This study shows that capping protein (CP) is essential for mammalian cell migration in vitro and in vivo. The authors also show that CP is present in filopodia of multiple cell types and that it regulates filopodial structure and function. Thus CP function in both lamellipodia and filopodia may contribute to efficient migration.
Abstract
Capping protein (CP) binds to barbed ends of growing actin filaments and inhibits elongation. CP is essential for actin-based motility in cell-free systems and in Dictyostelium. Even though CP is believed to be critical for creating the lamellipodial actin structure necessary for protrusion and migration, CP's role in mammalian cell migration has not been directly tested. Moreover, recent studies have suggested that structures besides lamellipodia, including lamella and filopodia, may have unappreciated roles in cell migration. CP has been postulated to be absent from filopodia, and thus its role in filopodial activity has remained unexplored. We report that silencing CP in both cultured mammalian B16F10 cells and in neurons of developing neocortex impaired cell migration. Moreover, we unexpectedly observed that low levels of CP were detectable in the majority of filopodia. CP depletion decreased filopodial length, altered filopodial shape, and reduced filopodial dynamics. Our results support an expansion of the potential roles that CP plays in cell motility by implicating CP in filopodia as well as in lamellipodia, both of which are important for locomotion in many types of migrating cells.
INTRODUCTION
Heterodimeric capping protein (CP) binds to actin filament barbed ends and thus inhibits further polymerization (Wear et al., 2003). Inhibiting CP activity in live organisms and cells severely compromises actin-based motility and lamellipodia formation (Hug et al., 1995; Rogers et al., 2003; Mejillano et al., 2004; Iwasa and Mullins, 2007). CP is thus thought to have a pivotal role in enabling leading-edge protrusion and net migration of cells (Pollard and Borisy, 2003; Le Clainche and Carlier, 2008). Specifically, CP activity is thought to limit the growth of lamellipodial actin filaments nucleated by the Arp2/3 complex, thus creating short and stiff filaments that can effectively push the membrane forward (Pollard and Borisy, 2003; Le Clainche and Carlier, 2008).
In addition, capping of actin filaments maintains actin monomer supply by preventing elongation of filaments that do not contribute to leading-edge motility (Carlier and Pantaloni, 1997; Hu and Papoian, 2010). These actin monomers may then increase overall cell motility by either increasing actin filament elongation (Carlier and Pantaloni, 1997) or by increasing nucleation by the Arp2/3 complex (Akin and Mullins, 2008) in regions with a relatively lower concentration of CP. However, the requirement for CP in mammalian cell migration has not been directly tested.
Recent work, moreover, has questioned the necessity of lamellipodia for cell migration (Gupton et al., 2005; Suraneni et al., 2012; Wu et al., 2012; Edwards et al., 2013). Fibroblast cells in which lamellipodium formation was inhibited following tropomyosin injection moved faster than did control cells (Gupton et al., 2005). In addition, studies from two different laboratories have demonstrated that cells lacking normal lamellipodia secondary to depletion of the Arp2/3 complex can migrate using a filopodia-based protrusion system (Suraneni et al., 2012; Wu et al., 2012). Finally, the migration defect in cells with silenced CARMIL protein could be substantially rescued without rescuing the concurrent lamellipodial defect, leading the authors to suggest that lamellipodia are not essential for cell migration in this setting (Edwards et al., 2013).
Filopodia are thin, actin-based protrusions found in many cell types, including migrating cells and neurons (Mattila and Lappalainen, 2008). They are integral for both chemosensing and motility (Gupton and Gertler, 2007; Mattila and Lappalainen, 2008) and are usually composed of 15–30 actin filaments (Small, 1981; Lewis and Bridgman, 1992). Models of filopodial initiation posit that a relative absence of CP allows filopodia to initiate and grow (Svitkina et al., 2003; Faix and Rottner, 2006). CP has been reported to be undetectable in filopodia (Svitkina et al., 2003), and most current models for filopodial structure and function do not include a role for CP (Wear and Cooper, 2004; Gupton and Gertler, 2007; Mattila and Lappalainen, 2008; Faix et al., 2009). Interestingly, however, some investigators have suggested that CP within filopodia may have an important role in controlling filopodial dynamics (Mallavarapu and Mitchison, 1999; Zhuravlev and Papoian, 2009).
In this study, we investigated the requirement of CP for mammalian cell migration in vitro and in vivo using a knockdown approach. Moreover, we discovered the presence of CP in filopodia and explored its functional impact. We suggest that while a decrease in capping activity stimulates emergence of filopodia, CP activity within filopodia is nonetheless necessary for their proper form and function.
RESULTS
CP depletion impairs mammalian cell migration in vivo
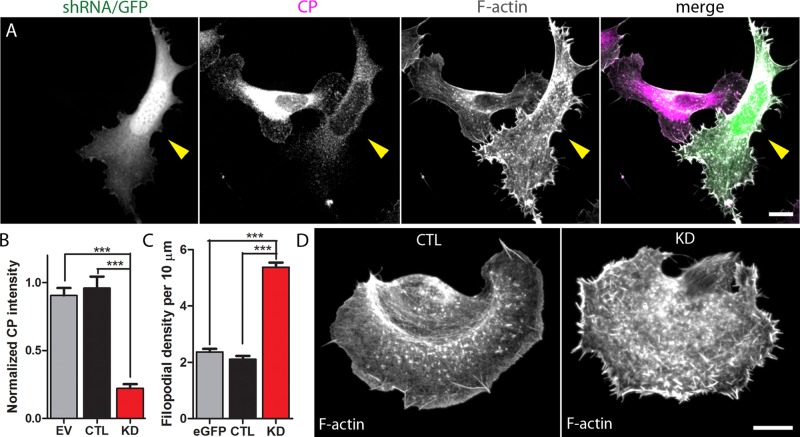
To understand the role of CP in cell migration, we first used a silencing strategy in B16F10 cells. We created a short hairpin RNA (shRNA) construct to CP based on a previously published shRNA sequence (Mejillano et al., 2004) and created a scrambled construct to use as control. In addition, we determined that a polyclonal antibody from Millipore (AB6017) recognizing the β2 subunit of CP is both sensitive and specific for immunostaining of multiple cell lines and primary neuronal cultures. CP immunoreactivity (CP-ir) using this antibody increased dramatically with CP overexpression (Figure S1A) and decreased with CP silencing in B16F10 cells (see Figure 1A). In addition, the signal decreased considerably with preadsorption by holoprotein (Figure S1, B and C).
FIGURE 1:
shRNA-mediated silencing of CP induces morphological changes. (A) CP-ir is robustly decreased in silenced B16F10 cells. Arrowhead indicates CP-silenced cell adjacent to a nontransfected cell. (B) Quantification of CP-ir decrease in silenced cells. For each category, the average CP fluorescence intensity of transfected cells was normalized to that of untransfected cells on the same coverslip that had been imaged and processed in an identical manner. EV, empty vector; CTL, Scramble-transfected cells; KD, CP-silenced cells. A total of 20–40 cells were analyzed for each condition; ***p < 0.001. (C) Filopodial density is increased in CP-depleted cells. A total of 47–69 cells per condition were analyzed across three different experiments and include 1600–3200 filopodia/condition; ***p < 0.001. Controls include eGFP-transfected (eGFP) and Scramble-transfected (CTL) cells. (D) A representative Scramble-transfected cell (left) and a CP-depleted cell (right). Cells are stained with phalloidin to visualize F-actin. Scale bar: 10 μm.
Transfection of B16F10 cells with CP shRNA effectively suppressed CP activity. In a representative experiment, shRNA treatment for 5 d reduced CP-ir by an average of 80% in transfected cells compared with untransfected neighboring cells (Figure 1, A and B). As reported previously (Mejillano et al., 2004), CP silencing led to a robust, more than twofold increase in filopodial density (Figure 1, C and D).
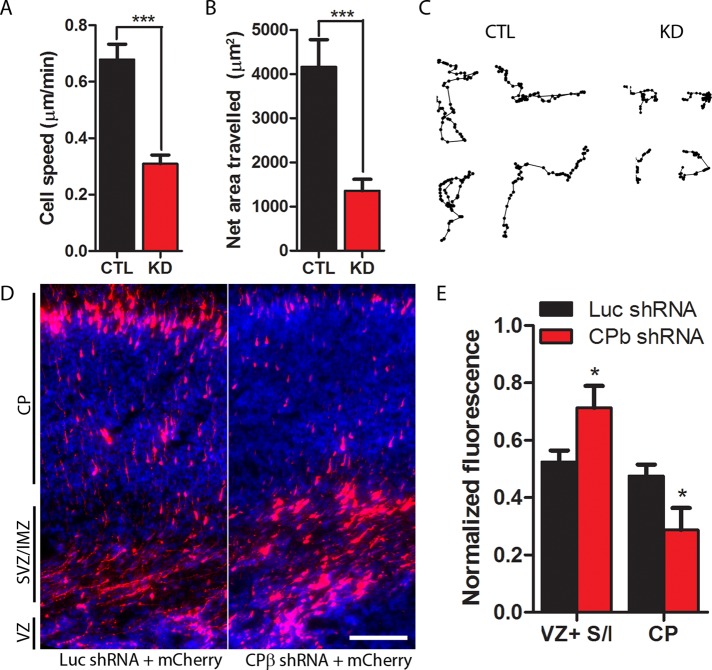
To test whether CP is required for mammalian cell migration, we used time-lapse imaging to measure rates of cell migration in control and CP-depleted cells. The average cell migration rate of Scramble-transfected cells was 0.68 μm/min (Figure 2A), a value similar to those reported previously for a related cell type, B16F1 cells (Hotulainen et al., 2005; Yang et al., 2007). Silencing of CP reduced rates of cell migration by more than half, from 0.68 μm/min to 0.31 μm/min (Figure 2A). It also reduced the net area traversed by cells over 6 h by 67% (Figure 2B). Representative tracks of Scramble-transfected and CP-depleted cells are shown in Figure 2C.
FIGURE 2:
Depletion of CP impairs cell migration in vitro and in vivo. (A) Average migration speeds of Scramble-transfected (CTL) and CP-depleted (KD) B16F10 cells. Cells were visualized by DIC imaging and tracked by their nuclei every 5 min for 6 h. (B) The average net area traversed by Scramble-transfected and CP-depleted cells in 6 h. Net area traversed was calculated for each cell by multiplying the greatest absolute difference between x-coordinates by the greatest absolute difference between y-coordinates. For (A) and (B), 34–36 cells were analyzed per condition; ***p < 0.001. (C) Representative migration tracks from Scramble-transfected and CP-depleted cells. (D) Neurons cotransfected with mCherry and CP shRNA (right) have impaired migration to the cortical plate compared with neurons cotransfected with mCherry and firefly luciferase shRNA (control; left). CP, cortical plate; SVZ/IMZ, subventricular zone/ intermediate zone; VZ, ventricular zone. Scale bar: 100 μm. (E) Quantification of cortical migration. For each condition, fluorescence intensity from each region was normalized to total fluorescence from all regions. Five animals from each category were analyzed; *p < 0.05. VZ+S/I, ventricular zone + subventricular/intermediate zone; CP, cortical plate.
We next investigated the effects of CP depletion on the migration of murine cortical neurons in vivo. After birth, cortical neurons migrate from the ventricular zone to the cortical plate (Bielas et al., 2004). Ninety-six hours after in utero electroporation, nearly half of control neurons, transfected with firefly luciferase shRNA, had reached the cortical plate, but less than a third of CP-depleted neurons had done so (Figure 2, D and E). Together these results demonstrate that CP is indeed critical for migration of mammalian cells in vitro and, more importantly, in vivo. Our results are consistent with a prior study in Dictyostelium (Hug et al., 1995) in which mutant cells underexpressing CP moved 35% slower than did control cells.
CP is detected in filopodia of multiple cell types
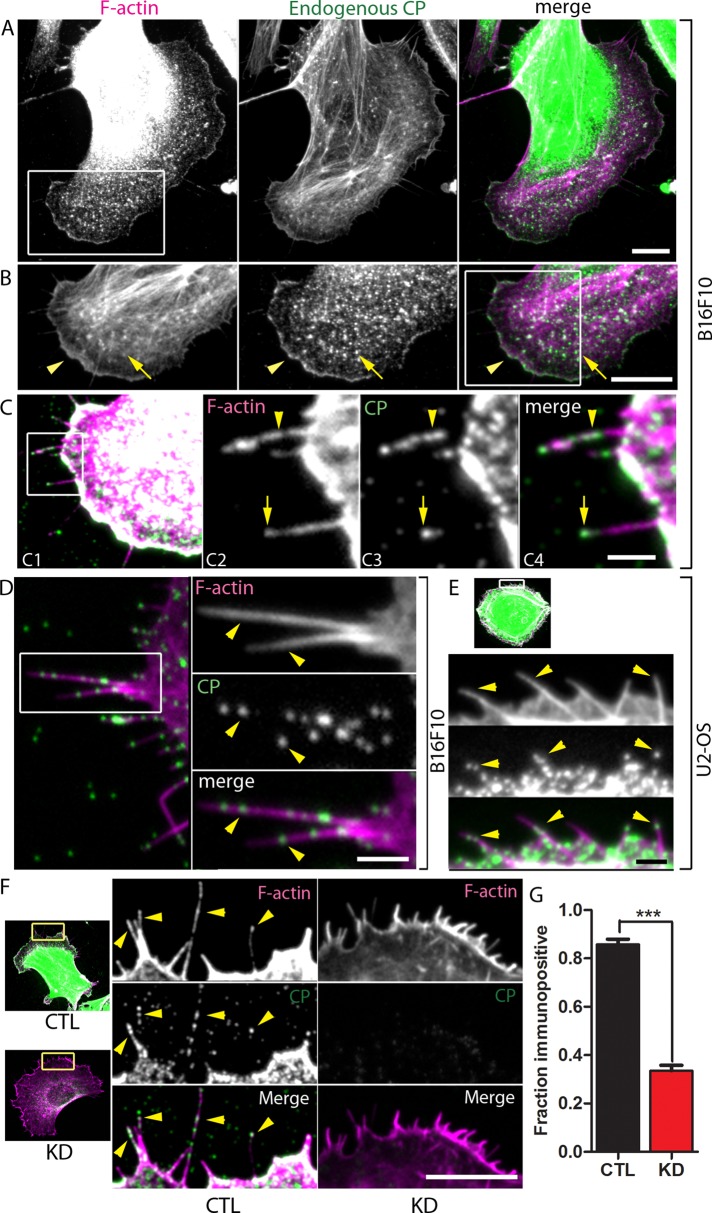
As expected, we clearly detected CP at the leading edge of migrating B16F10 mouse melanoma cells (Figure 3B, arrowhead). The punctate pattern of CP-ir we observed is similar to that observed previously by other investigators (Schafer et al., 1998; Rogers et al., 2003; Applewhite et al., 2007; Yang et al., 2007). CP has also been shown to be associated with sites of actin assembly in the lamella (Schafer et al., 1998), and we detected CP-ir in the lamella as well (Figure 3B, arrow).
FIGURE 3:
Endogenous CP is detected in filopodia of mammalian cells and filopodial CP-ir decreases with shRNA treatment. (A) B16F10 melanoma cell stained using antibody to endogenous CP. Note substantial CP signal in cell body. (B) A magnified view of the boxed region in (A). CP is present at the leading edge (arrowhead) and in the lamella (arrow). (C) Enlarging and increasing the contrast of the boxed region in (B) shows that CP-ir is detected in filopodia (C1). Further magnification of the boxed region in (C1) shows that CP-ir is detected both along the length (arrowhead) and at the presumptive tip (arrow) of filopodia (C2–C4). (D) An example of CP-ir in filopodia of a different B16F10 cell. (E) CP-ir is detected along filopodia of U2-OS cells. Arrowheads in (D) and (E) indicate CP-ir. (F) The CP signal is decreased in filopodia from CP-silenced cells (KD) compared with that in filopodia from Scramble-transfected cells (CTL). Arrowheads indicate some of the CP-ir puncta. (G) The proportion of CP-positive filopodia is reduced in CP-depleted cells compared with that in Scramble-transfected cells. For each condition, a total of 250–445 filopodia from 13 to 20 cells across two independent experiments were analyzed; ***p < 0.001. Scale bars: A, B, and F, 10 μm; C–E, 2 μm.
Interestingly, although CP's role has been studied most extensively at the leading edge, the majority of CP-ir was not at the leading edge but instead in the cell body (Figures 1A and 3A); this distribution persisted even after extracting soluble CP from live cells with 1% Triton before fixation (unpublished data). This pattern of immunoreactivity, in which the majority of the CP signal is in the cell body, is similar to that appearing in published images of endogenous CP in various cell types (Schafer et al., 1998; Rogers et al., 2003). CP has been shown to be an integral component of both the dynactin complex (Schafer et al., 1994) and the WASH complex (which associates with recycling endosomes; Derivery et al., 2009). Moreover, CP may have microtubule-associated functions (Davis et al., 2009; Bartolini et al., 2012). Therefore CP may fulfill other cellular roles besides leading-edge motility.
Unexpectedly, we also clearly detected low levels of CP-ir in a punctate distribution along filopodia of B16F10 cells (Figure 3, C, D, and F, CTL). This was surprising because CP has been postulated to be absent from filopodia (Svitkina et al., 2003; Faix and Rottner, 2006), and indeed, the relative absence of CP is thought to be important for filopodia to emerge from the dendritic actin network (Svitkina et al., 2003; Wear and Cooper, 2004).
We quantified CP-ir puncta along the protruding portion of filopodia (since puncta localization within the embedded portion is ambiguous). Out of 311 filopodia examined from B16F10 cells, 79% had at least one CP punctum, and 52% had at least two puncta. On average, there were 2.4 CP puncta detected per filopodia, corresponding to ∼1 punctum detected per micrometer length of filopodium. The majority of immunopositive filopodia (91%) had CP puncta along their length, but a large fraction of immunopositive filopodia (36%) also had CP puncta detectable at the presumptive tip.
To confirm whether filopodial CP-ir was a general phenomenon, we examined another cell line (U2-OS osteosarcoma cells; Figure 3E), as well as growth cones in primary hippocampal neuronal cultures (Figure S2). CP-ir was clearly detected in the filopodia of both these cell types. Moreover, knockdown of CP by shRNA significantly decreased CP-ir within filopodia (Figure 3F) and decreased the fraction of CP-positive filopodia (Figure 3G).
CP depletion reduces filopodial length and changes filopodial shape
Remarkably, filopodial length was significantly decreased by a third in CP-depleted cells (Figures 1D and 4A). For measurement of filopodial length, the entire actin bundle was quantified regardless of its position relative to the lamellipodium, since filopodia, retraction fibers, and microspikes are dynamically interchangeable (Svitkina et al., 2003). A frequency histogram comparing shRNA-transfected and Scramble-transfected cells shows that CP depletion results in selective loss of longer filopodia and therefore a higher proportion of shorter filopodia (Figure 4B). These novel findings imply that CP depletion directly or indirectly reduces filopodial length.
FIGURE 4:
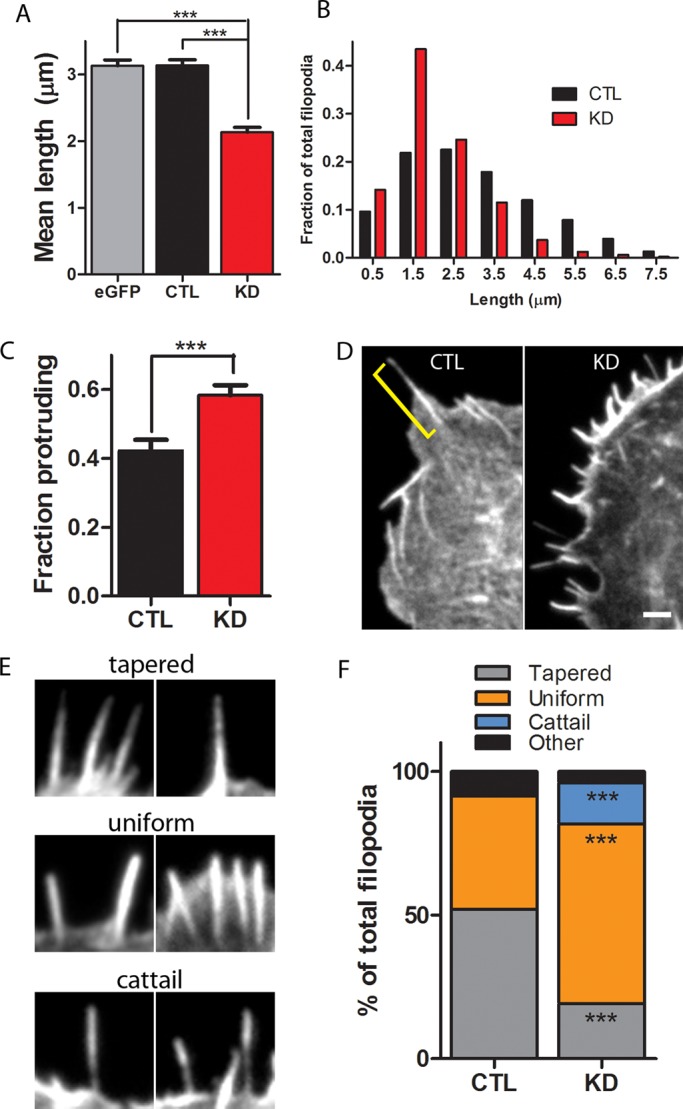
CP depletion reduces filopodial length and changes filopodial morphology. (A) Quantification of filopodial length in Scramble-transfected (CTL) and CP-knockdown (KD) cells. Length of the entire actin bundle was measured (as indicated in (D) by the yellow bracket), as described in Svitkina et al. (2003), and averaged per cell. A total of 47–69 cells per condition were analyzed across three different experiments and include 1600–3200 filopodia/condition; ***p < 0.001. (B) Frequency histogram comparing the lengths of Scramble-transfected and shRNA-transfected cells. (C) Knockdown of CP increases the fraction of filopodial length that protrudes beyond the cell margin. Filopodial values were averaged per cell, and 600–1150 filopodia from 30 to 35 cells across three independent experiments were analyzed; ***p < 0.001. (D) A representative Scramble-transfected (left) and CP-knockdown (right) cell. Note that a greater portion of each individual filopodium is embedded within the lamellipodium in the Scramble-transfected cell. (E) Categories of filopodial shapes found in Scramble-transfected and CP-depleted cells. See text for category descriptions. (F) CP knockdown alters the apparent shape of filopodia. A total of 325–355 filopodia from two independent experiments were analyzed for each group; ***p < 0.001. Scale bar: 2 μm.
Besides dramatically reducing filopodial length, other effects of CP depletion on filopodial morphology were apparent. First, nearly the entire length of individual filopodia in CP-depleted cells appeared to be protruding beyond the cell margin (Figure 4, C and D; see Materials and Methods for details on measurements). In contrast, filopodia from Scramble-transfected cells often had much of their length embedded within the cell lamellipodium (Figure 4, C and D).
Second, the apparent shapes of filopodia from CP-depleted cells, based on phalloidin staining, were visibly altered (Figure 4, E and F). More than 50% of the filopodia from Scramble-transfected cells had a cone-like or tapered appearance, with a smaller percentage having a more rod-like or uniform appearance (Figure 4, E and F). However, the majority of filopodia from shRNA-transfected cells had a rod-like appearance (Figure 4, E and F). In addition, a significant fraction of filopodia in CP knockdown cells had a “cattail” appearance, in which the base was visibly thinner than the shaft and tip regions (Figure 4E). This type of filopodium was rarely seen in Scramble-transfected cells. Of note, a similar filopodial morphology (“club-like filopodia”) was described with formin overexpression (a manipulation expected to decrease relative capping activity; Yang et al., 2007). Collectively our results demonstrate that CP activity is essential for normal length and morphology of filopodia. These effects of CP depletion on filopodial length, shape, and F-actin concentration (see next section) were confirmed by using shRNA directed against a second sequence of the CP β subunit (Figure S3; see Materials and Methods for sequence information).
CP depletion increases cellular and filopodial F-actin concentration
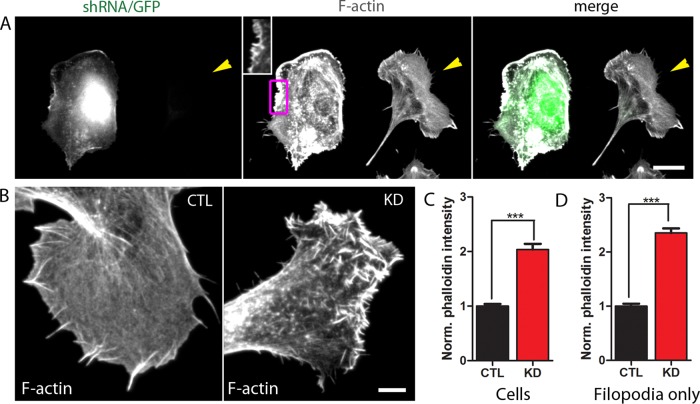
Strikingly, knockdown of CP caused a significant increase in F-actin concentration inside cells, as measured by phalloidin staining (Figure 5A). This increased staining was especially evident at cell margins at low magnification. At higher magnification (Figure 5, A, inset, and B), it was clear that the phalloidin staining of individual filopodia was also dramatically increased in CP-depleted cells. Quantification of phalloidin intensities demonstrated a greater than twofold increase in F-actin concentration globally in CP-depleted cells (Figure 5C) and within individual filopodia of CP-depleted cells (Figure 5D). These data suggest that filopodia from CP-depleted cells have a greater number of F-actin filaments than do those from Scramble-transfected cells. In other words, decreased capping leads to greater actin polymerization. These results are consistent with those in Dictyostelium (Hug et al., 1995), in which cells underexpressing CP had an increase in F-actin. CP silencing has also been shown to increase total actin levels in both Dictyostelium (Hug et al., 1995) and mammalian cells (Edwards et al., 2013).
FIGURE 5:
F-actin concentration is increased in CP-depleted cells. (A) F-actin concentration as determined by phalloidin staining is visibly increased in a CP-knockdown cell (left cell in image) compared with an untransfected cell (right cell, indicated by arrowhead). Inset is displayed with lower contrast to illustrate that much of the phalloidin intensity at the cell margin of the CP-depleted cell is associated with short filopodia. (B) Enlarged image of filopodia in a different Scramble-transfected cell (CTL, left) and CP-knockdown cell (KD, right). These two phalloidin-stained cells were imaged under identical conditions and are displayed with identical brightness and contrast settings. (C) Mean phalloidin intensity is increased globally in CP-knockdown cells. A total of 50–80 cells from two independent experiments were analyzed; ***p < 0.001. Phalloidin intensities were normalized to the mean intensity of Scramble-transfected cells. (D) Mean phalloidin intensity is increased in individual filopodia from CP-knockdown cells. A total of 180–340 filopodia from the same two independent experiments as in (C) were analyzed; ***p < 0.001. Phalloidin intensities were normalized to the mean intensity of filopodia from Scramble-transfected cells. Scale bars: A, 20 μm; B, 5 μm.
CP depletion reduces filopodial dynamics
To understand whether CP depletion has consequences for filopodial function, we examined filopodial behavior using time-lapse imaging. The average protrusion rate for Scramble-transfected filopodia was 95 nm/s, a value similar to that reported previously for B16F1 cells (Yang et al., 2007; Figure 6A and Supplemental Video S1). However, filopodia from CP-depleted cells had significantly reduced protrusion rates, on average only 61 nm/s (Figure 6A and Video S2). Rates of filopodial shortening were similar in control cells and CP-depleted cells (Figure 6A). In addition, filopodia from shRNA-transfected cells spent significantly less time growing and more time pausing (Figure 6B and Videos S1 and S2). Thus we conclude that CP depletion reduces filopodial dynamics and could therefore have functional consequences for cell-based activities that require proper filopodial function.
FIGURE 6:
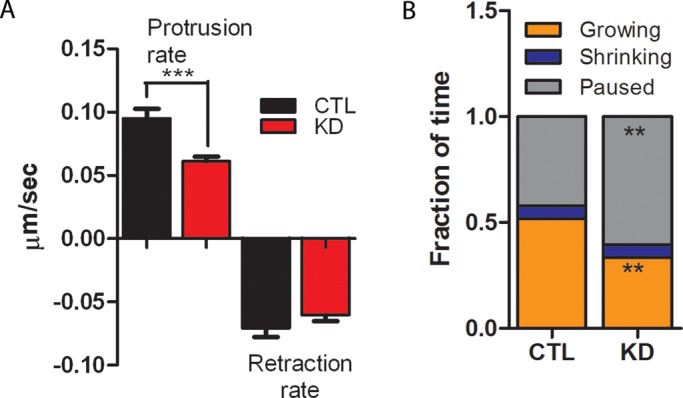
CP depletion reduces filopodial dynamics. (A) Quantification of filopodial protrusion and retraction rates. CTL, Scramble-transfected cells; KD, CP-knockdown cells. (B) Filopodia from CP-depleted cells spend more time pausing. For each condition in (A) and (B), 51–56 filopodia from 13 to 14 time-lapse sequences were analyzed; (A) ***p < 0.001; (B) **p < 0.01. See Materials and Methods for details on quantification.
DISCUSSION
CP is an essential player in creating the actin architecture of lamellipodia, which in turn has been thought necessary for cell protrusion and migration (Pollard and Borisy, 2003; Le Clainche and Carlier, 2008). CP is necessary for actin-based motility in vitro (Loisel et al., 1999) and for migration in vivo in Dictyostelium (Hug et al., 1995). Moreover, mammalian cells depleted of CP have a decreased lamellipodial protrusion rate (Mejillano et al., 2004). However, recent work has questioned the need for normal lamellipodial dynamics in actual cell migration (Gupton et al., 2005; Suraneni et al., 2012; Wu et al., 2012; Edwards et al., 2013). In addition, the requirement for CP in mammalian cell migration has not been established.
We report here that depleting CP impairs mammalian cell migration in vitro and in vivo. Therefore CP is necessary for proper locomotion. CP's role in migration could result from its proper function in the lamellipodia or in other cell compartments. For example, some investigators have proposed that the lamella, the region behind the lamellipodium, drives net cell displacement (Ponti et al., 2004; Gupton et al., 2005). CP is present in the lamella (see Figure 3, A and B) and has been shown to associate with sites of actin assembly in this region (Schafer et al., 1998). Thus CP function in the lamella may be vital for cell motility.
In addition, cells without normal lamellipodia are postulated to use a filopodia-based system to locomote (Suraneni et al., 2012; Wu et al., 2012). Though CP has been posited to be absent from filopodia (Svitkina et al., 2003; Mejillano et al., 2004; Faix and Rottner, 2006), we find that CP is detectable in filopodia from multiple mammalian cell types and is required for normal filopodial form and dynamics (see following discussion). Filopodia have important sensing and adhesion roles in cell migration (Mattila and Lappalainen, 2008; Arjonen et al., 2011), and it is possible that CP's role in filopodia is also necessary for proper cell migration.
Our findings regarding the immunolocalization of CP within filopodia and the effects of CP silencing yield new insights into filopodial structure. Given that CP is known to bind only to barbed ends of actin filaments (Wear et al., 2003), the punctate distribution of CP along the length of filopodia (as opposed to CP localizing exclusively to filopodial tips) suggests that actin filaments within filopodia are not necessarily of uniform length (Figure 7). Instead, it suggests that individual filaments may grow to different lengths before being stochastically capped by CP. Filopodia in which relatively few filaments extend the entire length from base to tip have a tapered appearance, whereas filopodia that contain actin filaments of equal lengths appear to be uniform. In control cells, most filopodia are tapered, with a smaller but significant fraction having a uniform morphology. In cells depleted of CP, decreased capping of individual filaments within filopodia would allow all of the filaments to grow uniformly. Thus there is a dramatic increase in the proportion of uniform filopodia, offset by a concomitant decrease in the proportion of tapered filopodia.
FIGURE 7:
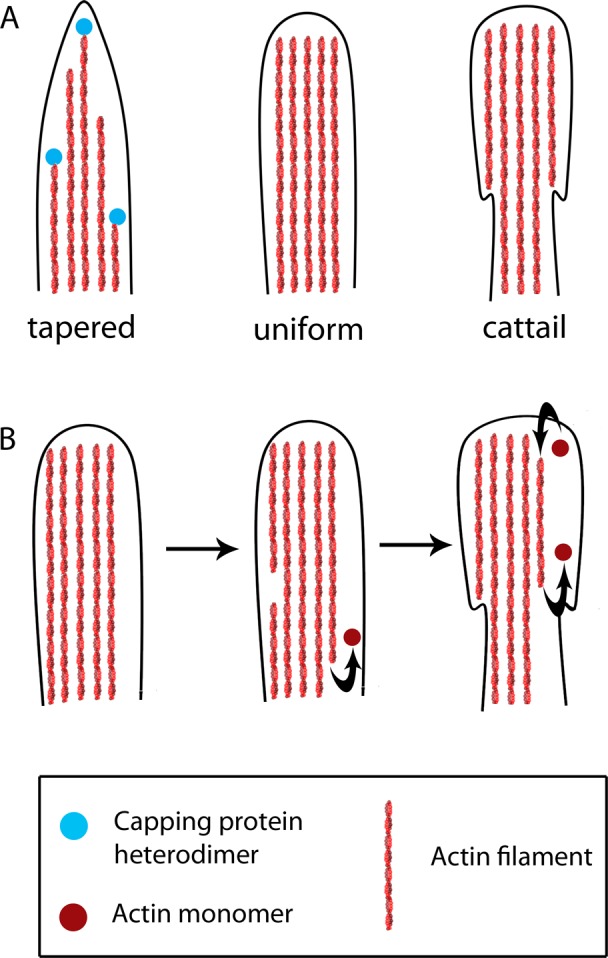
A model for how CP in filopodia influences filopodial shape. (A) We propose that stochastic capping of individual actin filaments within filopodia results in heterogeneity of filament lengths and consequently a tapered appearance (left). In filopodia with reduced capping activity, all filaments span the entire length of the filopodia, giving the filopodium a uniform appearance (middle). In CP-depleted cells, cattail filopodia are present, in which there is a greater density of actin filaments at the tip of the filopodia than at the base (right). (B) A possible mechanism for the creation of cattail filopodia. In cells depleted of CP, the majority of filopodia are of the uniform variety (left). Actin filaments that sever or begin to depolymerize from their pointed ends (middle) are normally capped and cleared away by retrograde flow. However, decreased capping within filopodia from CP-depleted cells results in these filaments treadmilling and persisting (right).
Reducing CP also induces the emergence of filopodia that have a thicker tip and a thinner base, which we have named “cattail” filopodia. One explanation for this morphology is that some filopodial actin filaments may normally sever or depolymerize from their pointed ends and begin to treadmill (Figure 7). In control filopodia, CP caps these treadmilling filaments, and retrograde flow moves them out of filopodia; hence, they are transient structures and not frequently detected. Fewer than 1% of control filopodia fell into this category (Figure 4F). In CP-depleted filopodia, however, these filaments may persist and thus become easily detected. Indeed, in time-lapse imaging of knockdown cells, we sometimes observed uniform filopodia morphing into cattail filopodia, indicating a temporal relationship between uniform and cattail filopodia (unpublished data). Our results suggest that CP within filopodia may directly influence filopodial morphology.
Why are filopodia in CP-depleted cells remarkably shorter than those in control cells? This observation may seem counterintuitive if one assumes that a decrease in capping allows actin filaments to polymerize for longer periods and attain greater lengths. However, unchecked polymerization may consume a limiting cellular factor, such as monomeric G-actin. CP silencing induced a 2.5-fold increase in filopodial density (Figure 1C). Moreover, individual filopodia from CP-silenced cells probably contain an increased number of actin filaments, as evidenced by their twofold increase in phalloidin intensity (Figure 5).
The burst of actin polymerization evidenced by increased filopodial density and increased F-actin content in individual filopodia conceivably depletes cellular G-actin and limits filopodial length. Indeed, mathematical modeling has suggested that growth of filopodia composed of more than ∼30 filaments would be limited by G-actin diffusion from the base to the growing filopodial tip (Mogilner and Rubinstein, 2005). Moreover, CP is thought to be essential for maintaining an adequate supply of actin monomers for actin-based motility (Carlier and Pantaloni, 1997; Loisel et al., 1999; Pollard and Borisy, 2003). Thus we suggest that inhibiting CP activity indirectly reduces filopodial length due to depletion of actin monomer.
Why does a greater part of individual filopodia in CP-depleted cells extend beyond the cell margin (Figure 4, C and D)? A possible explanation takes into account relative rates of retrograde flow, which exists in filopodia as well as in lamellipodia (Mallavarapu and Mitchison, 1999). A reduced rate of F-actin polymerization at filopodial tips in CP-depleted cells (as evidenced by a decreased protrusion rate) would be expected to reduce retrograde flow within filopodia. Thus a greater fraction of the filopodial length protrudes beyond the lamellipodia. Another explanation could be that, because lamellipodial protrusion is disrupted with CP depletion (Mejillano et al., 2004), the lamellipodium does not extend as far with respect to the filopodia as it does in control cells.
Why are filopodia in CP-depleted cells less dynamic than those in control cells? One possibility is that CP depletion indirectly reduces filopodial dynamics by limiting actin monomer supply. Limiting levels of G-actin reduce polymerization at filopodial tips and thus decrease protrusion rate. Consequently, filopodia spend less time growing and more time pausing. Interestingly, theoretical work has suggested that stochastic capping of individual actin filaments is necessary for macroscopic filopodial dynamics (Zhuravlev and Papoian, 2009), and this mechanism may also contribute to our observation of reduced filopodial dynamics in CP-depleted cells.
Consistent with other studies, we observe a dramatic increase in filopodial density upon inhibition of capping activity (Figure 1). We therefore agree with other investigators that a relative decrease in capping activity seems necessary for filopodia to emerge (Svitkina et al., 2003; Mejillano et al., 2004). We suggest, however, that low levels of CP within filopodia are essential for normal filopodial morphology and dynamic behavior. It is perhaps not so surprising that CP is detectable in filopodia: other actin-capping proteins, such as gelsolin and Eps8, are already known to be important components of axonal filopodia (Gallo, 2013).
Moreover, anti-capping proteins such as Ena/VASP (Lebrand et al., 2004; Mattila and Lappalainen, 2008) and the diaphanous formins (Yang et al., 2007; Faix et al., 2009) are present in filopodia and are thought to have important roles in filopodial form and function. These studies have shown that both excessive capping and anti-capping activity perturb filopodial morphology and dynamics. We therefore suggest that the anti-capping activity of Ena/VASP and formins within filopodia may be balanced by the capping activity of capping protein to produce normal filopodia. In sum, our data suggest that the filopodial functions of CP, in addition to other potential roles like microtubule regulation and membrane trafficking (Davis et al., 2009; Derivery et al., 2009; Bartolini et al., 2012), may contribute to the critical role of CP in cell migration in vivo.
MATERIALS AND METHODS
Reagents
Plasmids.
We created an shRNA construct to CPβ based on the T1 sequence described in Mejillano et al. (2004; CCTCAGCGATCTGATCGAC). We cloned this sequence into pSuper vector (a generous gift from T. Wittmann, University of California, San Francisco) that contained the polymerase-III H1-RNA promoter (Brummelkamp et al., 2002). We also created a Scrambled sequence (GCACCCGTCTTCAACAGGT) and cloned it into p-Super. We additionally used an shRNA directed against a second sequence in the CPβ subunit, GCACGCTGAATGAGATCTA. Both the construct containing this target sequence (T2) and one containing a mismatch control sequence (T2*), were generous gifts from T. Svitkina (University of Pennsylvania) and have been published previously (Mejillano et al., 2004). Enhanced green fluorescent protein (eGFP)-tagged versions of the α2 and β2 subunits of CP as well as CP holoprotein were generous gifts from D. Schafer (University of Virginia). Red fluorescent protein (RFP)-LifeAct was a generous gift from R. Truant (McMaster University, Canada).
Antibodies.
A polyclonal antibody against the CP β2 subunit was purchased from Millipore (AB6017; Billerica, MA) and was used at a dilution of 1:200 in B16F10 and U2-OS cells and a dilution of 1:50 in primary hippocampal neurons. Secondary antibodies (Alexa Fluor 488, 568, or 647) and Alexa Fluor 568 or 647-phalloidin were purchased from Life Technologies (Carlsbad, CA) and used at a dilution of 1:500.
Cell culture
B16F10 cells were a generous gift from S. Dowdy (University of California, San Diego), and U2-OS cells were a generous gift from G. Yeo (University of California, San Diego). Both cells lines were cultured in DMEM supplemented with 10% fetal bovine serum. Primary hippocampal neurons were cultured as described (Calabrese and Halpain, 2005).
B16F10 cells were transfected with Scramble or shRNA to CP for 5 d using Lipofectamine 2000 (Life Technologies) in 24-multiwell plates. For live-cell imaging experiments, cells were cotransfected with RFP-LifeAct to visualize the actin cytoskeleton. After 4 d, cells were trypsinized and replated onto glass coverslips that had been sequentially precoated with poly-l-ornithine (0.1 mg/ml overnight; Sigma-Aldrich, St. Louis, MO) and laminin (5 μg/ml for 3–4 h; Life Technologies) in preparation for imaging.
Immunostaining
For antibody staining, cells on glass coverslips were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) with 8% sucrose (wt/vol) for 20 min at 37°C. Cells were permeabilized in 0.2% Triton in PBS for 5 min, blocked with 2% bovine serum albumin for 45 min, incubated with primary antibody for 1 h at room temperature, washed with PBS, and incubated with secondary antibody and labeled phalloidin for 45 min at 37°C. After being washed, coverslips were mounted with Aquamount (Thermo Fisher Scientific, Lafayette, CO), dried overnight, and imaged.
In utero electroporation
In utero microinjection and electroporation was performed as previously described (Simó et al., 2010), in accordance with procedures approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. Briefly, timed pregnant CD1 mice (Charles River) were anesthetized on embryonic day (E) 14.5, and the uterine horns were exposed. Approximately 2 μg of a lentiviral vector DNA, containing an H1 promoter driving shRNA against CPβ or firefly luciferase (as a control) and a CAG promoter driving mCherry fluorescent protein, was microinjected into the lateral ventricles and introduced into the progenitor cells lining the lateral ventricle by electroporation. Development was allowed to continue until E18.5. Fetal brains were sectioned on a cryostat and imaged.
Image acquisition and analysis
Images for all fixed and live-cell analyses (except for cortical migration; see discussion later in this section) were acquired on an Olympus IX70 inverted microscope mounted with a Yokogawa CSU-X1 Spinning disk using either a 60× or 20× oil objective or a 20× dry air objective. Maximum projection images were acquired for all analyses, except for those related to cell migration, for which single-plane images containing the cell nucleus were obtained. For live imaging, samples were kept at 35°C in a humid chamber containing 5% CO2. For movies of filopodial dynamics, images were acquired every 5 s for 2 min. For cell migration experiments, cells were imaged using differential interference contrast (DIC) imaging every 5 min for 6 h. Cell tracking was accomplished by tracking the position of the nucleus in individual consecutive time-lapse images using the Manual Tracking plug-in in ImageJ.
For measurement of filopodial length, the entire actin bundle was quantified regardless of its position relative to the lamellipodium, since filopodia, retraction fibers, and microspikes are dynamically interchangeable (Svitkina et al., 2003). Measurements were based on the methods of Vignjevic et al. (2006). For the fractional length protruding beyond the cell margin (Figure 4C), the length protruding beyond the cell edge was divided by the total length. The segmented line function of ImageJ was used to manually measure all filopodia. Lamellipodial lengths of Scramble-transfected cells were also manually measured using ImageJ. CP-depleted cells often had abnormal lamellipodia, and so for these cells, the perimeter of the cell region containing filopodia was measured.
Filopodial morphologies were classified qualitatively from phalloidin images as follows: “tapered,” base detectably wider than tip; “uniform,” similar width along entire length, with base and tip not detectably different; “cattail,” base detectably thinner than tip; “other,” filopodial-like protrusions not readily classified. A second investigator blinded to the treatment analyzed a subset of the images using these criteria and obtained strikingly similar results.
For analysis of filopodial dynamics (Figure 6), 2-min time-lapse sequences of Scramble-transfected or shRNA-transfected cells cotransfected with RFP-LifeAct were analyzed. Lengths of individual filopodia were measured every 5 s until the filopodium disappeared or merged with a neighboring filopodium. Total growth (or total shrinkage) in micrometers was divided by total time spent growing (or shrinking) in seconds to obtain growth (or shrinkage) rate. The fraction of total time spent growing, shrinking, or pausing was also calculated by considering a filopodium to be pausing if its length did not change by more than 2 pixels between frames.
For cortical migration experiments, sections of fetal brain were imaged using a Nikon E800 scope and 10× objective. The fluorescence of mCherry-expressing neurons in each cortical region (ventricular zone, intermediate zone/subventricular zone, and cortical plate) was quantified using ImageJ and normalized to total fluorescence in all regions.
Statistical analysis was done using GraphPad Prism. Student's two-tailed unpaired t test or one-way analysis of variance (with Tukey's postcomparison test) was used for all statistical analyses, except for comparison of cortical migration (Figure 2), for which a one-tailed t test was used, and for filopodial morphologies (Figure 4), for which the binomial test was used.
Supplementary Material
Acknowledgments
We thank members of the Halpain lab for advice and for providing hippocampal cell cultures. We especially thank Soroosh Aidun for assistance with quantitative image analyses. We acknowledge Steven Dowdy, Dorothy Schafer, Tatyana Svitkina, Ray Truant, Torsten Wittmann, and Gene Yeo for providing reagents. We also thank Andres Santos (University of California, San Diego) for statistical advice and Alex Mogilner (University of California, Davis) for valuable input and critical reading of the manuscript. This work was supported by National Institutes of Health grants MH087823 (S.H.) and NS080194 (J.C.); S.A.S. was the recipient of a National Science Foundation Graduate GK–12 STEM fellowship.
Abbreviations used:
- CP
capping protein
- CP-ir
CP immunoreactivity
- DIC
differential interference contrast
- eGFP
enhanced green fluorescent protein
- PBS
phosphate-buffered saline
- RFP
red fluorescent protein
- shRNA
short hairpin RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0749) on May 14, 2014.
*Present address: Department of Pathology and Cell Biology, Columbia University Medical Center, 630 West 168th Street, New York, NY 10032.
REFERENCES
- Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adhes Migration. 2011;5:421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–4040. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol. 2004;20:593–618. doi: 10.1146/annurev.cellbio.20.082503.103047. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Calabrese B, Halpain S. Essential role for the PKC target MARCKS in maintaining dendritic spine morphology. Neuron. 2005;48:77–90. doi: 10.1016/j.neuron.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Carlier M-F, Pantaloni D. Control of actin dynamics in cell motility. J Mol Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- Davis DA, Wilson MH, Giraud J, Xie Z, Tseng H-C, England C, Herscovitz H, Tsai L-H, Delalle I. Capzb2 interacts with β-tubulin to regulate growth cone morphology and neurite outgrowth. PLoS Biol. 2009;7:e1000208. doi: 10.1371/journal.pbio.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Edwards M, Liang Y, Kim T, Cooper J. Physiological role of the interaction between CARMIL1 and capping protein. Mol Biol Cell. 2013;24:3047–3055. doi: 10.1091/mbc.E13-05-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TEB, Rottner K. Filopodia: complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Gallo G. Mechanisms underlying the initiation and dynamics of neuronal filopodia: from neurite formation to synaptogenesis. Int Rev Cell Mol Biol. 2013;301:95–156. doi: 10.1016/B978-0-12-407704-1.00003-8. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D. Cell migration without a lamellipodium translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci Signaling. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Papoian GA. Mechano-chemical feedbacks regulate actin mesh growth in lamellipodial protrusions. Biophys J. 2010;98:1375–1384. doi: 10.1016/j.bpj.2009.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA. Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell. 1995;81:591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Carlier M-F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Bridgman C. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier M-F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Mallavarapu A, Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton S, Waterman-Storer C, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Welch MD, Machesky LM, Bridgman PC, Meyer SM, Cooper JA. Visualization and molecular analysis of actin assembly in living cells. J Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simó S, Jossin Y, Cooper JA. Cullin 5 regulates cortical layering by modulating the speed and duration of Dab1-dependent neuronal migration. J Neurosci. 2010;30:5668–5676. doi: 10.1523/JNEUROSCI.0035-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J Cell Biol. 1981;91:695–705. doi: 10.1083/jcb.91.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S-i, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maéda Y, Cooper JA. How capping protein binds the barbed end of the actin filament. Curr Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan S, Berginski M, Haynes E, Sharpless N, Griffith J, Gomez S, Bear J. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S-i, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev PI, Papoian GA. Molecular noise of capping protein binding induces macroscopic instability in filopodial dynamics. Proc Natl Acad Sci USA. 2009;106:11570–11575. doi: 10.1073/pnas.0812746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.