The kinesin-3 family–specific, positively charged insert, the K-loop, in loop 12 of the motor domain plays a critical role in cargo transport by enhancing the initial interaction of cargo-bound dimeric motors with the microtubule. The replacement of the K-loop, however, does not abolish the superprocessive motion of this class of kinesin motors.
Abstract
The kinesin-3 family (KIF) is one of the largest among the kinesin superfamily and an important driver of a variety of cellular transport events. Whereas all kinesins contain the highly conserved kinesin motor domain, different families have evolved unique motor features that enable different mechanical and functional outputs. A defining feature of kinesin-3 motors is the presence of a positively charged insert, the K-loop, in loop 12 of their motor domains. However, the mechanical and functional output of the K-loop with respect to processive motility of dimeric kinesin-3 motors is unknown. We find that, surprisingly, the K-loop plays no role in generating the superprocessive motion of dimeric kinesin-3 motors (KIF1, KIF13, and KIF16). Instead, we find that the K-loop provides kinesin-3 motors with a high microtubule affinity in the motor's ADP-bound state, a state that for other kinesins binds only weakly to the microtubule surface. A high microtubule affinity results in a high landing rate of processive kinesin-3 motors on the microtubule surface. We propose that the family-specific K-loop contributes to efficient kinesin-3 cargo transport by enhancing the initial interaction of dimeric motors with the microtubule track.
INTRODUCTION
Long-distance intracellular transport is carried out by the microtubule-based motor proteins kinesin and dynein (Vale, 2003). Much of our knowledge about the mechanochemistry of kinesin motors is based on studies of kinesin-1, the founding member of the kinesin superfamily (Vale, 2003; Gennerich and Vale, 2009). Kinesin-1 is a dimeric molecule that uses the alternating catalysis of its two motor domains to undergo processive motion (take many steps along the microtubule surface; Gennerich and Vale, 2009; Sindelar, 2011). Although the kinesin motor domain is highly conserved across the superfamily, alterations to the core motor domain have evolved to allow family-specific kinetic and force-generating properties critical for motor function in cells. For example, the kinesin-5 family contains an insert in loop 5 on the motor surface that is proposed to relay nucleotide binding to motion and serves as the binding site for allosteric inhibitors of this motor class (Wojcik et al., 2013). Kinesin-13 motors contain an extension in loop 2 that endows these motors with microtubule depolymerase function rather than a transport function (Walczak et al., 2013).
The kinesin-3 family (KIF) is one of the largest among the kinesin superfamily and consists of five subfamilies in mammals (KIF1, KIF13, KIF14, KIF16, and KIF28). The founding member of kinesin-3 family, CeUNC-104, was identified in Caenorhabditis elegans due to a mutation that severely affects the transport of synaptic vesicles to the axon terminal (Hall and Hedgecock, 1991; Otsuka et al., 1991). A similar function has been ascribed to the mammalian homologue of CeUNC-104, murine KIF1A (MmKIF1A; Okada et al., 1995). Other kinesin-3 motors have been found to play important roles across species in cytokinesis and in the transport of endosomes, lysosomes, dense-core granules, viral particles, signaling molecules, and mitochondria (Hirokawa et al., 2010; Franker and Hoogenraad, 2013).
A defining feature of the kinesin-3 family is a conserved insert in loop 12 of the motor domain. This insert contains a number of positively charged lysine residues and is therefore referred to as the K-loop (Figure 1A). Loop 12 forms part of the microtubule-binding interface of the kinesin motor domain (Nitta et al., 2004; Marx et al., 2009) and is thus positioned to influence a variety of functional and mechanical outputs. Pioneering work from the Hirokawa lab demonstrated that the K-loop enables diffusive motion of monomeric MmKIF1A motors along the microtubule surface by acting as a mobile tether to the negatively-charged, glutamate-rich (E-hook), C-terminal tails of the tubulin subunits (Okada and Hirokawa, 1999, 2000; Kikkawa et al., 2000). Since that time, the Brownian motion of a variety of motors and microtubule-associated proteins (MAPs) along microtubules has been shown to depend on similar electrostatic interactions (Cooper and Wordeman, 2009).
FIGURE 1:
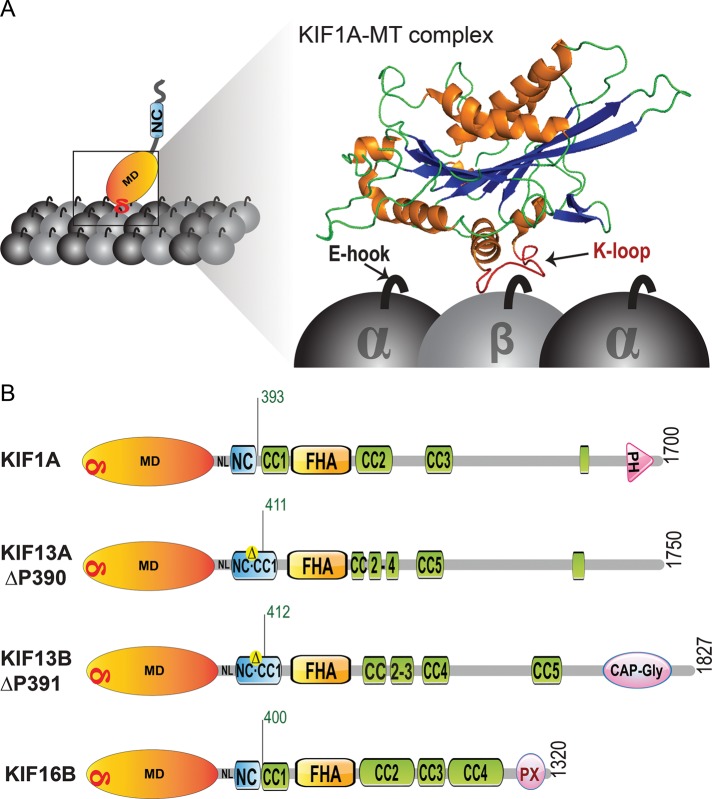
Schematic of kinesin-3 motors used in this study. (A) Schematic showing the location of the conserved K-loop of KIF1A (PDB 2ZFK) interacting with the E-hooks of the tubulin subunits. (B) Schematic of the domain organization of KIF1A, KIF13A, KIF13B, and KIF16B motors. Truncations that create the constitutively active, dimeric motors KIF1A(1-393), KIF13A(1-411ΔP), KIF13B(1-412ΔP), and KIF16B(1-400) are indicated by a black tick mark with the corresponding amino acid number in green text. δ, K-loop; CC, coiled coil; FHA, forkhead-associated; MD, motor domain; NC, neck coil; PH, pleckstrin homology; PX, Phox homology.
Whether the K-loop contributes to the mechanical and/or functional outputs of dimeric kinesin-3 motors has not been addressed. One possibility is that electrostatic interactions of the K-loop with the microtubule contribute to the diffusive motion of dimeric kinesin-3 motors. It is also possible that the K-loop evolved to endow kinesin-3 motors with family-specific kinetic and/or motility properties, such as the ability to generate superprocessive motility (Soppina et al., 2014). Here, using cellular and single-molecule assays, we examine the role of the K-loop sequence for three members of the kinesin-3 family, KIF1A, KIF13B, and KIF16B. We show that replacement of the K-loop does not abolish the superprocessive motion of this class of kinesin motor. Instead, the K-loop endows kinesin-3 motors with a high microtubule on-rate in their ADP-bound state.
RESULTS
The K-loop sequence facilitates the diffusion of dimeric kinesin-3 motors on the microtubule surface in the ADP-bound state
Although the exact sequence and number of lysine residues are not conserved, all mammalian kinesin-3 motors contain a K-loop insertion in loop 12 of their motor domain (Figure 2A). To examine the function of this family-specific insert, we used constitutively active, truncated versions of KIF1A, KIF13A, KIF13B, and KIF16B that are both dimeric and superprocessive (Soppina et al., 2014). Specifically, KIF1A(1-393), KIF13A(1-411ΔP), KIF13B(1-412ΔP), and KIF16B(1-400) (Figure 1B) were tagged with single or three tandem copies of a monomeric version of citrine (mCit), a variant of yellow fluorescent protein. In addition, a well-characterized dimeric and constitutively active version of kinesin-1 (KHC(1-560)) was analyzed in parallel as a motility control. Because the KIF13A and KIF13B motors are very similar (∼62% overall amino acid identity; Venkateswarlu et al., 2005) and their truncated versions showed similar motility properties and regulation mechanisms (Soppina et al., 2014), only the results for the KIF13B motor will be included for clarity. Constitutively active versions of the kinesin-3 motors KIF14 and KIF28 are not yet available, and thus these motors could not be included in our analysis.
FIGURE 2:
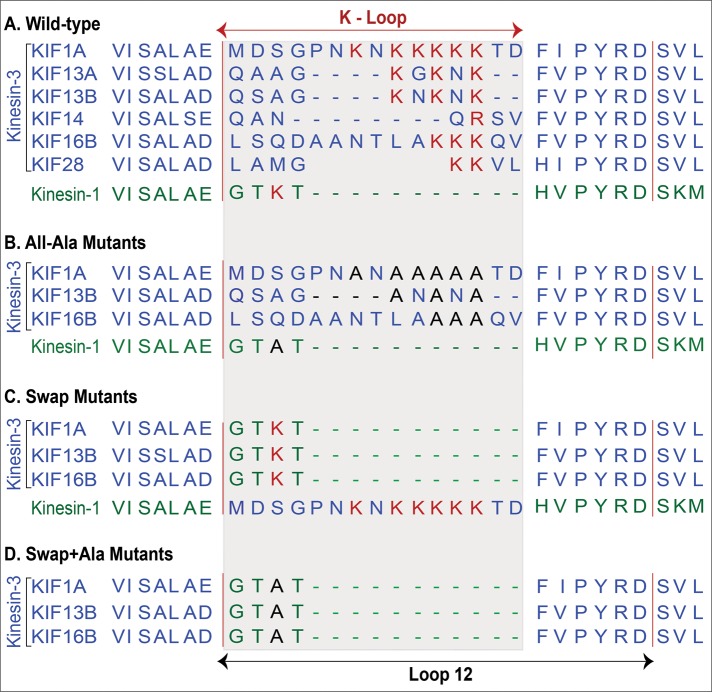
The kinesin-3 K-loop and mutants generated. (A) Amino acid sequence alignment of the loop 12 region of mammalian kinesin-3 family motors (blue text) and kinesin-1 (green text). (B) All-Alanine mutants, in which all lysine residues in the kinesin-3 and kinesin-1 K-loop regions were mutated to alanine. (C) Swap mutants, in which the K-loop of each kinesin-3 motor was replaced with that of kinesin-1 and the K-loop of kinesin-1 was replaced with that of KIF1A. (D) Swap+Alanine mutants, in which the single lysine residue remaining in the K-loop of the Swap mutants was mutated to alanine.
To dissect the functional role of the K-loop at a molecular level, we generated versions of KIF1A(1-393), KIF13B(1-412ΔP), and KIF16B(1-400) motors in which the K-loop sequences were either mutated or replaced by that of another processive motor. We first mutated all lysine residues in the K-loop of each motor to alanine residues (All-Ala mutants, Figure 2B). We next swapped the K-loop sequence of each kinesin-3 motor with that of kinesin-1 (Swap mutants, Figure 2C). In addition, we created a kinesin-1 Swap mutant by replacing the corresponding loop 12 of kinesin-1 with that of KIF1A (Swap mutant, Figure 2C). Finally, we generated a third set of mutants in which the single lysine residue in the kinesin-1 loop 12 was mutated to alanine in the kinesin-3 Swap mutant background (Swap+Ala mutants, Figure 2D).
We first tested whether the K-loop facilitates the diffusive motion of dimeric kinesin-3 motors along the microtubule surface. Previous work showed that both monomeric and dimeric KIF1A motors can weakly bind to and diffuse along the microtubule surface in their ADP-bound states (Okada and Hirokawa, 2000; Hammond et al., 2009). Although the K-loop sequence is required for diffusive motion of monomeric KIF1A motors (Okada and Hirokawa, 2000), whether the K-loop contributes to the diffusive motility of dimeric KIF1A motors has not been tested. Furthermore, it is not known whether dimeric KIF13 and KIF16 motors can undergo one-dimensional diffusion on the microtubule lattice and whether this is mediated by their K-loop sequences. We thus measured the motility properties of the wild-type, Swap, and Swap+Ala mutant motors in the presence of ADP at the single-molecule level using total internal reflection fluorescence (TIRF) microscopy. Wild-type motors were observed to undergo back-and-forth motion along the microtubule surface (Supplemental Figure S1). Mean square displacement analysis of these events demonstrated a linear increase in mobility with time (Soppina et al., 2014), consistent with diffusive motion. However, no motility was observed for either the Swap mutants or the Swap+Ala mutants in the presence of ADP (unpublished data and Table 1). Thus the K-loop sequence promotes diffusive motion of dimeric kinesin-3 motors in their ADP-bound state, presumably due to electrostatic interactions of the K-loop with the E-hooks of the tubulin subunits.
TABLE 1:
Diffusion properties of wild-type and K-loop mutant kinesin-3 motors in ADP.
| Motor protein | Construct | Diffusion in ADP? | Diffusion constant (μm2/s) | Coefficient of determination (R2) | Number of events |
| KIF1A(1-393) | Wild type | Yes | 3.53 | 0.967 | 1973 |
| Swap | No | – | – | – | |
| Swap+Ala | No | – | – | – | |
| KIF13B(1-412ΔP) | Wild type | Yes | 2.56 | 0.965 | 1750 |
| Swap | No | – | – | – | |
| Swap+Ala | No | – | – | – | |
| KIF16B(1-400) | Wild type | Yes | 1.0 | 0.990 | 2061 |
| Swap | No | – | – | – | |
| Swap+Ala | No | – | – | – |
Mutation of the K-loop does not abolish the superprocessive motion of dimeric kinesin-3 motors
We next examined the role of the K-loop in generating the superprocessive motion of dimeric kinesin-3 motors. Our recent work (Soppina et al., 2014; Supplemental Table S1) shows that expressed KIF1A(1-393) motors are weak dimers, resulting in a moderate processivity for the population (average run length, ∼2 μm). In contrast, KIF13B(1-412ΔP) and KIF16B(1-400) are strong dimers that undergo superprocessive motion (average run lengths, ∼10 μm). Stable dimers of KIF1A(1-393) are also capable of superprocessive motion (Soppina et al., 2014), indicating that superprocessivity may be a general property of the kinesin-3 family.
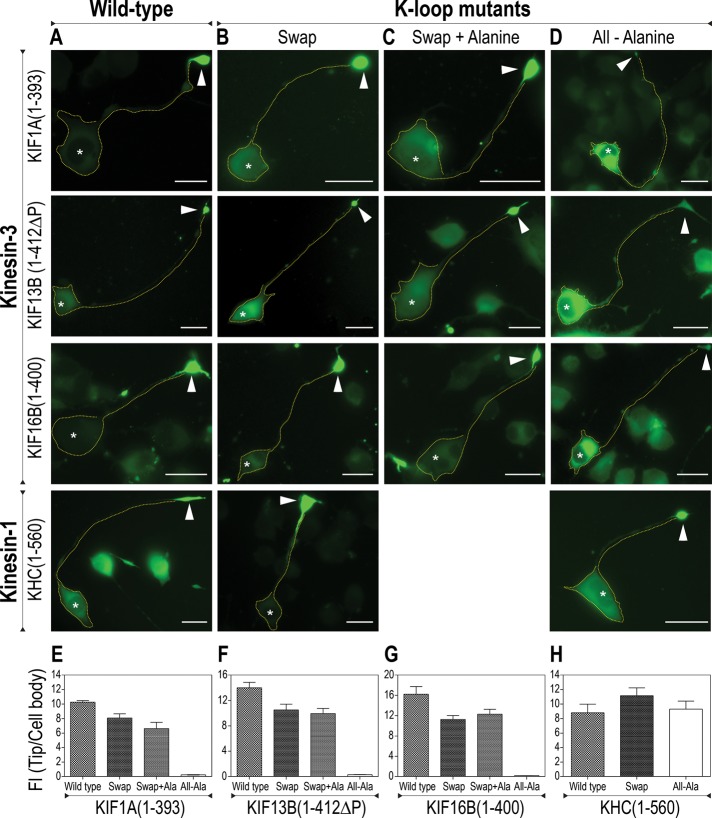
The ability of the wild-type and K-loop mutant motors to move processively along microtubules was first evaluated in neuronal Cath.-a-differentiated (CAD) cells (Figure 3), in which the separation of microtubule minus ends in the cell body and plus ends in the neurite tips enables the characteristic accumulation of processive motors in the distal neurite (Nakata and Hirokawa, 2003; Lee et al., 2004; Jacobson et al., 2006; Hammond et al., 2010; Huang and Banker, 2012; Huo et al., 2012). When expressed in CAD cells, the kinesin-3 All-Ala mutants remained in the cell body, suggesting that replacement of all lysine residues in the K-loop with alanines had severe detrimental effects on processive motility (Figure 3, D–G). In contrast, the kinesin-3 and kinesin-1 Swap mutants accumulated at the distal tips of neurite processes (Figure 3, B and E–H), similar to the wild-type motors (Figure 3, A and E–H), suggesting that these motors retained the ability to undergo processive motion. Identical results were obtained for the Swap+Ala mutants (Figure 3, C and E–G). These results imply that swapping of the K-loop sequences did not impose any adverse effect on either ATPase activity or processive motility of the dimeric motors.
FIGURE 3:
Replacement of the K-loop sequence does not affect motor processivity in CAD cells. (A–D) Differentiated CAD cells were transfected with plasmids for expressing (A) wild-type, (B) Swap, (C) Swap+Ala, or (D) All-Ala mutants of the indicated truncated kinesin-3 or kinesin-1 motors (all with C-terminal 3xmCit tag). The outline of each cell is indicated by a dotted yellow line, and the neurite tips are denoted with white arrowheads. Asterisks indicate nuclei. Scale bars, 20 μm. (E–H) Quantification of the tip accumulation as the ratio of the average fluorescence intensity (FI) in the neurite tip to that in the cell body (mean ± SD). N = 20 cells each.
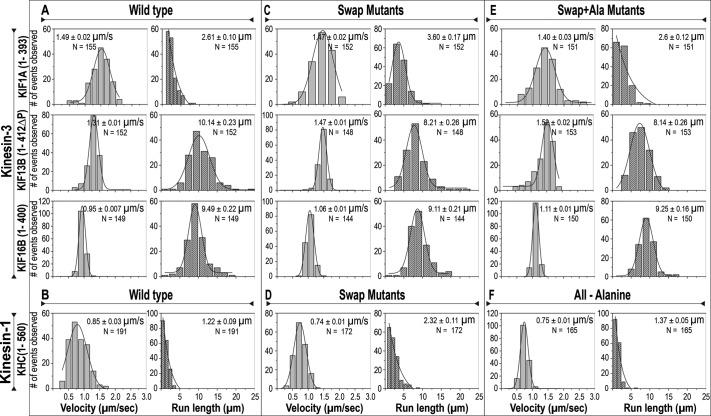
The motility properties of the processive motors were then determined at the single-molecule level (Figure 4 and Supplemental Figure S2). All Swap and Swap+Ala mutants showed unidirectional and smooth processive motion along microtubules. At similar protein levels, fewer events were observed for the Swap and Swap+Ala mutants than their wild-type counterparts, and thus, to directly compare the motility properties of the wild-type and mutant motors, we quantified ∼150 motility events for each motor. These results demonstrate that alteration of the K-loop sequence has no effect on motor velocity and to a large extent also has no effect on run length. For KIF13B, replacement of its K-loop sequence resulted in a modest decrease in processivity (Figure 4, A, C, and E, middle, and Supplemental Table S1), whereas for KIF16B, replacement of the K-loop sequence had no effect on this motor's high processivity (Figure 4, A, C, and E, bottom, and Supplemental Table S1). These results suggest that the K-loop sequence is not required for the superprocessivity of dimeric kinesin-3 motors. For moderately processive motors (the weakly dimeric kinesin-3 KIF1A(1-393) and the dimeric kinesin-1 KHC(1-560)), the K-loop is not required for processive motility but can modulate the motility properties. For KIF1A, decreasing the positive charge of loop 12 by swapping its K-loop with that of kinesin-1 surprisingly resulted in an increase in run length (Figure 4, A and C, top, and Supplemental Table S1). The reason for this is not clear, as Swap+Ala motors showed no change in run length compared with the wild-type motors (Figure 4E, top). For kinesin-1, addition of the positive charges from KIF1A's K-loop (kinesin-1 Swap mutant) resulted in an increase in run length (Figure 4, B and D, and Supplemental Table S1), perhaps by increasing the dwell time of this motor on the microtubule.
FIGURE 4:
Replacement of the K-loop sequence does not abolish the high velocity or superprocessivity of kinesin-3 family motors. (A–F) The indicated wild-type or K-loop mutant motors were C-terminally tagged with 3xmCit and expressed in COS-7 cells, and their single-molecule motility properties were measured in cell lysates. The velocities and run lengths of ∼150 events were counted for each motor and plotted as a histogram for the population. Data are the averages from at least two independent experiments. The averages (mean ± SEM) and N values are indicated in the top right corner of each graph. The data for the wild-type motors (A, B) were reported in Soppina et al. (2014) and are provided here for direct comparison.
Together these results demonstrate that the presence of the positively charged K-loop is not required for the superprocessivity of dimeric kinesin-3 motors. These results are in contrast to previous work studying a forced dimer of CeUNC-104, where deletion of the K-loop resulted in a decrease in processivity (Tomishige et al., 2002). The reason for this discrepancy is unclear, but it may be related to the presence of the kinesin-1 neck and stalk domains in the CeUNC-104 forced dimer.
The K-loop facilitates the initial interaction of kinesin-3 motors with microtubules
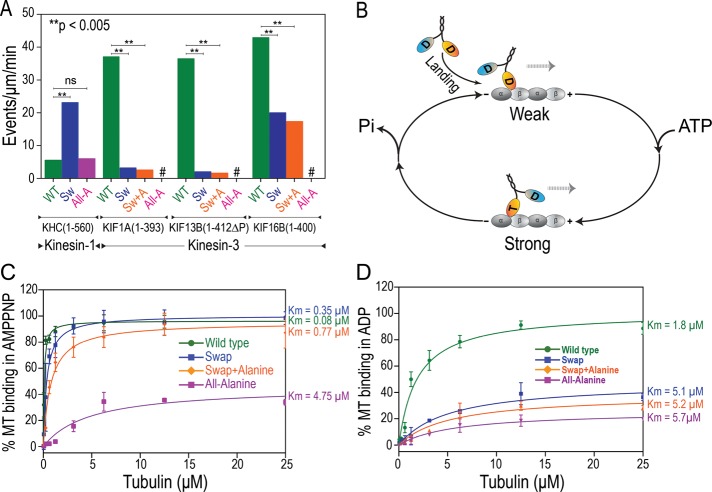
The strong conservation of the K-loop sequence within the kinesin-3 family suggests an important role in motor function. Our observation that the number of motility events observed for the Swap and Swap+Ala mutants in ATP was drastically decreased compared with the wild-type motors prompted us to hypothesize that the K-loop plays an important role in facilitating the initial interaction of kinesin-3 motors with microtubules. We thus carried out single-molecule motility assays, keeping the input protein levels the same for each motor and its mutants, and then quantified the motor landing rate as the frequency with which motors land and take processive steps along a given length of microtubule per unit time. For all kinesin-3 motors, replacement of the lysines in their K-loops with alanines (All-Ala mutants) abolished their ability to interact productively with the microtubule filament (Figure 5A; #, no events observed), whereas mutation of the single lysine in the kinesin-1 loop 12 (KHC(1-560) All-Ala mutant) had no effect on the ability of this motor to engage with the microtubule (Figure 5A, pink bar).
FIGURE 5:
The K-loop promotes the initial motor–microtubule interaction in the motor's ADP-bound state. (A) Quantification of the microtubule landing rates for the indicated wild-type and K-loop mutants (Sw, Swap; Sw+A, Swap+Ala; All-A, All-Ala). #, no motility events were observed. ns, not significant. (B) Simplified schematic of the kinesin ATPase cycle. In solution, both motor heads are bound to ADP (D). The initial interaction with the microtubule (landing) results in a weak microtubule-bound state. Release of ADP and subsequent binding of ATP (T) in the leading head converts the motor into a strong microtubule-bound state. (C, D) Microtubule-binding affinity of KIF1A wild-type and K-loop mutants. Cell lysates containing wild-type or K-loop mutant KIF1A(1-393) motors (tagged with mCit) were cosedimented with increasing amounts of microtubules in the presence of (C) AMPPNP to measure the strongly bound state or (D) ADP to measure the weakly bound state. The amount of KIF1A(1-393) motors in the microtubule pellet was determined by Western blotting with antibodies to the mCit tag. The affinity of each motor for microtubules (Km) is indicated on the right side of the graphs. Data are the average ± SD from two independent experiments.
For dimeric KIF1A and KIF13B motors, replacement of the K-loop with that of kinesin-1 (Swap mutants) or further mutation of the single remaining lysine in the Swap mutants (Swap+Ala mutants) resulted in a dramatic decrease (>10-fold) in the landing rate as compared with the wild-type motors (Figure 5A). For KIF16B, the Swap and Swap+Ala mutants also showed a significant decrease (p < 0.005) in the landing rate, albeit only a twofold difference as compared with the wild-type motors (Figure 5A). Differences in surface charge outside of the K-loop could explain the milder effect of K-loop deletion on the landing rates of KIF16B as compared with the other kinesin-3 motors (Grant et al., 2011). These results suggest that the K-loop in kinesin-3 motors plays an important role in promoting the initial interaction of dimeric motors with the microtubule. Consistent with this, replacement of the kinesin-1 K-loop with that of KIF1A (KHC(1-560) Swap mutant) resulted in a significant (p < 0.005) increase (approximately fivefold) in this motor's landing rate (Figure 5A). The mechanism by which the K-loop promotes the motor's interaction with the microtubule is likely to involve electrostatics, as the landing rate of the wild-type motors was decreased at higher ionic strength (Soppina et al., 2014). Taken together, these results indicate that positively charged residues on the kinesin-3 motor surface facilitate the initial interaction of the motor with the microtubule.
The K-loop contributes to microtubule affinity in the ADP-bound state
To further investigate the mechanistic role of the K-loop, we determined the effect of the K-loop mutations on the affinity of the dimeric KIF1A motor for the microtubule. During the ATPase cycle of a dimeric kinesin-1 motor, the two motor domains undergo a series of transition states in which the microtubule affinity is regulated by the nucleotide state (Hackney, 1988; Hancock and Howard, 1999). We considered a simple two-state model (Figure 5B) in which ADP-bound kinesin in solution interacts weakly with a microtubule (weak state) and then converts to a strongly microtubule-bound state upon binding of ATP in the nucleotide pocket (strong state). ATP hydrolysis then returns the motor to the weak state (Hackney, 1988; Crevel et al., 1996; Okada and Hirokawa, 2000; Figure 5B). We thus characterized the microtubule-binding affinity of wild-type KIF1A and its K-loop mutants in the ATP (strong) and ADP (weak) states. KIF1A motors were incubated with increasing concentrations of microtubules in the presence of 5′-adenylyl-β,γ-imidodiphosphate (AMPPNP; a nonhydrolyzable ATP analogue) or ADP, and the amount of bound motor was measured by microtubule cosedimentation.
In the AMPPNP (strongly bound) state, wild-type dimeric KIF1A motors showed a very high affinity for microtubules (Km = 0.08 μM), and this was only slightly decreased upon replacement of the KIF1A K-loop with that of kinesin-1 (Swap mutant, Km = 0.35 μM, p = 0.262) or further mutation of the single remaining lysine (Swap+Ala mutant, Km = 0.77 μM, p = 0.076; Figure 5C). However, mutation of all KIF1A K-loop lysines to alanine (All-Ala mutant, Km = 4.75 μM) nearly abolished the ability of the motor to bind strongly to microtubules (Figure 5C). These results indicate that the K-loop does not influence the interaction of the motor with the microtubule in the ATP-bound state.
In the ADP (weakly bound) state, wild-type dimeric KIF1A motors interacted with microtubules with a high affinity (Km = 1.8 μM), whereas the Swap and Swap+Ala mutants showed dramatic and significant (p < 0.05) decreases in microtubule affinity (Km = 5.1 and 5.2 μM, respectively; Figure 5D), such that their affinity for the microtubule in ADP was reduced to nearly that of the All-Ala mutant (Km = 5.7 μM). These results demonstrate that the unique K-loop sequence provides a distinct functional output for kinesin-3 motors during the initial interaction of the ADP-bound motor with the microtubule surface. For kinesin-1 motors, the ADP-bound state is a weakly microtubule-bound state, but for kinesin-3 motors, the presence of the K-loop converts the ADP-bound state to a strongly microtubule-bound state.
DISCUSSION
Although the K-loop sequence is a conserved and defining feature of kinesin-3 motors across species, the functional relevance of the K-loop to kinesin-3 motility was not known. We show that the K-loop sequence contributes to two aspects of kinesin-3 biology. First, the K-loop enables the diffusive motion of kinesin-3 motors along the microtubule surface. Whether diffusive motion has a physiological role in the transport of cellular cargoes requires further study. Second, the K-loop enables a high on-rate to the microtubule in the motor's ADP state. We propose that the K-loop thus enables kinesin-3 motors to be highly efficient cargo transporters in cells, particularly during long-distance transport in neurons. A high microtubule on-rate would ensure that vesicles containing only a few motors would readily engage with a microtubule and begin the transport process. The superprocessivity of kinesin-3 motors would ensure that once engaged, the motors and associated cargoes would efficiently reach their destination.
That the K-loop facilitates the diffusive motion of both monomeric (Okada and Hirokawa, 1999, 2000; Kikkawa et al., 2000) and dimeric (this study; Soppina et al., 2014) motors along the microtubule surface is consistent with studies demonstrating that electrostatic interactions facilitate the binding of a variety of molecules to microtubules, including members of the kinesin-5, kinesin-7, and kinesin-13 families (Kapitein et al., 2005, 2008; Helenius et al., 2006; Kim et al., 2008; Cooper et al., 2010; Weinger et al., 2011). Electrostatic-based diffusion has also been proposed to contribute to the functional output of other MAPs, such as the dynein/dynactin complex, the Ndc80 complex, Dam1, and tau (Wang and Sheetz, 1999; Westermann et al., 2005; Reck-Peterson et al., 2006; Ciferri et al., 2008; Rosenberg et al., 2008; Cooper and Wordeman, 2009; Li and Zheng, 2011; Ramey et al., 2011; Eckert et al., 2012; Hinrichs et al., 2012). Electrostatics also contributes to the interaction of myosin motors with actin filaments (Yengo and Sweeney, 2004; Geeves et al., 2005; Hodges et al., 2007) and, of interest, enables myosin Va to diffuse along the microtubule surface while searching for a kinesin–cargo complex (Ali et al., 2007, 2008; Zimmermann et al., 2011). It thus appears that charged-based diffusion along the microtubule surface is a common property for many cytoskeleton-associated proteins.
Yet, beyond a simple electrostatic-based interaction, the K-loop sequence has evolved to allow kinesin-3 motors to functionally engage with the microtubule filament at a higher rate than that of other kinesin families. Such high microtubule association rates were predicated based on the high net positive charge of kinesin-3 motors at the microtubule-binding interface (Grant et al., 2011). The functional output of the K-loop can be measured at the single-molecule level under ATP conditions (Figure 5A) and in ensemble assays under ADP conditions (Figure 5D), where motors are in dynamic interactions with the track. In contrast, no significant effect was seen in ensemble assays under AMPPNP conditions (Figure 5C), as the replacement of the KIF1A K-loop with that of kinesin-1 (Swap mutants) or further mutation of single lysine residue to alanine in the K-loop of kinesin-3 Swap mutants (Swap+Ala mutants) caused a small but not significant decrease in microtubule affinity (from Km = 0.08 to 0.35 μM for Swap and 0.77 μM for Swap+Ala mutants, with p > 0.1) under conditions in which motors cannot dissociate from the track and thus accumulate over time. That the K-loop primarily affects the motor–microtubule interaction in the motor's ADP-bound state is consistent with structural studies of monomeric KIF1A, which showed that in the motor's ADP-bound state, loop 12 is tilted toward the microtubule and forms weak interactions with the C-terminal tails of the tubulin subunits, whereas in the ATP-bound state, loop 11 is primarily responsible for the motor–microtubule interaction (Nitta et al., 2004). We thus propose that the positively charged K-loop plays a critical role in kinesin-3 cargo transport by enhancing the initial interaction of cargo-bound dimeric motors with the microtubule track (Figure 6A). The K-loop is not, however, required for superprocessive motility once kinesin-3 motors are engaged in a transport event (Figure 6B).
FIGURE 6:
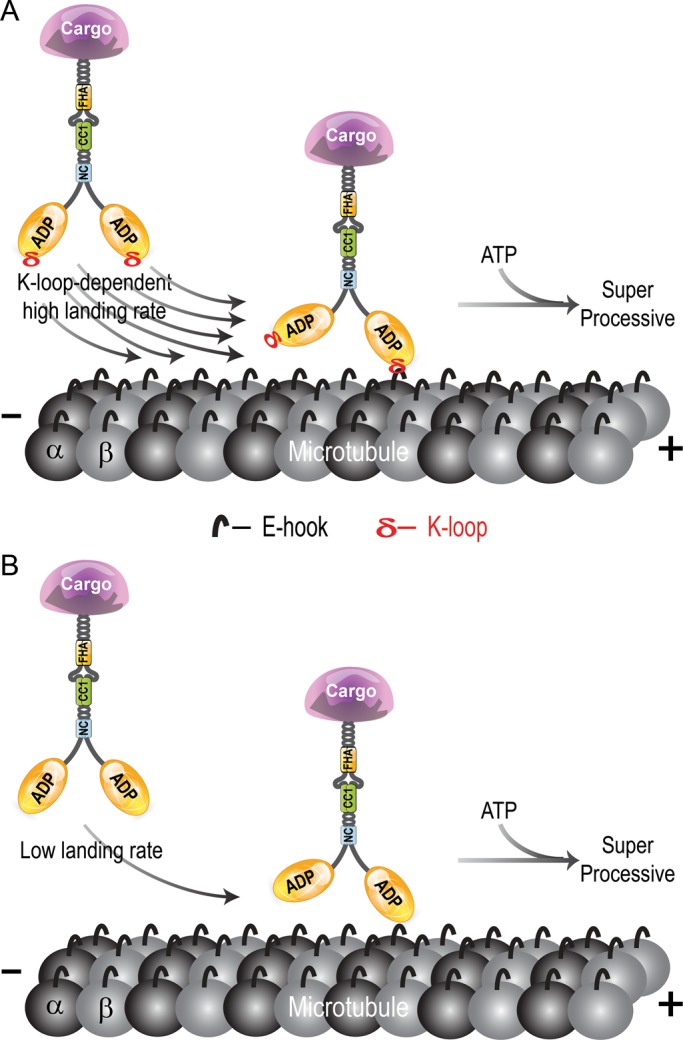
The family-specific K-loop increases the microtubule on-rate of cargo-bound kinesin-3 motors. (A) Wild-type kinesin-3 motors containing a positively charged K-loop display a high affinity for the microtubule (high landing rate), followed by ATP-dependent superprocessive motility. (B) Mutation of the K-loop results in a decreased affinity for the microtubule (lower landing rate) but does not affect the processivity of the bound motor. CC, coiled coil; FHA, forkhead homology-associated; NC, neck coil.
That mutation of the K-loop does not abolish the superprocessivity of dimeric kinesin-3 motors was surprising. The mechanical features that enable the superprocessive motility of kinesin-3 motors thus remain to be determined. For other kinesin families, two features have been suggested to contribute to processivity. First, positively charged residues in the neck coil (NC) or C-terminal tail regions were found to increase motor residence time on a microtubule (Thorn et al., 2000; Kim et al., 2008; Grissom et al., 2009; Rosenfeld et al., 2009; Varga et al., 2009; Mayr et al., 2011; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011). However, this mechanism seems unlikely to contribute to the high processivity of kinesin-3 motors, as the net charge of the NC region of dimeric kinesin-3 motors is predicted to be neutral or net negative. Second, the length of the neck linker, a highly flexible segment that connects the kinesin motor domain to the NC, regulates the processivity of kinesin motors by communicating mechanical strain and/or chemical state information between the two catalytic heads (Vale and Milligan, 2000; Yildiz et al., 2008; Miyazono et al., 2010; Shastry and Hancock, 2010, 2011; Clancy et al., 2011; Duselder et al., 2012). However, the kinesin-3 neck linker length is identical to that of kinesin-2, a mildly processive motor (run lengths, ∼1 μm), indicating that neck linker length does not determine the unusual processivity of the kinesin-3 family. Clearly, further work is needed to determine the molecular and mechanical features that contribute to the superprocessive motion of kinesin-3 family motors.
MATERIALS AND METHODS
Plasmids
Mammalian KIF1A and KIF13A expression plasmids (Jacobson et al., 2006; Huang and Banker, 2012) were a kind gift of Gary Banker (Oregon Health & Science University, Portland, OR), and KIF13B (Yoshimura et al., 2010) and KIF16B (Hoepfner et al., 2005) expression plasmids were generous gifts from Hiroaki Miki (Osaka University, Osaka, Japan) and Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany), respectively. Fluorescent protein–tagged versions of full-length or truncated motors were generated by subcloning into mCitrine-N1/C1 and/or 3xmCitrine-N1/C1 vectors (based on Clontech's EYFP-N1/C1 vectors) either using appropriate restriction sites or by PCR amplification. The K-loop mutations were generated by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) or by overlapping PCR. All plasmids were verified by DNA sequencing. The constitutively active dimeric kinesin-1 construct KHC(1–560)-3xmCitrine has been described (Cai et al., 2009).
CAD cell assay
The mouse catecholaminergic cell line CAD (Qi et al., 1997) was grown in a 1:1 mixture of F12:DMEM (BioWhittaker, Radnor, PA) plus 10% (vol/vol) fetal bovine serum (FBS; Hyclone, Logan, UT) at 37°C with 5% CO2. Cells were induced to differentiate by transfer to serum-free medium and then transfected with 1.0 μg of plasmid DNA using TransIT-LT1 (Mirus Bio LLC. Madison, WI). After 2 d, the cells were fixed in 4% (vol/vol) paraformaldehyde (PFA) in phosphate-buffered saline (PBS), quenched with 50 mM ammonium chloride in PBS, permeabilized with 0.02% (vol/vol) Triton X-100 in PBS, incubated in primary and secondary antibodies, and then mounted in Prolong Gold (Life Technologies, Grand Island, NY). Images were obtained on an inverted Nikon TE2000E epifluorescence microscope equipped with 60×/1.40 numerical aperture (NA) objective and a Photometrics CoolSnap HQ camera (Photometrics, Tucson, AZ). To quantify motor distribution, the average fluorescence intensity of the cell body and its neurite tip were measured using ImageJ software (National Institutes of Health, Bethesda, MD). The data are reported as the ratio of average fluorescence intensity in the neurite tip to that in the cell body. The mean and SD for each motor were plotted using Prism 6 software (GraphPad Software, La Jolla, CA).
COS-7 cell lysates
COS-7 (monkey kidney fibroblast; American Type Culture Collection, Manassas, VA) cells were grown in DMEM plus 10% (vol/vol) FBS and 2 mM l-glutamine at 37°C with 5% CO2. Cells were transfected with 1.0 μg of plasmid DNA using Expressfect (Denville Scientific, Metuchen, NJ). The next day, cells were trypsinized and harvested by low-speed centrifugation at 4°C. The pellet was washed once with DMEM and lysed in ice-cold lysis buffer (LB; 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM ethylene glycol tetraacetic acid [EGTA], 1% [vol/vol] Triton X-100, pH 7.4) freshly supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitors (10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastatinal, 1 mg/ml pepstatin). The lysate was clarified by centrifugation at 16,000 × g at 4°C and either it was used fresh for assays or aliquots were frozen in liquid nitrogen and stored at −80°C until further use.
We believe that analysis of motors in cell lysates can provide important information about structure–function aspects of kinesin motors. The motors are synthesized under physiological conditions, and the cell lysates are highly diluted in motility buffer, making secondary effects unlikely. For kinesin-1 motors, we found no differences in motility under a wide variety of conditions for motors expressed and imaged in mammalian cell lysates versus those expressed and purified from bacteria (unpublished data). For kinesin-3 motors, structure–function analysis of the neck coil region revealed changes in processivity in cell lysates (Soppina et al., 2014). In addition, the superprocessivity of truncated KIF1A motors that we observe in cell lysates was noted for a forced dimer of C. elegans UNC-104 in a purified state (Tomishige et al., 2002).
Single-molecule assays
All single-molecule assays were performed using a Nikon Ti-E objective-type TIRF microscope with Perfect Focus System, a 100×/1.49 NA CFI APO TIRF objective, an Agilent 3-Line (488, 561, and 640 nm; Agilent Technologies, Santa Clara, CA) Monolithic Standard Power Laser Launch with acousto-optical tunable filter, and an electron-multiplying charge-coupled device camera (Andor iXon+ DU897; Andor Technology USA, South Windsor, CT), controlled by Nikon Elements image acquisition software (Nikon Instruments, Melville, NY). All assays were performed at room temperature in a narrow flow cell (∼10 μl volume) prepared by attaching a clean #1.5 coverslip to a microscope slide with double-sided adhesive tape. Microtubules were polymerized from purified tubulin (TL238; Cytoskeleton, Denver, CO) in BRB80 buffer (80 mM 1,4-piperazinediethanesulfonic acid [PIPES]/KOH, pH 6.8, 1 mM MgCl2, 1 mM EGTA) supplemented with 1 mM GTP at 37°C for 20 min. Polymerized microtubules were stored at room temperature after addition of five volumes of prewarmed BRB80 containing 20 μM Taxol and additional 5-min incubation at 37°C. Polymerized microtubules (20–40 μm in length) were diluted in P12 buffer (12 mM PIPES/KOH, pH 6.8, 1 mM MgCl2, and 1 mM EGTA freshly supplemented with 10 μM Taxol) and then infused into a flow cell and incubated for 5 min at room temperature to adsorb onto the coverslip. Subsequently, 50 μl of blocking buffer (10 mg/ml bovine serum albumin in P12 buffer with 10 μM Taxol) was introduced and incubated for 20 min to prevent nonspecific binding of kinesin motors onto the coverslip surface. Finally, kinesin motors in a 50 μl of Motility Mix (0.1–2.0 μl of COS-7 cell lysate with 30 μl of blocking buffer, 15 μl of P12 buffer, 2 mM nucleotide, 0.5 μl of 100 mM dithiothreitol, 0.5 μl of 100 mM MgCl2, and 0.5 μl each of 20 mg/ml glucose oxidase, 8 mg/ml catalase, and 1 M glucose) was added to the flow chamber, and the ends were sealed with molten paraffin wax.
For motility assays, 3xmCit-tagged motors in Motility Mix with 2 mM ATP were imaged at 20 frames/s without binning and at low laser power to avoid photobleaching during processive motor runs. The position of fluorescent motor spots was manually tracked frame by frame using a custom-written plug-in in ImageJ as described previously (Cai et al., 2007). The velocities and run lengths of individual motors were binned, and histograms were generated for the population by plotting the number of events in each bin. The average velocities and run lengths were then obtained by fitting either a single Gaussian peak (superprocessive motors) or an exponential (moderately processive motors) to the population histogram. These run lengths are likely to be an underestimate, as some motors reached the end of the microtubule track before ending their run. The measurements for each construct come from at least two independent protein preparations and include motile events lasting at least 10 frames (500 ms). All data are presented as mean ± SEM. All p values were calculated by using a two-tailed unpaired Student's t test.
For diffusion assays, the single-molecule motility experiments were performed in the presence of 2 mM ADP and imaged at 60 frames/s. Events that lasted >10 frames were analyzed by mean square displacement (MSD) plots. The curves were fitted with both linear (diffusive) and parabolic (processive) fits, and the coefficient of determination (R2) was used to determine the better fit. In ADP, the movement increased linearly with time, indicating a diffusional process. The MSD plots are provided in Figure S9 in Soppina et al. (2014).
For landing assays, the amount of 3xmCit-tagged wild-type or K-loop mutant motors in the COS-7 lysates was first normalized by a dot-blot in which increasing volumes of COS-7 lysates were spotted onto nitrocellulose membrane. The membrane was air-dried and immunoblotted for the mCit tag (anti–green fluorescent protein [GFP]; A6455; Invitrogen Life Technologies, Grand Island, NY). The spots were quantified to normalize the motor concentration. Equal amounts of motors were added to flow cells and imaged at 20 frames/s. The number of motors landing on a microtubule was counted and then divided by the total length of the microtubules and the recording time in order to obtain a landing rate with the units of events/micrometer/minute.
Microtubule-binding assay
Lysates of COS-7 cells expressing mCit-tagged motors were incubated with increasing concentrations of Taxol-stabilized microtubules in the presence of 2 mM AMPPNP or 2 mM ADP for 30 min at room temperature with constant agitation. The motor–microtubule complexes were sedimented through a glycerol cushion (BRB80 containing 60% glycerol and 20 μM Taxol) at 90,000 rpm for 10 min at 25°C in a TLA100 rotor. The pellets were dissolved in SDS–PAGE sample buffer, and the amount of motor in the pellets was determined by Western blotting with an anti-GFP antibody (A6455; Invitrogen). Digital images of the blots were quantified using ImageJ software.
Supplementary Material
Acknowledgments
We thank members of the Verhey lab for helpful discussions. This work was supported by the National Institutes of Health under award number R01GM070862.
Abbreviations used:
- AMPPNP
5′-adenylyl-β,γ-imidodiphosphate
- CAD
Cath.-a-differentiated
- KIF
kinesin family
- mCit
mCitrine
- NC
neck coil
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0696) on May 21, 2014.
Both authors designed the study. V.S. performed experiments and analyzed data. Both authors wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- Ali MY, Krementsova EB, Kennedy GG, Mahaffy R, Pollard TD, Trybus KM, Warshaw DM. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM. Myosin V and kinesin act as tethers to enhance each others’ processivity. Proc Natl Acad Sci USA. 2008;105:4691–4696. doi: 10.1073/pnas.0711531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Verhey KJ, Meyhofer E. Tracking single kinesin molecules in the cytoplasm of mammalian cells. Biophys J. 2007;92:4137–4144. doi: 10.1529/biophysj.106.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wordeman L. The diffusive interaction of microtubule binding proteins. Curr Opin Cell Biol. 2009;21:68–73. doi: 10.1016/j.ceb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel IM, Lockhart A, Cross RA. Weak and strong states of kinesin and NCD. J Mol Biol. 1996;257:66–76. doi: 10.1006/jmbi.1996.0147. [DOI] [PubMed] [Google Scholar]
- Duselder A, Thiede C, Schmidt CF, Lakamper S. Neck-linker length dependence of processive kinesin-5 motility. J Mol Biol. 2012;423:159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- Eckert T, Le DT, Link S, Friedmann L, Woehlke G. Spastin's microtubule-binding properties and comparison to katanin. PLoS One. 2012;7:e50161. doi: 10.1371/journal.pone.0050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker MA, Hoogenraad CC. Microtubule-based transport—basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Fedorov R, Manstein DJ. Molecular mechanism of actomyosin-based motility. Cell Mol Life Sci. 2005;62:1462–1477. doi: 10.1007/s00018-005-5015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Gheorghe DM, Zheng W, Alonso M, Huber G, Dlugosz M, McCammon JA, Cross RA. Electrostatically biased binding of kinesin to microtubules. PLoS Biol. 2011;9:e1001207. doi: 10.1371/journal.pbio.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO, Howard J. Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hinrichs MH, Jalal A, Brenner B, Mandelkow E, Kumar S, Scholz T. Tau protein diffuses along the microtubule lattice. J Biol Chem. 2012;287:38559–38568. doi: 10.1074/jbc.M112.369785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hodges AR, Krementsova EB, Trybus KM. Engineering the processive run length of myosin V. J Biol Chem. 2007;282:27192–27197. doi: 10.1074/jbc.M703968200. [DOI] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Huang CF, Banker G. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 2012;13:549–564. doi: 10.1111/j.1600-0854.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Yue Y, Ren J, Yu J, Liu J, Yu Y, Ye F, Xu T, Zhang M, Feng W. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 2012;20:1550–1561. doi: 10.1016/j.str.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49:797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Okada Y, Hirokawa N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, et al. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 2004;23:1506–1515. doi: 10.1038/sj.emboj.7600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zheng W. Probing the structural and energetic basis of kinesin-microtubule binding using computational alanine-scanning mutagenesis. Biochemistry. 2011;50:8645–8655. doi: 10.1021/bi2008257. [DOI] [PubMed] [Google Scholar]
- Marx A, Hoenger A, Mandelkow E. Structures of kinesin motor proteins. Cell Motil Cytoskeleton. 2009;66:958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS One. 2011;6:e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono Y, Hayashi M, Karagiannis P, Harada Y, Tadakuma H. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta R, Kikkawa M, Okada Y, Hirokawa N. KIF1A alternately uses two loops to bind microtubules. Science. 2004;305:678–683. doi: 10.1126/science.1096621. [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Otsuka AJ, Jeyaprakash A, Garcia-Anoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- Qi Y, Wang JK, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey VH, Wang HW, Nakajima Y, Wong A, Liu J, Drubin D, Barnes G, Nogales E. The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol Biol Cell. 2011;22:457–466. doi: 10.1091/mbc.E10-10-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg KJ, Ross JL, Feinstein HE, Feinstein SC, Israelachvili J. Complementary dimerization of microtubule-associated tau protein: Implications for microtubule bundling and tau-mediated pathogenesis. Proc Natl Acad Sci USA. 2008;105:7445–7450. doi: 10.1073/pnas.0802036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, van Duffelen M, Behnke-Parks WM, Beadle C, Corrreia J, Xing J. The ATPase cycle of the mitotic motor CENP-E. J Biol Chem. 2009;284:32858–32868. doi: 10.1074/jbc.M109.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV. A seesaw model for intermolecular gating in the kinesin motor protein. Biophys Rev. 2011;3:85–100. doi: 10.1007/s12551-011-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, Verhey KJ. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Hanada T, Chishti AH. Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- Wang Z, Sheetz MP. One-dimensional diffusion on microtubules of particles coated with cytoplasmic dynein and immunoglobulins. Cell Struct Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Wojcik EJ, Buckley RS, Richard J, Liu L, Huckaba TM, Kim S. Kinesin-5: cross-bridging mechanism to targeted clinical therapy. Gene. 2013;513:133–149. doi: 10.1016/j.gene.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo CM, Sweeney HL. Functional role of loop 2 in myosin V. Biochemistry. 2004;43:2605–2612. doi: 10.1021/bi035510v. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206–2219. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann D, Abdel Motaal B, Voith von Voithenberg L, Schliwa M, Okten Z. Diffusion of myosin V on microtubules: a fine-tuned interaction for which E-hooks are dispensable. PLoS One. 2011;6:e25473. doi: 10.1371/journal.pone.0025473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.