of HUVECs incubated at 37°C for either 1 h or 3 h with anti-ICAM NDs or anti-ICAM NPs. Both formulations are labeled with a green fluorophore (Alexa Fluor 488-anti-ICAM for NDs or FITC-NPs, respectively). Surface-bound carriers are then labeled using a Texas-red-conjugated secondary antibody, hence they appear yellow (red + green), while internalized counterparts are green. Perinuclear region refers to the area comprised ≤ 2 μm around the nucleus. Scale bar = 10-μm. *Compares NDs vs. NPs. Three symbols represent p < 0.001. For each formulation, no significant difference was found between 1 h and 3 h.
However, despite the similarities with regard to binding, uptake, and endocytic mechanism, intracellular distribution of anti-ICAM NDs was found to be different from that of anti-ICAM NPs (Figure 2). Anti-ICAM NPs largely accumulated in the perinuclear region by 1 h (~70% of all cell-associated carriers), in agreement with their known lysosomal routing.[36] In contrast, anti-ICAM NDs appeared scattered throughout the cell body even after 3 h (~40% perinuclear). This suggets that NDs provide for a different trafficking route, likely escape from endosomal compartments, as it has been reported for other DNA devices.[23,24,27]
2.2. Endosomal Escape
To explore if anti-ICAM NDs mediate disruption of endocytic vesicles, this formulation was incubated for 15 min at 37°C with cells concomitantly to the presence of Texas-red dextran in the cell medium (Figure 3 and Supplementary Figure S1). This protocol has been previously shown to allow dextran incorporation within the fluid-phase (lumen) of endocytic vesicles engulfing anti-ICAM carriers.[36] Since mammalian cells cannot degrade dextran, this polysaccharide can be used as a trackable marker regardless of its intracellular destination.
Figure 3.
Subcellular distribution of a fluid-phase marker mediated by targeted nucleodendrimers. Fluorescence microscopy (top) of HUVECs co-incubated at 37°C with anti-ICAM NDs (full panels) or anti-ICAM NPs (insets) in the presence of Texas-red dextran, followed by fixation and nuclear staining with DAPI (blue). Scale bar = 10-μm. Dashed lines = cell borders. The number of Texas red-positive endocytic vesicles and total area occupied by fluorescence pixels were quantified (bottom). Δ = fold change between 3 h and 1 h, where * p < 0.05.
After uptake, unbound materials were washed and internalized materials were allowed to traffic intracellularly. Texas-red dextran was similarly incorporated into endocytic vesicles for both anti-ICAM NPs and anti-ICAM NDs: ~60 vesicles/cell in 1 h (Figure 3 and Supplementary Figure S1A). Yet, the total area occupied by dextran was greater in cells incubated with anti-ICAM NDs (1.5-fold; Supplementary Figure S1B), which paires well with the more scattered distribution of NDs through the cell body (Figure 2). The number of dextran-positive vesicles and area occupied by this marker were stable over time for anti-ICAM NPs (Δ ~1 comparing 3 h over 1 h; Figure 3), indicating that dextran does not leave endocytic vesicles. This was expected based on previous reports showing trafficking of anti-ICAM NPs to endo-lysosomal compartments.[37] In contrast, with time, anti-ICAM NDs caused dextran to appear less punctate and more diffuse throughout the cell body: by 3 h the number of dextran-positive vesicles was reduced by ~2-fold and the area occupied by this marker approximately doubled (Figure 3 and Supplementary Figure S1A-B). These results indicate that anti-ICAM NDs may allow dextran to escape into the cytosol following endocytosis, as the case for other DNA-based systems.[23,24]
To validate this, NDs where incubated in vitro with membranous artificial vesicles (liposomes) at pH 4.5 to mimic interactions with membranous endo-lysosomes of cells. Liposomes were degraded in the presence of NDs (a detergent was used as a positive control), which was dependent on the dose of both liposomes and NDs (Figure 4) and did not occur at neutral pH (Supplementary Figure S2).
Figure 4.
Effect of nucleodendrimers on membranous vesicles. Percent degradation of liposomes (% lip. degr.) quantified by measuring their optical density (OD; 595 nm) in a pH 4.5 solution, after 30 min incubation at room temperature with NDs or with 2% Triton X-100 (TX100) used as a positive control.
2.3. Intracellular Delivery and Effects
To explore if NDs provide intracellular delivery of molecules other than dextran, a series of tests were performed (Figure 5 and Supplementary Figure S3). First, cells were co-incubated for 30 min with anti-ICAM NDs and phalloidin, a drug known to interact with F-actin in permeabilized cells but unable to penetrate intact cell membranes per se, as verified in Supplementary Figure S3A. In agreement with NDs mediating endosomal escape following endocytosis, phalloidin allowed visualization of F-actin when co-incubated with anti-ICAM NDs at 37°C (arrows in Figure 5A), but not at 4°C, a temperature precludes endocytosis (Supplementary Figure S3A). In contrast, a lack of F-actin staining was found for anti-ICAM NPs (Figure 5A), since they do not provide endo-lysosomal escape.[36]
Figure 5.
Intracellular delivery of various compounds by targeted nucleodendrimers. (A-B) Fluorescence microscopy of HUVECs co-incubated at 37°C in a pulse-chase mode (see Methods section) with anti-ICAM NDs or anti-ICAM NPs along with (A) biotin-phalloidin or (B) rhodamine-albumin nuclear localization sequence chimera (albumin-NLS). Biotin-phalloidin was visualized with Texas-red streptavidin (note: cell nuclei naturally contain biotin). Arrows = distribution to the intended intracellular targets, including F-actin in (A) or the nucleus in (B). Arrowheads = distribution to intracellular off-target sites, such as lack of F-actin staining in (A) or perinuclear distribution in (B). (C) Fluorescent protein expression 48 h after co-incubation of HUVECs for 5 h at 37°C with anti-ICAM NDs or anti-ICAM NPs and EGFP-RhoA-encoding plasmid. Inset = phase-contrast verifying presence of cells. (A-C) Scale bar = 10-μm.
Next, similar experiments were conducted to examine if agents delivered into the cytosol by mediation of NDs could be re-directed to other subcellular compartments. For this purpose, cells were co-incubated with anti-ICAM NDs and a recombinant protein (albumin), which was tagged by a nuclear localization sequence (NLS). As a positive control, this recombinant protein could label the cell nucleus of permeabilized cells (Supplementary Figure S3B), since this grants direct access across the plasmalemma. As a negative control, no nuclear label was found in cells incubated at 37°C with anti-ICAM NPs (Figure 5B), since these carriers do not exit endo-lysosomal compartments.[36] Lack of nuclear localization was observed for anti-ICAM NDs incubated at 4°C (Supplementary Figure S3B), since no endocytosis occurs at this temperature (Figure 1B). However, co-incubation with anti-ICAM NDs at 37°C to allow endocytic transport resulted in albumin-NLS reaching the cell nucleus (arrows in Figure 5B).
Next, functional activity after redistribution from the cytosol to the nucleus was assessed by evaluating the transfection potential of NDs. As shown in Figure 5C, co-incubation of a plasmid encoding for EGFP-RhoA with anti-ICAM NDs at 37°C (but not 4°C; Supplemental Figure S3C) provided expression of this recombinant protein, while transfection was undetectable in the case of anti-ICAM NPs (Figure 5C) or plasmid alone (Supplementry Figure S3C).
To demonstrate activity of a model therapeutic (biological) agent, intracellular delivery of catalase was tested. Catalase is an antioxidant enzyme unable to transverse cellular membranes.[36] Indeed, incubation of cells with catalase for 3 h did not prevent cell death caused by H2O2-mediated oxidative injury after catalase delivery (Supplelentary Figure S3D). Neither cells co-incubated with anti-ICAM/catalase NPs were protected against this pathological insult (Figure 6), which was expected since catalase is known to traffic and degrade within lysosomes 3 h after delivery by anti-ICAM NPs.[36] However, co-delivery with anti-ICAM NDs resulted in significant antioxidant protection (~90%), in agreement with endosomal escape into the cytosol.
2.4. Applicability and Cell Viability
To evaluate whether this strategy could be adapted to target other cells types, anti-ICAM NDs were incubated with human mesothelioma REN cells, human skin fibroblasts, or human colorectal adenocarcinoma Caco-2 cells (Figure 7A). As expected based on known ICAM-1 expression on these cell types under pathological-like conditions,[32] anti-ICAM NDs specifically associated to these cells compared to control IgG NDs (22-, 38-, and 47-fold, respectively).
Figure 7.
Targeting of nucleodendrimers to various receptors and cell types. (A) Fluorescence microscopy of REN cells, skin fibroblasts, Caco-2 cells after 1 h incubation at 37°C with anti-ICAM NDs. (B) Fluorescence microscopy of HUVECs after 1 h incubation at 37°C with NDs coupled to antibodies recognizing PECAM-1, transferrin receptor (TfR), or mannose-6-phosphate receptor (M6PR). NDs were visualized after permeabilization and staining with FITC-secondary antibody. Scale bar = 10-μm. Dashed lines = cell borders.
Furthermore, NDs could also be targeted to other cell-surface molecules (Figure 7B). These included platelet-endothelial adhesion molecule 1 (PECAM-1), another Ig-like CAM expressed on the endothelium.[38] PECAM-1 is functionally involved in leukocyte extravasation[38] and associated with CAM-mediated endocytosis.[33] Targeting NDs to PECAM-1 resulted in a 60-fold increased binding compared to control IgG NDs (shown in Figure 1B). This was also the case when targeting NDs to alterative receptors, including the transferrin receptor (TfR; 32-fold over IgG NDs) and mannose-6-phosphate receptor (M6PR; 24-fold over IgG NDs), both of which are structurally and functionally unrelated to one another or to endothelial CAMs, and are associated with clathrin-mediated uptake.[39,40] Thus, specific interaction of NDs with receptors and cell types can be tailored by selecting appropriate targeting ligands.
Finally, binding, uptake, or endosomal escape by targeted NDs did not seem to affect the cell viability. This was demonstrated by lack of an effect on endothelial cell monolayer appearance, the number of cells, or the cell viability measured 48 h after incubation with these carriers for 5 h, which was comparable to that of anti-ICAM NPs (Figure 8).
Figure 8.
Effect of nucleodendrimers on the cell viability. Fluorescence microscopy (left) and quantification of the number of HUVECs (middle) and their viability (right) 48 h after incubation with anti-ICAM NDs for 5 h. Green = calcein-positive live cells. Red = ethidium homodimer-positive dead cells. Scale bar = 500-μm.
3. Discussion
This study demonstrates that targeted NDs can provide receptor/pathway- specific and efficient cytosolic delivery of a variety of cargoes, which can then be re-directed to other subcellular compartments without apparent cytotoxicity. Specific binding and endocytosis seems ruled by the targeting moiety coupled to the carrier, since NDs behaved similar to NPs regarding these parameters. However, intracellular transport appears to be provided by the carrier itself, where NDs allow endosomal contents to escape into the cytosol compared to NPs, where contents remain within vesicular compartments. The degree of endosomal escape is relevant considering that the examples studied here as a proof-of-concept represent suboptimal configurations: cargoes were not linked to NDs and their incorporation into cells simply relied on passive co-internalization within endocytic vesicles induced by NDs. However, it is possible to directly couple cargoes to a ND. This can be achieved either by chemical linkage to outter or inner DNA branches, by coupling cargoes to oligonucleotides whose sequence is complementary to single-stranded DNA branches on the ND, or certain drugs can intercalate within DNA.[21,27] In this regard, an important question to optimize the system is whether NDs themselves escape endocytic vesicles or whether they distabilize the vesicle membrane but remain within its interior, which must to be investigated. Nevertheless, even if NDs would remain within vesicular compartments, cargoes could be linked to NDs cleavable linkers, separating both components and allowing the cargo to escape into the cytosol,[4] similar to non-linked cargoes shown here.
This endosomal escape property provided by NDs is likely due to the nature of the material composing these carriers, DNA. Indeed pristine (not targeted) NDs disrupted membranous liposomes in vitro when incubated at endo-lysosomal-like pH. This is in accord with the fact that several DNA-devices have recently shown potential to bypass membranous cellular barriers. This is the case for elegant studies employing DNA origami, tetrahedric cages, or DNAsomes to deliver doxorubicin in drug resistant cancer cells,[20,26,27] as well as spherical-DNA particles or DNAsomes proven to deliver oligonucleotide payloads intracellularly.[23,24,27] This is yet a poorly understood property of these DNA-devices, which may arise as a consequence of the display and packing of DNA in these architectures: it is known that DNA strands “packed” in an oriented brush border-like conformation display pKa shifts toward pH 4-6 and, hence, could protonate at physiological endo-lysosomal pH.[41,42] DNA protonation changes pairing of bases and strand conformation, it causes volume changes, and also leads to interaction with lipid membranes, altogether providing a possible mechanism for endosomal escape.[18,19,41-44] While previous studies showed endosomal escape ability in the context of delivery of oligonucleotides or doxorubicin,[19,20,23,24,27] oligonucleotides were a part of the DNA component of those carriers and doxorubicin is a DNA intercalating agent. Hence, the amenability of DNA to be used as a material for intracellular delivery of other cargo molecules had been a question. The present study expands this knowledge, showing the potential of DNA-devices to deliver a broad spectrum of cargoes within cells with no apparent disruption of the plasmalemma and bypassing endocytic compartments.
Highlighting the proterties of DNA as a promising material for drug delivery, several DNA structures have been reported capable of selective loading and release of cargo molecules, including DNA hydrogel networks, DNAsomes, origami, nanotubes, nanospheres, pyramidal or tetrahedric cages, etc.[14-16,26,27] Some of these strategies have not been tested in biological systems and their potential to interact with cells in ways amenable to intracellular transport are unknown, yet they seem promising.[18,19,21] On the other hand, although still very scarce, some studies have evaluated DNA-based systems for intracellular transport in biological systems, providing fundamental information to this relatively new area of research. This is the case for DNA origami, tetrahedric cages, DNAsomes, or DNA assembled on spherical nanoparticles, which were shown to enter cells.[20,22-24,27] The approach shown here expands previous knowledge in several ways. For instance, with the exception of silica nanoparticles displaying a DNA shell or liposome-like DNAsomes which formed structures ~80-120-nm in diameter,[23,27] all other studies of intracellular transport of DNA-devices employed relatively small nanoassemblies ≤ 20-nm.[20,22,24] This size may pose concerns of penetration through certain tissues in the body, e.g. extravasation into perivascular spaces through endothelial fenestrae or caveoli.[4] This could impair site-specificity of action and restrict potential applications. Also, this size is well below that of endocytic vesicles that form on the plasmalemma, e.g. caveoli are ~ 50-80-nm, clathrin pits are ~100-150-nm, and macropinosomes or phagasomes are ≥ 1-μm.[3] Hence, such DNA-devices may enter these compartments in a passive manner as cells continuously display endocytic activity by concomitant means.
This was the case for spherical-DNA carriers and DNA cages of such small size, which entered cells via non-specific scavenger receptors and/or non-selective macropinocytosis,[20,24] making selective intracellular delivery difficult. This remained the case even after targeting selected cell-surface receptors, such as the example of anti-HER2 conjugated spherical-DNA devices.[24] Although conjugation of this antibody to DNA strands did not affect its affinity, this was reduced 10-fold after conjugation on the spherical nanoparticle,[24] so that these particles displayed only 3-8-fold increased cell binding as compared to non-targeted counterparts when tested at physiological temperature.[24] As a result, uptake was largely mediated by scavenger receptors instead of HER2, resulting in no differences in uptake of targeted vs. untargeted particles by 24 h.[24] Spherical-DNA particles and DNAsomes of larger size (~80-120-nm) were speculated to access into cells via scavenger receptors or lipid-raft-like caveolae domains, respectively, although this may be due to the fact that such particles were not targeted to any particular marker.[23,27]
Hence, size and targeting valency are key elements to optimize the potential of DNA-devices designed to facilitate intracellular access. These limitations were overcome in this study by employing a model with larger size (~200-nm) and higher valency (≥20), which resulted in markedly greater selectivity: ~190-fold enhanced cellular binding for anti-ICAM NDs over untargeted counterparts and efficient (82%) uptake by a selected endocytic mechanism (e.g. CAM-mediated endocytosis for this formulation). Given this, DNA-devices such as those shown here are preferable to achieve selective drug delivery while previous configurations may show greater performance where non-selective targeting is sufficient (e.g. enhanced permeability retention in tumors, etc.).[4]
4. Conclusion
DNA-devices of size viable for endocytosis and sufficient targeting valency can provide: (a) specific binging to selected cell-surface markers, (b) efficient endocytosis into cells via the associated pathway, (c) cytosolic delivery of a broad range of compounds (drugs, sugars, proteins, nucleic acids), (d) subsequent access to other intracellular compartments (e.g. the nucleus), and (e) functional intracellular effects, (f) without apparent cytotoxicity. The properties and translational implications of the strategy presented here add significantly to the knowledge and potential applications of DNA-devices. Modulable physicochemical properties, versatility of assembly, potential for manipulation by chemical, enzymatic, and physical means, and full degradability in the physiological environment, make DNA an interesting material for this purpose.[24,44] Furthermore, DNA-devices can be designed to delay degradation by DNases and recognition by the immune system due to steric hindrance.[24,44] Although in vivo performance and safety remain to be investigated and optimized, this study shows that DNA-devices hold potential to bypass cellular membranous barriers for delivery of a broad spectrum of compounds intracellularly, further shifting the now questionable paradigm of DNA being unable to enter cells.
5. Experimental Section
Antibodies and Reagents
Antibodies recognized human ICAM-1 (ATCC; Manassas, VA), PECAM-1 (a gift from Centocor, Malvern, PA), transferrin receptor (TfR; EMD Millipore, Billerica, MA), or mannose-6-phosphate receptor (M6PR; Abcam, Cambridge, MA). Non-specific IgGs were from Jackson Immunoresearch (Pike West Grove, PA). Biotin-phalloidin was from Molecular Probes (Eugene, OR). Rhodamine-albumin-NLS and catalase were from Sigma Aldrich (St. Louis, MO). Plasmid pEGFP-RhoA was from Addgene (Cambridge, MA). Live/Dead® cell viability kit was from Live Technologies (Grand Island, NY). Liposomes were from Encapsula Nano Sciences (Brentwood, TN). UltraAmp™ signal amplifiers used as NDs were from Genisphere (Hatfield, PA) and polystyrene NPs from Polysciences (Warrington, PA). Other reagents were from Sigma-Aldrich (St. Louis, MO).
Nanocarrier Preparation
Model polymer NDs or NPs were prepared by incubating generation 4 UltraAmp™ DNA dendrimers (displaying ~20 anti-mouse IgG molecules) or 100-nm FITC-polystyrene particles, respectively, with anti-ICAM, anti-PECAM, anti-TfR, or anti-M6PR, or IgG[33]. This allows antibody binding to secondary IgG on NDs (Figure 1A) or antibody adsorption on the surface of polystyrene NPs.[33] Uncoated antibodies were removed from NP preparations by centrifugation and coated counterparts were resuspended in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA), and sonicated to avoid aggregation.[33] Size and polydispersity were measured by DLS (Malvern Zetasizer, Worcestershire, UK). As a note, polystyrene particles were selected as model polymer carriers since after coating with antibodies, they have been shown to provide similar targeting, endocytosis, and in vivo biodistribution as biodegradable poly(lactic-co-glycolic acid) NPs, allowing for comparison with historical data.[33,34]
Cell Culture
Human umbilical vein endothelial cells (HUVECs; Lonza, Walkersville, MD), mesothelioma REN cells, skin fibroblasts, or epithelial colorectal adenocarcinoma Caco-2 cells (American Type Culture Collection, Manassas, VA) were seeded on coverslips in 24-well plates and treated with TNF-α 16 h prior to experiments. Cells were cultured with antibiotics at 37°C, 5% CO2, and 95% relative humidity in: (HUVECs) M199 supplemented with 15% fetal bovine serum (FBS), 15 μg/mL endothelial cell growth supplement, 2 mM L-glutamine, and 100 μg/mL heparin; (REN) RPMI 1640 containing 10% FBS, and 2 mM L-glutamine; or (fibroblasts and Caco-2) DMEM supplemented with 10% FBS and 4.50 g/L glucose (GibcoBRL, Grand Island, NY). In all cases, incubation of NDs or NPs with cells was conducted in the presence of complete cell medium (containing 10-20% serum) supplemented with 1% BSA.
Binding and Uptake
Cells were incubated for the indicated times with NDs or FITC-labeled NPs coated with non-specific IgG or with targeting antibodies at 4°C or 37°C. For inhibition experiments, cells were incubated with anti-ICAM NDs in the presence of anti-ICAM in the medium or with endocytic inhibitors, including 50 μM MDC (an inhibitor of clathrin coated pits), 1 μg/ml filipin (an inhibitor of caveolar pathways), 3 mM amiloride (an inhibitor of ICAM-1 pathway and macropinocytosis), or 0.5 μM wortmannin (an inhibitor of macropinocytosis). Non-bound carriers were washed and cells were fixed with 2% paraformaldehyde. To quantify binding, FITC-labeled NPs were directly imaged by fluorescence microscopy,[33] while ND samples were permeabilized with 0.2% Triton X-100 and incubated with FITC (green)-labeled secondary antibodies to stain all cell-associated NDs (see microscopy settings below).
For internalization, NDs coated with Alexa Fluor 488 (green)-labeled antibody or antibody-coated FITC (green)-labeled NPs were used. After cell fixation (without permeabilization), samples were incubated with Texas-red secondary antibodies. This allows staining of the antibody coat of carriers located on the cell surface in Texas-red.[33] Samples were analyzed by fluorescence microscopy to quantify total carriers (which appear green) and surface-bound carriers (which appear yellow given co-localization of red + green), from which internalized carriers were calculated.[33] Cell borders were identified by phase-contrast. Samples were analyzed using a 60X PlanApo objective and Olympus IX81 microscope (Olympus Inc., Center Valley, PA) with fluorescence filters (Semrock, Rochester, NY) for Texas-red (ex. BP360-370 nm, em. BA590-800+ nm) and FITC (ex. BP460-490 nm, em.BA515-550 nm). Images were taken with Orca-ER camera (Hamamatsu, Bridgewater, NJ) and analyzed using Image-Pro 6.3 (Media Cybernetics Inc., Bethesda, MD). To minimize potential artifacts due to slightly different focal planes, ND vs. NP intensities, fluorescent spots containing more than one carrier, etc., all pictures were taken at the nucleus midsection, the number (not intensity) of spots whose fluorescence surpassed a threshold background was quantified, and finally then the number of carriers within that spot was based on the intensity expected for a ND or NP and the 2-D area to be occupied by a 200-nm object.
Intracellular Delivery
For co-internalization of carrier and cargo within common endocytic vesicles, cells were incubated at 37°C with anti-ICAM NDs or anti-ICAM NPs along with the agents indicated below (referred to as pulse).[35] Cells were then washed to stop further binding and uptake, and incubated at 37°C with cell medium to enable intracellular trafficking of the internalized agents (referred to as chase).[35] Cargoes were: (a) 2 mg/ml Texas-red dextran (15 min pulse, 1 h or 5 h chase), (b) biotin-phalloidin (30 min pulse, 5 h chase), (c) rhodamine-albumin-NLS (30 min pulse, 5 h chase), (d) plasmid pEGFP-RhoA (5 h pulse, 48 h chase), or (e) catalase (3 h continuous incubation). For dextran, biotin-phalloidin, albumin-NLS, and EGFP-RhoA plasmid, cells were fixed and visualized by fluorescence microscopy (using additionally Texas-red streptavidin for biotin-phalloidin; note that due to presence of biotin in the nucleus, Texas-red streptavidin also stains the nucleus). The number of dextran-positive objects ≥ 100-nm and total pixel area occupied by the signal were quantified. Phalloidin staining of F-actin or nuclear vs. endocytic distribution of albumin-NLS were qualitatively assessed. For catalase, after 3 h incubation, cells were challenged with 50 μM H2O2 (16 h), washed, and viability was tested using the Live/Dead® kit and fluorescence microscopy to quantify calcein-positive (live, green) vs. ethidium homodimer-1-positive (dead, red) cells.[35]
Cytotoxicity
Cells were incubated at 37°C for 5 h with anti-ICAM NDs or anti-ICAM NPs, then washed. Viability was estimated 48 h later, as described above.[35]
Liposomes
Liposomes suspended in PBS at pH 7.4 or 4.5 were incubated for 30 min at room temperature in the absence or presence of untargeted NDs. Triton X-100 (2%) was used as a positive control for liposome disruption. The resulting solutions were analyzed by DLS to obtain scattered counts per second and spectroscopy at 595 nm for optical density, both of which decay as a consequence of liposomal disruption.
Statistics
Data are mean ± standard error of the mean (SEM). For in vitro experiments N was ≥ 4. For cell culture, experiments were repeated at least twice, using ≥ 105 cells/condition, from which ~10 areas distributed throughout each coverslip were randomly selected for imaging, each area containing ~7-10 cells, and cells were analyzed individually. Significance was assessed by Student’s t-test (α p < 0.05).
Supplementary Material
Figure 1.
Binding and endocytosis of targeted nucleodendrimers. (A) Nucleodendrimers consist of DNA modules containing a central dsDNA region (waist) with four ssDNA ends (arms), where modules assemble by complementarity into layers surrounding a central core.[28] Outer arms are functionalized with mouse IgG secondary antibody, to which a targeting antibody can be coupled by affinity. [28] (B) Fluorescence microscopy (top) and quantification (bottom) of HUVECs incubated for 1 h with Alexa Fluor 488-(green)-labeled anti-ICAM NDs or IgG NDs, followed by staining of surface-bound NDs with Texas-red-conjugated secondary antibody. Surface-bound NDs appear yellow (red + green) and internalized counterparts are green. Inset = phase-contrast verifying presence of cells. Scale bar = 10-μm. *Compares 4°C vs. 37°C. #Compares IgG NDs vs. anti-ICAM NDs. Dashed lines = cell borders. (B) Endocytosis of anti-ICAM NDs (1 h - 37°C) in the presence of amiloride (Aml), filipin (Fil), monodansylcadaverine (MDC), or wortmannin (Wrt). *Compares inhibitors to control cells. (A-B) Two or three symbols represent p < 0.01 or p < 0.001, respectively.
Acknowledgements
Thanks to Dr. Vladimir Muzykantov (University of Pennsylvania Medical School, Philadelphia, PA) for sharing anti-PECAM (a gift from Dr. Nakada, Centocor, Malvern, PA) and REN cells (a gift from Dr. Steven Albelda; University of Pennsylvania Medical School, Philadelphia, PA), as well as constructive discussions on this topic. Special thanks to Dr. Carmen Garnacho (University of Seville, Seville, Spain), Zois Tsinas (Bioengineering, University of Maryland, College Park, MD), and Daniel Serrano (Cell Biology and Molecular genetics, University of Maryland College Park, MD) for technical help regarding optical density, dynamic light scattering, and schematic representation of nucleodendrimers. This study was funded by NIH grant R21-HL85533 and the Nanobiotechnology Program of the Maryland Department of Business and Economic Development (to S.M.).
Footnotes
Supporting Information
Supporting Information is available online from the Wiley Online Library or from the author.
References
- 1.Rajendran L, Knolker HJ, Simons K. Nat. Rev. Drug. Discov. 2010;9:29. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 2.Adler AF, Leong KW. Nano Today. 2010;5:553. doi: 10.1016/j.nantod.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan R, Richardson SC. Mol. Pharm. 2012;9:2380. doi: 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]
- 4.Muro S. J. Control. Release. 2012;164:125. doi: 10.1016/j.jconrel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho YW, Kim JD, Park K. J. Pharm. Pharmacol. 2003;55:721. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 6.Choi SH, Lee SH, Park TG. Biomacromolecules. 2006;7:1864. doi: 10.1021/bm060182a. [DOI] [PubMed] [Google Scholar]
- 7.El-Sayed ME, Hoffman AS, Stayton PS. Expert. Opin Biol. Ther. 2005;5:23. doi: 10.1517/14712598.5.1.23. [DOI] [PubMed] [Google Scholar]
- 8.Jones AT. Int. J. Pharm. 2008;354:34. doi: 10.1016/j.ijpharm.2007.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Mastrobattista E, Hennink WE, Schiffelers RM. Pharm. Res. 2007;24:1561. doi: 10.1007/s11095-007-9349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroder T, Niemeier N, Afonin S, Ulrich AS, Krug HF, Bräse S. J. Med. Chem. 2008;51:376. doi: 10.1021/jm070603m. [DOI] [PubMed] [Google Scholar]
- 11.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J. Control. Release. 2011;151:220. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Nicol F, Szoka FC., Jr Adv. Drug Deliv. Rev. 2004;56:967. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 13.Kakimoto S, Hamada T, Komatsu Y, Takagi M, Tanabe T, Azuma H, Shinkai S, Nagasaki T. Biomaterials. 2009;30:402. doi: 10.1016/j.biomaterials.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 14.Bath J, Turberfield AJ. Nat. Nanotechnol. 2007;2:275. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 15.Campolongo MJ, Tan SJ, Xu J, Luo D. Adv. Drug Deliv. Rev. 2010;62:606. doi: 10.1016/j.addr.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh YH, Ruiz RCH, Peng S, Lee JB, Luo D. Chem. Soc. Rev. 2011;40:5730. doi: 10.1039/c1cs15162b. [DOI] [PubMed] [Google Scholar]
- 17.Alemdaroglu FE, Herrmann A. Org. Biomol. Chem. 2006;5:1311. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, Maeda M. Biomacromolecules. 2005;6:2927. doi: 10.1021/bm0504330. [DOI] [PubMed] [Google Scholar]
- 19.Keum JW, Bermudez H. Chem. Commun. (Cham.) 2012;48:12118. doi: 10.1039/c2cc37471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KR, Kim DR, Lee T, Yhee JY, Kim BS, Kwon IC, Ahn DR. Chem. Commun.(Cham.) 2013;49:2010. doi: 10.1039/c3cc38693g. [DOI] [PubMed] [Google Scholar]
- 21.Lo PK, Karan P, Aldaye FA, McLaughlin CK, Hamblin GD, Cosa G, Sleiman HF. Nat. Chem. 2010;2:319. doi: 10.1038/nchem.575. [DOI] [PubMed] [Google Scholar]
- 22.Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ. ACS Nano. 2011;5:5427. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 23.Young KL, Scott AW, Hao L, Mirkin SE, Liu G, Mirkin CA. Nano Lett. 2012;12:3867. doi: 10.1021/nl3020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K, Hao L, Hurst SJ, Mirkin CA. J. Am. Chem. Soc. 2012;134:16488. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo D, Li Y, Um SH, Cu Y. In: in Methods in Molecular Medicine. Saltzman WM, Shen HM, Brandsma JL, editors. Humana Press; Totowa, New Jersey, USA: 2006. Ch 10. [Google Scholar]
- 26.Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang ZG, Zou G, Liang X, Yan H, Ding B. J. Am. Chem. Soc. 2012;134:13396. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 27.Roh YH, Lee JB, Kiatwuthinon P, Hartmen MR, Cha JJ, Um SH, Muller DA, Luo D. Small. 2011;7:74. doi: 10.1002/smll.201000752. [DOI] [PubMed] [Google Scholar]
- 28.Nilsen TW, Grayzel J, Prensky W. J. Theor. Biol. 1997;187:273. doi: 10.1006/jtbi.1997.0446. [DOI] [PubMed] [Google Scholar]
- 29.Mora JR, Getts RC. Expert. Rev. Mol. Diagn. 2007;7:775. doi: 10.1586/14737159.7.6.775. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Gordo M, Lin J, Bardina L, Pastor-Vargas C, Cases B, Vivanco F, Cuesta-Herranz J, Sampson HA. Int. Arch. Allergy Immunol. 2012;157:31. doi: 10.1159/000324677. [DOI] [PubMed] [Google Scholar]
- 31.Rothlein R, Dustin ML, Marlin SD, Springer TA. J. Immunol. 1986;137:1270. [PubMed] [Google Scholar]
- 32.Serrano D, Muro S. In: in Mechanobiology of the Endothelium. Aranda-Espinoza H, editor. CRC Press; accepted. [Google Scholar]
- 33.Muro S, Wiewrodt R, Thomas A, Koniaris L, Albelda SM, Muzykantov VR, Koval M. J. Cell. Sci. 2003;116:1599. doi: 10.1242/jcs.00367. [DOI] [PubMed] [Google Scholar]
- 34.Muro S, Dziubla T, Qiu W, Leferovich J, Cui X, Berk E, Muzykantov VR. J. Pharmacol. Exp. Ther. 2006;317:1161. doi: 10.1124/jpet.105.098970. [DOI] [PubMed] [Google Scholar]
- 35.Garnacho C, Dhami R, Simone E, Dziubla T, Leferovich J, Schuchman EH, Muzykantov V, Muro S. J. Pharmacol. Exp. Ther. 2008;325:400. doi: 10.1124/jpet.107.133298. [DOI] [PubMed] [Google Scholar]
- 36.Muro S, Gajewski C, Koval M, Muzykantov VR. Blood. 2005;105:650. doi: 10.1182/blood-2004-05-1714. [DOI] [PubMed] [Google Scholar]
- 37.Hsu J, Serrano D, Bhowmick T, Kumar K, Shen Y, Kuo YC, Garnacho C, Muro S. J. Control. Rel. 2011;149:323. doi: 10.1016/j.jconrel.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman PJ, Berndt MC, Gorski J, White GC, 2nd, Lyman S, Paddock C, Muller WA. Science. 1990;247:1219. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 39.Jefferies WA, Brandon MR, Hunt SV, Williams AF, Gatter KC, Mason DY. Nature. 1984;312:162. doi: 10.1038/312162a0. [DOI] [PubMed] [Google Scholar]
- 40.Kornfeld S. Annu. Rev. Biochem. 1992;61:307. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 41.Banville DL, Marzilli LG, Wilson WD. Biochem. 1986;25:7393. doi: 10.1021/bi00371a022. [DOI] [PubMed] [Google Scholar]
- 42.Rappaport SM, Medalion S, Rabin Y. Soft Matter. 2009;5:3010. [Google Scholar]
- 43.Horkay F, Basser PJ, Hecht AM, Geissler E. Macromol. Symp. 2010;291-292:354. doi: 10.1002/masy.201050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee O-S, Schatz GC. Methods Mol. Biol. 2011;726:283. doi: 10.1007/978-1-61779-052-2_18. [DOI] [PubMed] [Google Scholar]
- 45.Cutler JI, Auyeung E, Mirkin CA. J. Am. Chem. Soc. 2012;134:1376. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.