Abstract
T cell development is marked by the loss of alternative lineage choices accompanying specification and commitment to the T cell lineage. Commitment occurs between the CD4 and CD8 double-negative (DN) 2 and DN3 stages in mouse early T cells. To determine the gene regulatory changes that accompany commitment, we sought to distinguish and characterize the earliest committed wild type DN adult thymocytes. A transitional cell population, defined by the first down-regulation of surface c-Kit expression, was found to have lost the ability to differentiate into dendritic cells (DC) and natural killer (NK) cells when cultured without Notch-Delta signals. In the presence of Notch signaling, this subset generates T lineage descendants in an ordered precursor-product relationship between DN2, with the highest levels of surface c-Kit, and c-Kit-low DN3 cells. These earliest committed cells show only a few differences in regulatory gene expression as compared to uncommitted DN2 cells. They have not yet established the full expression of Notch-related and T cell differentiation genes characteristic of DN3 cells before β-selection. Instead, the downregulation of select stem cell and non-T lineage genes appears to be key to the extinction of alternative lineage choices.
Introduction
Early T cell development is a prolonged process in the thymus marked by loss of alternative lineage choices accompanying T cell specification and commitment to the γδ or αβ T lineages. While much progress has been made in understanding the gene expression patterns and regulatory networks involved in the early stages of T cell development (1–5), the specific regulatory events that result in T lineage commitment have not yet been elucidated.
To identify the critical gene expression changes leading to T cell commitment, the event must be accurately timed. Commitment is scored by loss of alternative developmental potential, and is thus dependent on refined purification of staged precursor cells. Mouse T cell precursors are CD4, CD8 double negative (DN) cells lacking a TCR, which were initially subdivided into DN1, DN2, DN3 and DN4 stages based upon CD25 and CD44 expression (6). Subsequently, additional markers were found to more precisely define these populations, especially for the DN1 stage, which includes both T cell precursors and irrelevant cells when defined solely based on CD44 and CD25 expression (7). High levels of c-Kit (CD117), the receptor for stem cell factor, were found to mark the pluripotent early thymic precursor (ETP) subset of DN1 cells as well as DN2 progenitor cells (8). Furthermore, cell size and CD27 expression were found to subdivide adult DN3 cells into pre- and post-β-selection cells (DN3a and DN3b respectively), allowing clear delineation of the earliest events in β- and γδ-selection and the manipulation of these cells (9–11).
Multiple studies have shown that DN3 cells are committed to the T lineage but the DN2 population is not, with possible alternative lineages including myeloid, DC, NK, and mast cells (12–16). Fetal thymic DN2 cells from pLck-GFP transgenic mice were found to be heterogenous in developmental potential: GFP+ DN2 cells could not differentiate into DCs and made NK cells very poorly in comparison to GFP− DN2 cells (17), implying that regulatory events establishing T lineage commitment may occur close to the DN2 stage, at least in fetal cells, well before the cell cycle arrest characteristic of the DN3 stage β-selection checkpoint (9).
Between the DN2 to DN3 stages is a period of major developmental change at molecular and cellular levels, triggered by an unknown intrinsic and/or extrinsic events. This is a time of markedly decreased proliferation, increased RAG-mediated TCR gene rearrangement, upregulation of Notch target gene expression, shifted growth and survival requirements crucial for β-selection checkpoint enforcement, and upregulated expression of many critical T cell genes (rev. in (18)). DN2 cells also express many “legacy” stem cell genes and genes associated with alternative lineages many of which decline precipitously between the DN2 and DN3 stages (1–3, 5, 16, 19). Although the expression of many transcription factors, T cell identity genes, and non-T cell genes changes dramatically from the ETP to the DN3 stage, it is not clear which of these gene changes are causally linked to T lineage commitment. For this, a more precise ordering of the gene expression changes in relationship to the commitment event is required.
In this study we found that the first phenotypic shift in c-Kit surface expression can be used in wild type adult mouse thymocytes as a marker to reveal a the earliest committed cells which precedes the phenotypic and functional changes that define the DN3a stage. Two inbred mouse strains were used to confirm the generality of these findings: C57BL/6 (B6) mice, the most commonly studied strain for immunological studies, and non-obese diabetic (NOD) mice, an autoimmunity-prone strain with a defect in early T cell checkpoint control (20). Gene expression analysis reveals that commitment is most closely associated, in both strains, with downregulation of a specific set of progenitor cell or non-T lineage regulatory genes, that it precedes major increases in Notch-dependent gene expression, and that only a select group of T lineage transcription factors is upregulated before commitment.
Materials and Methods
Mice
C57Bl/6J, NOD.Shi/J, NOD.B6-(D6Mit254-D6Mit289)/CarJ (NOD.NK1.1), and Eμ-Bcl-2-25 transgenic (Bcl2Tg) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). These strains were bred and maintained in the Caltech Laboratory Animal Facility. Female mice were used at 4–6 weeks of age. All animal protocols were reviewed and approved by the Animal Care and Use Committee of the California Institute of Technology.
Antibody staining, cell sorting, and flow cytometric analysis
To isolate DN populations, mice were sacrificed, their thymuses were removed and single cell suspensions made. Mature cells were depleted by staining with biotinylated antibodies to CD8a (53–6.7), TCRγδ (eBioGL3), TCRβ(H57–597), Gr1 (R86.8C5), Ter119 (Ter119), CD122 (5H4), NK1.1 (PK136), and CD11c (N418), after which the cells were incubated with streptavidin coated magnetic beads and the passed through a magnetic column (Miltenyi Biotec, Auburn, CA). Eluted DN cells were stained with c-Kit-APC (2B8), CD25-APC-AlexaFluor750 (PC61.5), CD44-PacficBlue (CIM7), CD27-FITC (LG.7F9), plus the viability dye, 7-Aminoactinmycin D (Invitrogen, Eugene, OR), and γδTCR-PE (eBioGL3) and CD11c-PE (N418) to deplete any remaining γδT cells or DCs. Purified ETP (CD25−CD44hi c-Kithi), DN2a (CD25+CD44hic-Kit+++), DN2b (CD25+CD44hi c-Kit++), and DN3a ((CD25+CD44lo c-Kitlo CD27loFSChi) precursor cells were sorted using a FACSAria with Diva software (Becton Dickinson Immunocytometry Systems [BDIS], Mountain View, CA). For analysis, cultures were forcefully pipetted, filtered through nylon mesh to remove stromal cell clumps, and stained with CD45-APC or - PECy5.5 (30-F11), to identify input cells, plus various combinations of lineage identifying antibodies: c-Kit-PE (2B8), CD44-PECy5.5 (CIM7), CD25-FITC (PC61.5), CD4-PECy5.5 (L3T4), CD8a-FITC (53–6.7), TCRγδ-PE (eBioGL3), NK1.1-PE (PK136), CD11c-APC (N418), CD11b (Mac1)-PE (M1/70), MHC I-A/I-E-FITC (M5/114.15.2, for B6 cells) and I-Ak (10–3.6 for NOD cells, Biolegend, San Diego, CA), and analyzed using a FACSCalibur (BDIS) and FlowJo software (Treestar, Ashland, OR). Antibodies were purchased from eBiosciences (San Diego, CA) except as noted. For intracellular PU.1 staining, DN thymocytes were stained with surface markers, c-Kit-PE (Clone 2B8) and CD25-FITC (Clone PC61.5), fixed and permeabilized using Cytofix/Cytoperm Kit (BD) and stained with antibody to PU.1-AlexaFluor647 or an isotype control- AlexaFluor647 (Cell Signaling Technology, Danvers, MA).
Cell culture
OP9 co-cultures were carried out as previously described (10, 21). For T cell development, sorted cells were placed on monolayers of OP9-DL1, supplemented with 2.5 ng/ml IL-7 and 5 ng/ml Flt3L. For DC and NK cell development OP9-Control (Ctrl) cells were used, supplemented with 2.5 ng/ml IL-7 and 10 ng/ml Flt3L. Human forms of the cytokines were used and obtained from Peprotech (Rocky Hill, NJ). Tissue culture media and fetal calf serum were obtained from Invitrogen/Gibco (Carlsbad, CA). In some experiments varying doses of γ-secretase inhibitor X (GSI, Calbiochem) was added at the start of the cultures to attenuate Notch signals. Stocks of GSI were dissolved in DMSO and diluted in tissue culture medium for addition to cell cultures. For proliferation assays, DN cell subsets were FACS sorted and then stained for 8 min at 37°C with 5 μM CFSE (Cell Trace CFSE Proliferation Kit, Invitrogen), in accordance with the manufacturer’s instructions, before culture.
RNA extraction and quantitative RT-PCR analysis
RNA was isolated from sorted DN cells from B6 and NOD mice and qRT-PCR carried out as previously described (16, 20). Briefly cells were lysed in Qiazol (Qiagen, Valencia, CA), RNA isolated using RNAeasy extraction kits (Qiagen), and cDNA reverse transcribed using random hexamers and SuperscriptIII (Invitrogen) following manufacterer’s instructions. cDNA samples were diluted and mixed with gene-specific primers and SyBr GreenER (Invitrogen) and run on an ABI Prism 7700 Sequence Detector (ABI, Mountain View, CA). Primers used for this study were synthesized by Eurofins MWG Operon (Huntsville, AL) and have been published previously (2, 5, 19, 20), with the exception of Lyl1 (FOR-CAGGACCCTTCAGCATCTTC; REV-ACGGCTGTTGGTGAACACTC). Results were calculated using the ΔCt method normalizing all samples to Actb expression.
Results
Surface c-Kit levels distinguish a developmentally intermediate population between DN2 (DN2a) and DN3 cells
DN2 cells are CD25-positive cells that also express hematopoietic progenitor-associated CD44 and c-Kit, both of which decline as the cells progress to the DN3 stage (22) (Fig. 1A and 1B). High c-Kit expression is critical to define the DN2 stage (23, 24), but we had previously noted that CD25+ cells with a small shift in c-Kit levels were distinguishable in their expression of several developmentally important genes (3). We sought to determine if this subset could provide a useful developmental-intermediate population to stage the loss of non-T cell potential and for analysis of the accompanying gene expression changes.
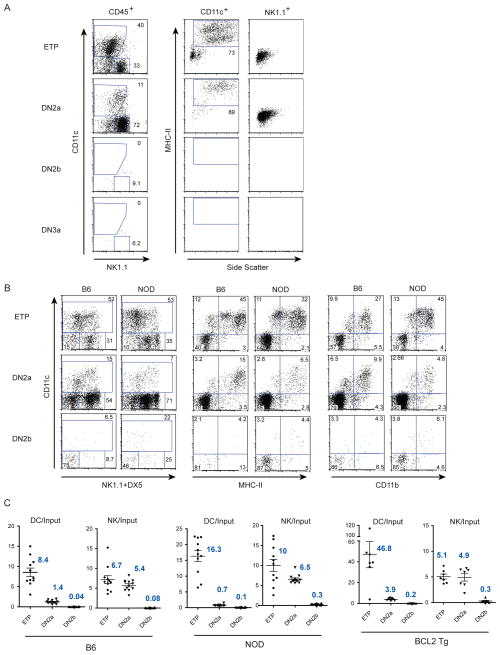
Figure 1. Surface phenotypes of early thymic DN populations from adult B6 and NOD mice.
FACs plots showing surface receptor expression and gates used for definition of thymic DN cell populations enriched by depletion of DP, SP, γδT, DC and NK cells and stained as described in Materials and Methods. (A) CD44 vs. CD25 and Kit vs. CD25 plots for B6 and NOD DN cells with quadrant lines indicating typical designations for “DN1-DN4”. (B) ETP, DN2a and DN2b populations gated first by CD44 and Kit and then by Kit and CD25, as indicated with lines and arrows, were used throughout this study. (C) Histograms showing surface expression of Kit and CD44 for the gated DN populations.
Figure 1 shows the phenotypic dissection and gating of T cell precursor stages from B6 and NOD mouse thymuses. Thymocytes were rigorously depleted of DP, TCRβ+ and TCRγδ+ cells, DCs and NK cells, as described in Materials and Methods, before staining and purification by cell sorting. CD25 plus CD44 or c-Kit expression have previously been used to define the DN populations, (Fig. 1A). As shown in Fig 1B, selection of DN thymocytes expressing high levels of c-Kit and CD44 can be subdivided into CD25− ETPs and two CD25+ c-Kithi cell populations. However, whereas all the CD25+ cells appear to fall into a single distribution in terms of CD44 expression (upper dot plots), two levels of c-Kit can readily be distinguished for both B6 and NOD DN cells (lower dot plots). The CD25+ cells with the highest c-Kit, even higher than in ETP, were designated as DN2a (the most stringently defined DN2 in (23)), and those with slightly lower c-Kit levels as DN2b. DN2b cells exhibit a lower but overlapping distribution of surface CD44 compared to DN2a cells, and are gated to be clearly distinct from DN3 cells (Fig. 1C). It is worth noting that the c-Kit differences are quite small relative to the overall levels of c-Kit, and that gated DN2 cells in mutational and gene expression analysis studies have often included DN2b and other cells (23).
To test whether these subsets represent a sequence of successive T cell precursor stages, purified ETP, DN2a, DN2b, and DN3a cells were co-cultured with OP9-delta-like 1 (DL1) stromal cells, which provide the Notch ligands required for T cell development (25). Similar results were seen for B6, NOD, and Bcl2 transgenic mice when analyzed for development using c-Kit, CD44 and CD25 expression (Fig. 2A, B). After 4 days of culture, Bcl2-transgenic ETPs generated both DN2a and DN2b-like CD25+ cells (high to intermediate c-Kit and CD44), DN2a cells shifted to phenotypes typical of DN2b and DN3a cells (intermediate c-Kit and CD44), DN2b cells progressed to the DN3 stage (low c-Kit and CD44), and about half of the DN3a cells down regulated CD25, reaching DN4 or beyond (low c-Kit and CD44) (Fig. 2A; right-side histograms compare c-Kit and CD44 expression for each population after culture). DN subsets from B6 and NOD mice gave similar results (Fig. 2B). In each case, DN2b cells also generated intermediate percentages of DN3 and DN4 cells, based on c-Kit and CD25 expression, between that of DN2a and DN3a cells.
Figure 2. DN2b cells are a developmentally intermediate population between DN2a and DN3.
Purified ETP, DN2a, DN2b, and DN3a precursors were plated and co-cultured with OP9-DL1 cells and analyzed for developmental progression. (A) Development of Bcl2 transgenic mouse DN cells after 4 days, showing changes in Kit vs. CD25 (top row) and CD44 vs. CD25 (bottom row). Histograms show direct comparisons of Kit and CD44 surfaces levels between the different cultured populations. (B) Kit vs. CD25 expression for purified DN populations from B6 (top row) and NOD (bottom row) mice, after 4 days of culture. (C) Plots showing development, after 13 days of culture, of NOD and B6 CD4+CD8+ DP cells (top panels), and CD45+ γδTCR+ T cells from the same cultures (bottom panels). Results are representative of 2–4 similar experiments.
Later, after 13 days of culture, all DN populations from B6 and NOD thymus produced more mature CD4+CD8+ DP cells and/or γδTCR+ cells (Fig. 2C). DN2b cells generated proportions of DP and γδ T cells that were intermediate between those of DN2a and DN3a cells. The proportion of γδTCR+ cells was consistently higher from DN2a than from DN2b or DN3a cells of both mouse strains, suggesting that differentiation from DN2a to DN3a may also be associated with a progressive reduction in γδT lineage potential (26–28). Furthermore, these results show that DN2b cell population is more similar to DN2a cells than DN3a cells in their retention of a DN population along with production of DP cells, and their ability to produce abundant γδT cells. NOD DN2 subsets were also noted to consistently produce higher proportions of γδT cells and lower proportions of DP than their B6 counterparts, a result which is addressed in depth in a separate report (manuscript in preparation). Overall, these data show that all of these DN subsets are T lineage precursors, and that DN2b cells develop in vitro at a rate intermediate between those of DN2a and DN3a cells in an ordered precursor-product sequence and can thus be used in subsequent experiments as a post-DN2(a) transitional population.
DC and NK developmental potential is extinguished between the DN2a and DN2b stages
Early T cell precursors can develop into DCs, myeloid cells, and NK cells in the absence of Notch signaling (7, 13–15, 29–31). To determine exactly when DC and NK potentials are lost, ETP, DN2a, DN2b and DN3a cells were purified from B6 thymuses and co-cultured with OP9-Control (Ctrl) cells. After 7 days, cultures were assayed for DC and NK cells using CD11c and MHC Class II (MHC-II) as markers for DCs and NK1.1 and DX5 (CD49b) for NK cells. Both ETP and DN2a cells produced distinct populations of CD11c+ and NK1.1+ cells (Fig. 3A, left panels). Most of the CD11c+ cells were also positive for surface MHC-II molecules and were high in side scatter, supporting their identification as DCs (Fig. 3A, middle panels), in contrast to the NK1.1+ cells which were negative for MHC-II and low in side scatter, as expected for NK cells (Fig. 3A, right panels). DN populations from NOD.NK1.1 mice gave similar results to those from B6 mice, producing both DCs and NK cells from ETPs and DN2a cells (Fig 3B). A high proportion of the CD11c+ cells from both mouse strains expressed surface MHC-II and CD11b (Mac1) (middle and right panels).
Figure 3. DC and NK lineage potential is lost in the DN2a to DN2b transition.
Sorted DN cells were co-cultured with OP9-Ctrl cells for 7 days and assayed for surface markers of DC and NK cells. (A) Left panels: CD11c and NK1.1 expression on CD45+ cells generated from ETP, DN2a, DN2b and DN3a cells. CD11c+ (DC, middle panels) and NK1.1+ (NK, right panels) gated subsets, further analyzed for MHC-II expression and side scatter. (B) DC and NK cell production from B6 and NOD.NK1.1 DN cells, showing phenotypes of CD45+ gated cells using CD11c with NK1.1 plus Dx5 (left), with MHC-II (middle), and with CD11b (right). Plots are representative of 12 independent wells of each cell type in the same experiment, as well as two additional independent experiments. (C) Graphs showing the numbers of DCs and NK cells produced per input cell from independent ETP, DN2a and DN2b cultures, starting with 100, 500 and 500 sorted input cells/well respectively, from B6, NOD and Bcl2 transgenic (Tg) mice. Lines indicate mean ± SD for each group and mean values are as shown.
In marked contrast, DN2b and DN3a cells produced few if any non-T cells of either type (Fig. 3A, B), and overall CD45+ cell recovery from DN2b and DN3a cells was drastically reduced in the absence of Notch-Delta signaling. The numbers of DC and NK cells produced as calculated per input cell from multiple independent cultures of DN cells from B6 and NOD mice are shown in Fig. 3C. ETPs consistently produced 5–20 times more DC and >5 times more NK cells than the number of input cells. DN2a cells also produced about 5 times more NK cells than input cells, but lower numbers of DCs, approximately equal to the number of input cells. The drop in both NK cell and DC potential in DN2b cells was severe, especially in B6 cultures where these lineages were virtually undetectable.
To test whether loss of viability in the absence of DL1 might preempt the detection of alternative lineage potentials of DN2b cells, DN cells from Bcl2-transgenic mice were also assayed for non-T cell potential. We have previously shown that this transgene preserves viability of T cell precursors under various experimental regulatory perturbations (16, 32). There was indeed some evidence that viability support may be a limiting factor for the generation of DCs, but not NK cells, since many more DCs were generated from Bcl2 transgenic ETP and DN2a cells than from wild type cells (Fig. 3C, right panels). However, Bcl2 transgenic DN2b cells showed the same loss of alternative potentials as DN2b cells from nontransgenic animals. Thus, the failure of DN2b cell differentiation in the absence of Notch signals reflects an intrinsically programmed loss of non-T cell developmental potential and suggests an acute increase in Notch dependence for viability, as compared to DN2a cells.
DN2b cells are more sensitive than DN2a cells to Notch signal attenuation
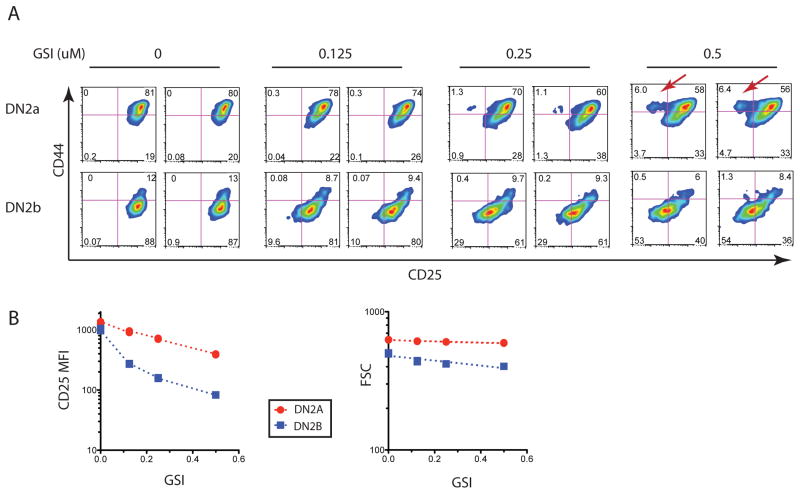
To test if the greater dependence of DN2b cells on Notch-Delta interactions is due to lower sensitivity to Notch signaling than in DN2a cells, we investigated the short term responses of B6 DN2a and DN2b cells to decreasing amounts of Notch signals. Graded doses of a γ-secretase inhibitor X (GSI) was added to normal OP9-DL1 co-cultures of DN2a and DN2b cells and development was assayed after 3 days, while the DN2b cells were still viable and readily characterized. In agreement with the OP9-Ctrl co-culture results, DN2a cells generated a CD44hiCD25lo population of cells by day 3 at the highest dose of GSI (red arrows), while DN2b cultures did not produce this population (Fig. 4A). These cells are likely to include the NK and DC precursors, which have this phenotype, although NK and DC lineage-specific markers were not yet expressed in this subset. CD25 expression, which is directly regulated by Notch signaling (33), declined in both DN2a and DN2b cells treated with GSI in a dose-dependent manner (Fig 4A and 4B). However, DN2b cells appear to be more sensitive than DN2a cells to reduced Notch signals as seen in their more rapid loss of CD25 with increasing GSI dose (Fig. 4B, left histogram), not attributable solely to changes in cell size (Fig. 4B, right histogram), again indicating a functional distinction between DN2a and DN2b cells.
Figure 4. DN2a and DN2b cell responses to attenuation of Notch signaling.
Sorted B6 DN2a and DN2b cells were co-cultured with OP9-DL1 in the presence or absence of different doses of Notch inhibitor, GSI, for 3 days. (A) Plots showing CD25 and CD44 for two independent cultures at each GSI concentration as indicated. Red arrows show the CD44hi CD25lo non-T cells generated from DN2a cells, but not DN2b cells, in response to decreasing Notch signals. (B) Graphs showing the mean geometric MFI for CD25 and the mean height for FSC comparing responses of the DN2a and DN2b cells to the varying doses of GSI. Data shown are two separate wells of each GSI dose from one of two independent experiments giving similar results.
DN2b cells are distinct from DN3 cells: continuing proliferation and delayed β-selection
These results are still consistent with DN2b cells being simply a phenotypic variant of DN3 cells. To assess whether the DN2b cells are also developmentally distinct from DN3 cells, we compared the number of cell divisions each subset must undergo before β- or γδ-selection, as measured by CD25 downregulation, when cultured in the presence of Notch signals (9, 10). Sorted ETP, DN2a, DN2b, and DN3a cells from B6 mice were stained with CFSE and cultured on OP9-DL1 for 2–4 days. As shown in Fig. 5, all populations show proliferation in vitro as tracked by CFSE dilution. DN3a cells were distinguished by their dual phenotypic response. Most of the proliferating DN3a cells began CD25 downregulation in their first cell divisions by day 2, with substantial CD25 loss by day 3, after approximately 4 divisions, indicating that these are cells that have successfully rearranged and expressed TCR genes and are undergoing β- or γδ-selection (10). Another population of DN3a descendants retained CD25 and divided no more than 1–2 times by days 2–3 (right top quadrants). In addition, these cells were smaller in size than the more rapidly dividing cells (right hand FSC histogram), and they disappeared by day 3, both characteristics consistent with TCR-negative checkpoint-arrested cells, which have limited life spans. In contrast, ETP and DN2a cells showed extensive proliferation while retaining CD25, consistent with a requirement for extended development before β-selection. DN2b cells were clearly unlike both DN2a and DN3a cells, proliferating over the 4-day period, although at a slower rate than ETP and DN2a cells. None of these cells downregulated CD25 as quickly as DN3a cells, sustaining CD25 expression even after 5–8 cell divisions. Thus DN2b cells represent a coherent intermediate subset that can be distinguished functionally both from DN2a cells that precede them and from DN3a cells that follow them in the developmental sequence.
Figure 5. Proliferation and differentiation of DN subsets.
Sorted ETP, DN2a, DN2b and DN3a cells from B6 mice were stained with CFSE and placed in co-cultures with OP9-DL1 cells. Dot plots of CD25 vs. CFSE after 2, 3 and 4 days, showing the downregulation of CD25 with cell division. Summary histograms are shown at the bottom comparing the CFSE levels through time for each DN population. The histogram at the right compares the forward scatter (FSC) values of CD25+ cells that have undergone only 1–2 cell divisions (upper right quadrant) vs. the CD25+ cells that have undergone >2 divisions (upper left quadrant) from the DN3a cell cultures on day 3. Results are representative of 3 similar experiments.
These results separate T lineage commitment from the cell cycle arrest that facilitates TCR gene rearrangement and checkpoint control in the DN3 stage. It further establishes the DN2b population of the adult thymus as the closest normal equivalent of the proliferating pLck-GFP+ DN2 population reported in transgenic fetal thymus (17), and confirms that commitment prior to checkpoint arrest in the DN3 stage is a normal integral aspect of T cell development.
Major changes in Notch-related genes follow declines in non-T cell genes at commitment
Identification of DN2b cells as a committed subset distinct from pluripotent DN2a cells and pre-β-selection DN3a cells made it possible to reassess the gene expression changes most closely linked to commitment. Therefore, we used qRT-PCR to compare expression of a panel of signaling response genes, differentiation genes, and major hematopoietic transcription factor genes in ETP, DN2a, DN2b, DN3a, and more mature cells that have either passed through β-selection (DN3b) or γδ-selection (γδT), sorted from both B6 and NOD thymocytes (Fig. 6).
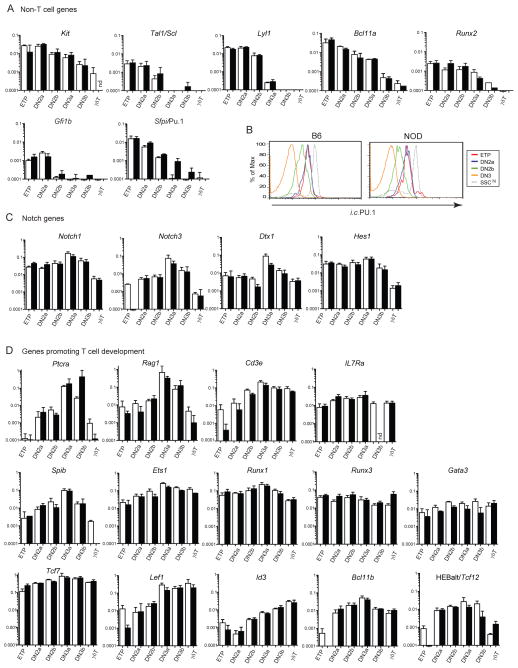
Figure 6. T lineage commitment precedes major changes in expression of genes related to Notch signaling and early T cell development.
(A, C, D) Quantitative RT-PCR results showing relative expression levels of T and non-T cell genes in ETP, DN2a, DN2b, DN3a cells, plus DN3b (β-selected T cells), and γδT cells for comparison, sorted from B6 (black bars) and NOD (white bars) mice. Values for each sample were calculated relative to Actb controls and presented as mean ± SE, n=2–5, nd = not determined. Graphs show the relative expression of non-T cell genes (A), Notch and related genes (C), and T lineage identity and required transcription factor genes (D). NOD ETP populations analyzed here are likely to include a small percentage of more mature T cells, which is made apparent by the use of a log scale. (B) Intracellular PU.1 in ETP, DN2a, DN2b and DN3a cell populations from B6 and NOD mice. Large granular cells (SSChi) included for positive controls (gray dashed line). Data are representative of 3 independent experiments.
The loss of stem or multipotent progenitor-associated gene expression accompanied commitment (Fig. 6A). Gfi1b and Sfpi1 (PU.1) were steeply downregulated between DN2a and DN2b, falling tenfold between the ETP and DN2b stages overall. Downregulation of PU.1 RNA as a landmark for commitment was also seen in the pLck-GFP transgenic system (17). Proto-oncogenes Lyl1 and Tal1, and Kit followed similar patterns, with further decrease at DN3a. Runx2 and Bcl11a fell more gradually between ETP and DN3a and were then deeply repressed during β-selection (Fig. 6A). For PU.1, these changes in RNA expression reflect stepwise decreases in transcription factor protein in precursors transitioning from DN2a to DN2b, as shown by intracellular staining. Fig. 6B shows that ETP and DN2a cells from both B6 and NOD thymuses express similarly high levels of PU.1 protein, with much lower levels in DN3a cells. Importantly, the DN2b cells from both genotypes show a uniform distribution of PU.1 at intermediate levels between those of DN2a and DN3a, not a bimodal distribution, confirming the discrete nature of this stage (Fig. 6B, and D. D. Scripture-Adams & E.V.R., unpublished data). Thus, the first decrease in PU.1 protein occurs after Kit downregulation in individual, newly-committed DN2b cells, consistent with arguments that loss of Sfpi1/Pu.1 and Gfi1b may be needed to cross an early threshold for T lineage commitment (4). However, PU.1 protein, at reduced levels, and the progenitor-cell factors Lyl1 and Tal1 remain available to these cells during this period of continued IL-7-dependent expansion.
Notch signaling is implicated directly in inducing the T cell program and in protecting uncommitted T cell precursors from lineage diversion, as well as in viability support for committed cells as shown above (rev. in (3, 34–36)). Notch target gene expression peaks at the DN3 stage, including autostimulation of Notch1 and Notch3 (2, 10, 20, 37, 38), and a key question has been whether intensified Notch signaling directly triggers commitment. We therefore asked whether this increase in expression of Notch target genes occurs from DN2a to DN2b, at the time of commitment, or from DN2b to DN3a, after cells are already committed. The results show that expression of Notch target genes changes very little from DN2a to DN2b with the major upregulation occuring between DN2b and DN3a. Three of four Notch target transcripts analyzed remained constant or even decreased (Dtx1) from DN2a to DN2b, before increasing from DN2b to DN3a stages. The fourth well-defined Notch target gene, Hes1, was already highly expressed from the earliest stages and showed little change from ETP to DN3a. Also, Ptcra, a T-lineage gene that responds strongly to Notch signaling in vitro (4, 16, 32), was dramatically upregulated in the DN2b to DN3a transition, after little or no increase from DN2a to DN2b (Fig. 6D). These results imply that increases in Notch signaling do not cause, but rather follow, the loss in alternative lineage potential in the DN2a to DN2b transition.
T lineage identity genes: regulatory changes after and before commitment
Crucial T lineage differentiation genes Ptcra, Rag1, and Cd3e are fully turned on in the DN3 stage (Fig. 6D). In earlier searches for commitment factors, upregulation of Ets1, Lef1, and Spib distinguished DN3a cells from unseparated DN2 cells, suggesting these as candidate genes for roles in commitment (5). However, as with Notch target genes, the sharpest upregulation of DN3-associated genes, including Ptcra, Rag1, Lef1, and Spib, occurred from DN2b to DN3a, not in the DN2a to DN2b transition (Fig. 6D). Thus, this cluster of gene expression changes also appears to occur primarily after, not during, commitment. The same general pattern was seen in cells from both B6 and NOD mice.
Several key factors required for T cell development are turned on long before commitment. IL7ra changes little across this interval, while Runx1 is sustained at high levels, both of which slightly increase to a DN3a peak. Tcf7 and Gata3 are already upregulated in ETP cells as compared to prethymic progenitor cells (2, 21) and we found transcripts of these genes, as well as Runx3, increased little to none from DN2a to DN2b (Fig. 6D). These factors may be required for T lineage commitment, but they do not appear to be responsible for the abrupt loss in alternative lineage potential.
Only a select group of transcription factor genes showed major switch-like shifts in expression from ETP to DN2b. Id3 was one of the only genes found to be upregulated significantly, although at low levels, from DN2a to DN2b. Two T-lineage regulatory genes undergo dramatic upregulation between ETP and DN2, Bcl11b and the alternative promoter form of Tcf12 (HEBalt) (2, 3, 39). We confirmed that the major phase of upregulation for both genes occurred between the ETP and DN2a stages, with only small increases in expression from DN2a to DN2b. Although this RNA induction is not itself initially sufficient to commit the cells, the accumulation of products of these genes may contribute to induction of the process and making commitment irreversible.
Discussion
Commitment is an important landmark in any developmental process, as it defines the crucial transition in a cell’s internal regulatory state that makes its identity self-perpetuating. The uncommitted cell has access to a number of alternative fates, which a committed cell does not. Recently there has been much discussion of whether these alternatives, and by extension the commitment process itself, are physiologically significant. For example, lineage-tracing evidence shows that the latent myeloid and dendritic cell potentials of early intrathymic precursors may not generally be used in vivo (40). However, this is a separate question from the significance of commitment. In most developmental systems, it is normal for cells to give rise only to a subset of the fates they might adopt if they were transplanted to a different environment before commitment. The thymic microenvironment is specialized to provide highly coercive signals, especially from Notch ligands, to antagonize non-T differentiation pathways from the ETP stage on (32, 41). These alternative fates for uncommitted cells remain only latent possibilities as long as the thymic microenvironment is intact. Conversely, it is important to note that even hematopoietic stem cells, which certainly do have T lineage potential, would not generate any T cells at all if all their descendents were prevented from reaching the thymus. In this paper, we deliberately expose cells to broadly permissive environmental conditions in order to determine the point at which it is the cell’s intrinsic regulatory apparatus that becomes the limitation on its developmental fate. Understanding the steps involved in this commitment process is necessary to perceive the order and the nature of the particular intrinsic regulatory changes that provide a cell with an irreversible T cell identity.
The DN2 to DN3 transition is a crucial turning point in the differentiation of early thymic T cells. DN2 cells maintain an ability to divert to alternative lineages under various conditions including loss of Notch signaling and alterations in key transcription factor genes (reviewed in (3)) and they express a wide variety of genes including stem cell genes, T cell specific genes, and genes associated with alternative lineages (1, 2, 5, 16, 19). Cells at this stage are sensitive to environmental factors that can affect later T lineage choices including αβ vs. γδ and regulatory T cells (42–44). For these reasons a more detailed analysis of the regulatory events around the transition from the DN2 to DN3 stage is critical to our understanding of T cell developmental networks. This study shows that the initial slight downregulation of c-Kit expression defines a developmentally intermediate population of cells in nontransgenic, adult mouse thymus that are committed to the T lineage but have not fully differentiated to the DN3 stage based on their continuing cell divisions and gene expression pattern. This transition marks a dramatic increase in dependence on Notch-Delta signals for viability as well as for maintenance of CD25+ phenotype. Not only are these DN2b cells distinct from the uncommitted DN2a cells that precede them, but also they are distinct from the DN3a cells that follow them, in terms of cell cycle programming, γδ-T lineage potential, and expression of regulatory genes that may contribute to these functional differences. Thus, our results also show that T lineage commitment occurs before the cell cycle arrest that facilitates TCR gene rearrangement and checkpoint control in the DN3 stage. In fact, these latter events are more tightly associated with the loss of γδ-T cell developmental potential.
The repression of critical progenitor regulatory genes, Lyl1, Tal1, Sfpi1 (PU.1) and especially Gfi1b, is the most direct indicator of commitment. The protooncogenes Lyl1 and Tal1 are closely related basic-helix-loop-helix (bHLH) factors with critical overlapping but distinct roles in fetal and adult hematopoetic stem cell survival and differentiation (45). Tal1 is a known regulator of Kit expression (46), although it is not known if Lyl1 also functions in this role, especially in adult thymocytes, as it is the more abundantly expressed gene in these cells.
The abrupt increase in Notch dependence for survival at the DN2b stage may also contribute to a committed state as progenitor-type survival pathways are blocked by loss of these factors. This may be related to the reduction in expression of these stem cell legacy genes, including the decrease in expression of surface c-Kit, the stem cell growth factor receptor. However, our results argue against a proximal role for increased Notch signaling in this repression itself. Although Notch signaling is required throughout early T cell development, the peak upregulation of Notch-related genes occurs after the DN2b stage, after repression of Gfi1b and Sfpi1 begins. This implies a separation between Notch-dependent events involved in ETP mobilization, long before commitment, and Notch-dependent events at the DN2b to DN3a transition, which follows it. Rather than intensifying their expression of Notch target genes, DN2b cells appear to require stronger Notch signals in order to maintain the same levels of expression of Notch target genes such as Cd25.
Many of the key T cell transcription factor genes, including Gata3, Tcf7, Ets1, Runx1, and Lef1, are already expressed in the ETP stage and only gradually increase to their maximal levels in the DN3a stage, with no major changes occurring around the time of commitment. While expression of these factors may be required for commitment, they are only likely to be rate-limiting if they are needed to titrate out an antagonist or competitor. However, the quantitative balance between these positive T lineage regulators and the regulators promoting alternative fates may play a more direct role than previously appreciated in the kinetics of upregulation of T lineage identity genes in individual cells. T cell identity genes like Ptcra, Rag1, and CD3 are initially turned on by the DN2a stage but their major upregulation occurs only after commitment, between the DN2b and DN3 stages, and their expression does not necessarily exclude lineage diversion. For example, DCs and mast cells differentiating from pro-T cells, but not from bone marrow, still express CD3e as proof of their T cell origins (16, 47). SCL (Tal1) and Lyl1 have been shown to be able to repress Ptcra and Rag1 expression (48, 49), thus it may be necessary to remove conflicting regulatory inputs from factors like Gfi1b, SCL (Tal1), Lyl1, and PU.1 to unleash the potential of the T cell factors already in place and to complete full T cell specification.
The only transcription factor genes that are both known to be important for T cell development and are sharply induced around the DN2 stage are an alternative splice variant of the bHLH factor Tcf12, HEBalt (39), and the zinc finger transcription factor Bcl11b (2). Both genes clearly undergo their major upregulation between the ETP and DN2a stages, just before commitment, making these attractive as potential candidate inducers of the T cell specification program and/or repressors of non-T lineage genes. Id3, notably alone, is upregulated between DN2a and DN2b. Id3 is activated in response to β-selection and positive selection signals (50, 51), suggesting that the DN2a to DN2b transition may also depend on a specific signal triggered by an environmental factor or by newly expressed signaling pathway intermediates. The reciprocal changes in genes encoding bHLH proteins (HEBalt, Id3, Tal1, and Lyl1), all potentially competing as partners for E2A (52), as well as the drop in Gfi1b, a Tal1/E2A target gene (53–55), suggest the possibility that a shift in the composition of bHLH heterodimers in pro-T cells could play a major role in commitment.
This fine-scale ordering of commitment and specification events is a prerequisite for accurate elucidation of the gene regulatory networks controlling early T cell development. These results on the developmental potential and gene expression in the first fully committed transitional cell population will provide a framework for investigating possible commitment signals as well as the mechanisms of repression and altered survival that result in T lineage commitment.
Acknowledgments
We thank Robert Butler, Koorosh J. Elihu, and Marion Duprilot for outstanding technical assistance, Diana Perez, Shelley Diamond and Pat Koen of the Caltech Flow Cytometry Facility for cell sorting, Scott Washburn and Natasha Bouey for excellent animal care, and Deirdre Scripture-Adams for helpful discussions and Long Li for providing Lyl1 primers.
This work was supported by NIH grant R01 AI064590 (to M.A.Y.), the Louis A. Garfinkle Memorial Laboratory Fund, and the Al Sherman Foundation. E.V.R. is supported by the Albert Billings Ruddock Professorship.
References
- 1.Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- 2.Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgescu C, Longabaugh WJ, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci U S A. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 7.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 10.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gamma delta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed alpha beta versus gamma delta lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas K, Vremec D, Wu L, Shortman K. A linkage between dendritic cell and T-cell development in the mouse thymus: the capacity of sequential T-cell precursors to form dendritic cells in culture. Dev Comp Immunol. 1998;22:339–349. doi: 10.1016/s0145-305x(98)00012-3. [DOI] [PubMed] [Google Scholar]
- 13.Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, Katsura Y, Kawamoto H. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol. 2003;171:3401–3406. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 16.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda K, Kakugawa K, Nakayama T, Minato N, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCR beta gene rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 18.Rothenberg EV, Yui MA. Development of T Cells. In: Paul WE, editor. Fundamental Immunology. 6. Lippincott Williams & Wilkins; Philadelphia, PA: 2008. pp. 376–406. [Google Scholar]
- 19.David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- 21.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 23.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol. 2002;2:888–897. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- 24.Ceredig R, Bosco N, Rolink T. Problems defining DN2 thymocytes. Immunol Cell Biol. 2008;86:545–547. doi: 10.1038/icb.2008.44. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 26.Bruno L, Scheffold A, Radbruch A, Owen MJ. Threshold of pre-T-cell-receptor surface expression is associated with alpha beta T-cell lineage commitment. Curr Biol. 1999;9:559–568. doi: 10.1016/s0960-9822(99)80259-0. [DOI] [PubMed] [Google Scholar]
- 27.Kang J, Volkmann A, Raulet DH. Evidence that gamma delta versus alpha beta T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alpha beta and gamma delta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRbeta chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 30.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 31.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 32.Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciofani M, Zuniga-Pflucker JC. A survival guide to early T cell development. Immunol Res. 2006;34:117–132. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 35.Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Dias S, Xu W, McGregor S, Kee B. Transcriptional regulation of lymphocyte development. Curr Opin Genet Dev. 2008;18:441–448. doi: 10.1016/j.gde.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 40.Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Pennington DJ, Silva-Santos B, Shires J, Theodoridis E, Pollitt C, Wise EL, Tigelaar RE, Owen MJ, Hayday AC. The inter-relatedness and interdependence of mouse T cell receptor gamma delta+ and alpha beta+ cells. Nat Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 43.Kang J, Coles M, Raulet DH. Defective development of gamma/delta T cells in interleukin 7 receptor-deficient mice is due to impaired expression of T cell receptor gamma genes. J Exp Med. 1999;190:973–982. doi: 10.1084/jem.190.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 45.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 47.Corcoran L, Ferrero I, Vremec D, Lucas K, Waithman J, O’Keeffe M, Wu L, Wilson A, Shortman K. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol. 2003;170:4926–4932. doi: 10.4049/jimmunol.170.10.4926. [DOI] [PubMed] [Google Scholar]
- 48.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 49.Zhong Y, Jiang L, Hiai H, Toyokuni S, Yamada Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene. 2007;26:6937–6947. doi: 10.1038/sj.onc.1210494. [DOI] [PubMed] [Google Scholar]
- 50.Bain G, Quong MW, Soloff RS, Hedrick SM, Murre C. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J Exp Med. 1999;190:1605–1616. doi: 10.1084/jem.190.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 52.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 53.Wilson NK, Miranda-Saavedra D, Kinston S, Bonadies N, Foster SD, Calero-Nieto F, Dawson MA, Donaldson IJ, Dumon S, Frampton J, Janky R, Sun XH, Teichmann SA, Bannister AJ, Gottgens B. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 54.Xu W, Kee BL. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]