Dynactin is an adaptor complex that binds cytoplasmic dynein to cellular cargoes. Its dynamitin (p50) component plays a key role in dynactin stability. Affinity chromatography, direct binding assays, and RNAi demonstrate that the unstructured dynamitin N-terminus binds the dynactin component Arp1 directly.
Abstract
Dynactin is a multiprotein complex that works with cytoplasmic dynein and other motors to support a wide range of cell functions. It serves as an adaptor that binds both dynein and cargoes and enhances single-motor processivity. The dynactin subunit dynamitin (also known as p50) is believed to be integral to dynactin structure because free dynamitin displaces the dynein-binding p150Glued subunit from the cargo-binding Arp1 filament. We show here that the intrinsically disordered dynamitin N-terminus binds to Arp1 directly. When expressed in cells, dynamitin amino acids (AA) 1–87 causes complete release of endogenous dynamitin, p150, and p24 from dynactin, leaving behind Arp1 filaments carrying the remaining dynactin subunits (CapZ, p62, Arp11, p27, and p25). Tandem-affinity purification–tagged dynamitin AA 1–87 binds the Arp filament specifically, and binding studies with purified native Arp1 reveal that this fragment binds Arp1 directly. Neither CapZ nor the p27/p25 dimer contributes to interactions between dynamitin and the Arp filament. This work demonstrates for the first time that Arp1 can directly bind any protein besides another Arp and provides important new insight into the underpinnings of dynactin structure.
INTRODUCTION
First identified as an activity required for dynein to move membrane vesicles on microtubules in vitro (Schroer and Sheetz, 1991), dynactin has emerged as an essential component of the cytoplasmic dynein motor complex. Dynactin in most species contains 11 different polypeptide components in stoichiometries ranging from 1 to ≥5. Dynactin's largest structural domain is a 37 × 5 nm copolymer of Arp1, actin, and Arp11 capped with other subunits (Schroer, 2004). Its conspicuous 24-nm-long projecting arm (p150Glued amino acids [AA] 1 to ∼350), which can bind microtubules at the distal tip, extends from a V-shaped “shoulder” structure that is docked at one end of the Arp filament (Imai, Narita, Maeda, and Schroer, unpublished data). The shoulder contains the remainder of p150Glued (AA ≈ 350–1280), including the dynein-binding site (Siglin et al., 2013), plus dynamitin and p24 (reviewed in Schroer, 2004; Cheong and Schroer, 2011). For dynein to be able to transport cargo, p150Glued must be anchored to the cargo-binding Arp filament domain, but the structural features that underlie this critical interface remain undefined. It has been proposed that p150Glued binds Arp1 directly (Waterman-Storer et al., 1995), and in yeast, the dynamitin homologue (Jnm1p) was reported to interact with Arp1 in a two-hybrid analysis (Clark and Rose, 2005), suggesting that multiple protein–protein interactions may be involved.
The importance of dynamitin to dynactin's structural integrity was first revealed through protein overexpression experiments, and, as a result, dynamitin overexpression has become a widely used tool for interfering with dynein-based events in vivo. The name “dynamitin” was coined based on the “explosion” of mitotic spindle poles, Golgi complex, and dynactin molecules seen in overexpressing cells (Echeverri et al., 1996; Burkhardt et al., 1997). Dynamitin also disintegrates dynactin structure when the purified proteins are mixed in vitro (Eckley et al., 1999; Wittman and Hyman, 1999; Melkonian et al., 2007). Dynamitin's remarkable ability to separate the dynactin molecule into its cargo- and dynein-binding components suggests that its associations with other dynactin subunits is of fundamental importance to dynactin structure. Purified dynamitin can form complexes with itself and the shoulder components p24 (Maier et al., 2008), but it has never been shown to bind directly to any other dynactin subunit in a biochemical assay. An intriguing and obvious possibility is that dynamitin also binds directly to Arp1 to anchor the shoulder/arm complex to the Arp filament. In support of this notion, an N-terminal fragment of dynamitin tagged with glutathione S-transferase was seen to pull down the p62 and Arp1 components of dynactin from cultured cell lysates (Jacquot et al., 2010). Whether dynamitin was binding Arp1 directly or whether another dynactin subunit or extrinsic protein was required was not determined, leaving open the structural details of dynamitin–shoulder anchoring.
We previously proposed that dynamitin's ability to disassemble dynactin depends upon binding of exogenous protomers to endogenous protomers via an interaction involving a series of self-association motifs (Melkonian et al., 2007; Maier et al., 2008). However, fragments lacking some or all of these motifs are sufficient for disruption (Maier et al., 2008; Jacquot et al., 2010), indicating that dynamitin may be able to trigger dynactin disassembly via more than one mechanism. In the present study, we used direct protein–protein binding assays to characterize interactions between dynamitin's N-portion and the self-associating C-terminal portion with other dynactin subunits. Tandem-affinity purification (TAP) revealed that dynamitin AA 1–87 binds directly to the Arp1 filament but not components of the shoulder and projecting arm. Biochemical and RNA interference (RNAi)–based assays were used to demonstrate that the dynamitin N-terminus binds directly and specifically to Arp1. A complementary dynamitin fragment (AA 100–403) that does not disrupt dynactin was found to bind p150Glued and p24 but not Arp filament components. Our findings indicate that dynamitin is a divalent scaffold that tethers dynactin's dynein and microtubule-binding subunit to its cargo-binding domain.
RESULTS
In silico and structural analysis of dynamitin AA 1–87
The dynamitin N-terminus is well conserved from human to Caenorhabditis elegans (Figure 1A). It is enriched in charged amino acids and has an overall acidic pI (4.2–4.4) but also contains a number of basic residues. It is highly susceptible to phosphorylation in vitro (Cheong, 2010) and can be phosphorylated in vivo at multiple sites (Figure 1A; PhosphoSitePlus, www.phosphosite.org), suggesting that it may be a regulatory site. The remainder of dynamitin (AA ≈ 100 to end) is predicted to fold into a series of α-helices with coiled-coil and multicoil propensity that support oligomerization (Maier et al., 2008). We reasoned that the dynamitin N-terminus might be structurally and functionally distinct from the rest of the molecule. In keeping with this idea, multiple primary sequence analysis programs (DisoPred, Ward et al., 2004; FOLDINDEX, Prilusky et al., 2005; DomPred, Marsden et al., 2002) predict that the dynamitin N-terminus is an intrinsically disordered domain that is distinct from the rest of the polypeptide. We found AA 1–100 to be readily degraded when dynamitin was subjected to limited proteolytic digestion (Tanimoto, Maeda, Imai, and Maeda, personal communication; Maier and Schroer, unpublished observations), indicative of a lack of stable secondary and tertiary structure.
FIGURE 1:
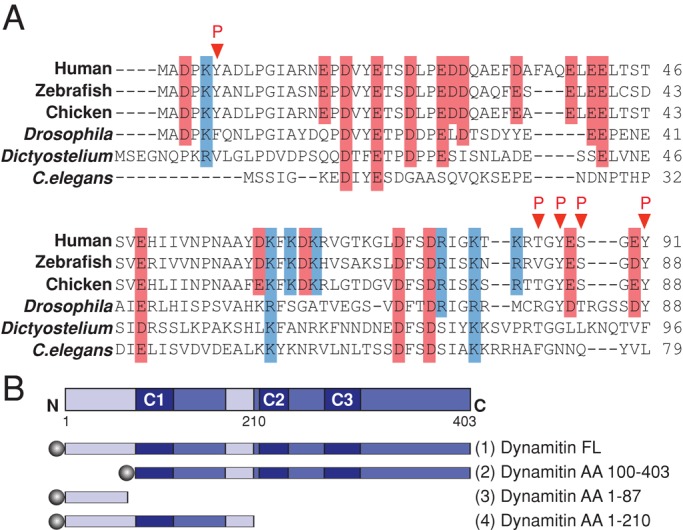
Dynamitin primary structure. (A) Sequence alignment of the dynamitin N-terminus of selected nonfungal species. Acidic residues are shaded red, basic residues are shaded blue, and identified in vivo phosphorylation sites (www.phosphosite.org) are indicated by red arrowheads. (B) Top, the dynamitin primary sequence, indicating predicted structural motifs, including unstructured motifs (light blue; AA 1–105 and AA 186–213) and multicoil motifs C1 (AA 105–135), C2 (AA 219–251), and C3 (281–308). Bottom, cartoons of the dynamitin constructs used in this study. Each construct was tagged (shaded circles) at the N-terminus with TAP, 6X-His, myc, or mCherry as specified in the Results. Note that the fragment that is the focus of this work (chicken dynamitin AA 1–87) is different from the previously published chicken AA 1–78 fragment (Maier et al., 2008; Valetti et al., 1999).
We used circular dichroism (CD) to determine the extent of secondary structure in the dynamitin N-terminus. The AA 1–87 chicken dynamitin fragment was engineered, and, along with three other N-terminal dynamitin fragments, was expressed in bacteria, purified, and subjected to CD. Spectra were compared with p150Glued AA 217–548 (also known as coiled-coil 1 [CC1]), which has extensive α-helical character (Siglin et al., 2013). Dynamitin AA 1–210, a fragment that contains dynamitin's first predicted α-helical coiled-coil (AA 105–135), yielded a complex spectrum that indicated a mixture of α-helix and disorder (15.9% structured vs. 72% structured for CC1). The AA 1–210 variant L118P, in which one of the hydrophobic heptad repeat residues is replaced with proline (Maier et al., 2008), exhibited a spectrum similar to wild type (wt) AA 1–210, with a slightly lower α-helix content (11.7%), consistent with the expected effect of the mutant. By contrast, the spectra obtained for dynamitin AA 1–78 and AA 1–87 were consistent with minimal secondary structure, with 2.8 and 1.8% α-helix content, respectively. This analysis verifies that the dynamitin N-terminus is intrinsically disordered. It further suggests that the reason dynamitin AA 1–78 does not disrupt dynactin structure in vivo (Valetti et al., 1999) or in vitro (Maier et al., 2008) is that it adopts an aberrant structure that is not exhibited by the longer 1–87 fragment. To learn more about the activities of dynamitin's N- and C-terminal domains, we used AA 1–87 and a complementary fragment, AA 100–403 (Figure 1B), in overexpression and biochemical studies.
Dynamitin (AA 1–87) is sufficient to cause dynactin disruption
Overexpression of chicken dynamitin AA 1–87 in Cos-7 cells had effects on cell architecture that were indistinguishable from those of full length dynamitin (Figure 2, A–C, and Supplemental Figure S1). We observed scattered Golgi complexes (Figure 2A) and endocytic recycling compartment/trans-Golgi network components (stained for TGN46; Supplemental Figure S1). Cells overexpressing dynamitin AA 1–87 also contained disorganized, unfocused mitotic spindles (Figure 2, B and C, and Supplemental Figure S1) and showed an increased mitotic index (control, 4.6 ± 0.3%; full-length dynamitin, 10.3 ± 0.6%; N-terminus, 9.9 ± 0.2%; n = 1000 cells per condition per experiment in three independent experiments), with most cells arrested in pseudoprometaphase. Because another N-terminal fragment, AA 1–78, could interfere with dynactin function in vivo without triggering p150Glued release (Valetti et al., 1999), we did not expect AA 1–87 to affect dynactin structure. To our surprise, sucrose gradient sedimentation revealed that AA 1–87 did, in fact, cause p150Glued and endogenous dynamitin to be released from the Arp1 filament (Figure 2, D and E). A comparable fragment of human dynamitin (AA 1–90) has a similar effect on dynactin integrity when overexpressed (Jacquot et al., 2010). We conclude that chicken dynamitin AA 1–87, which lacks the oligomerization motifs we showed previously to correlate with dynamitin–dynamitin binding and subunit release, can indeed bind and destabilize dynactin. This suggests a different mechanism of dynamitin-mediated disruption.
FIGURE 2:
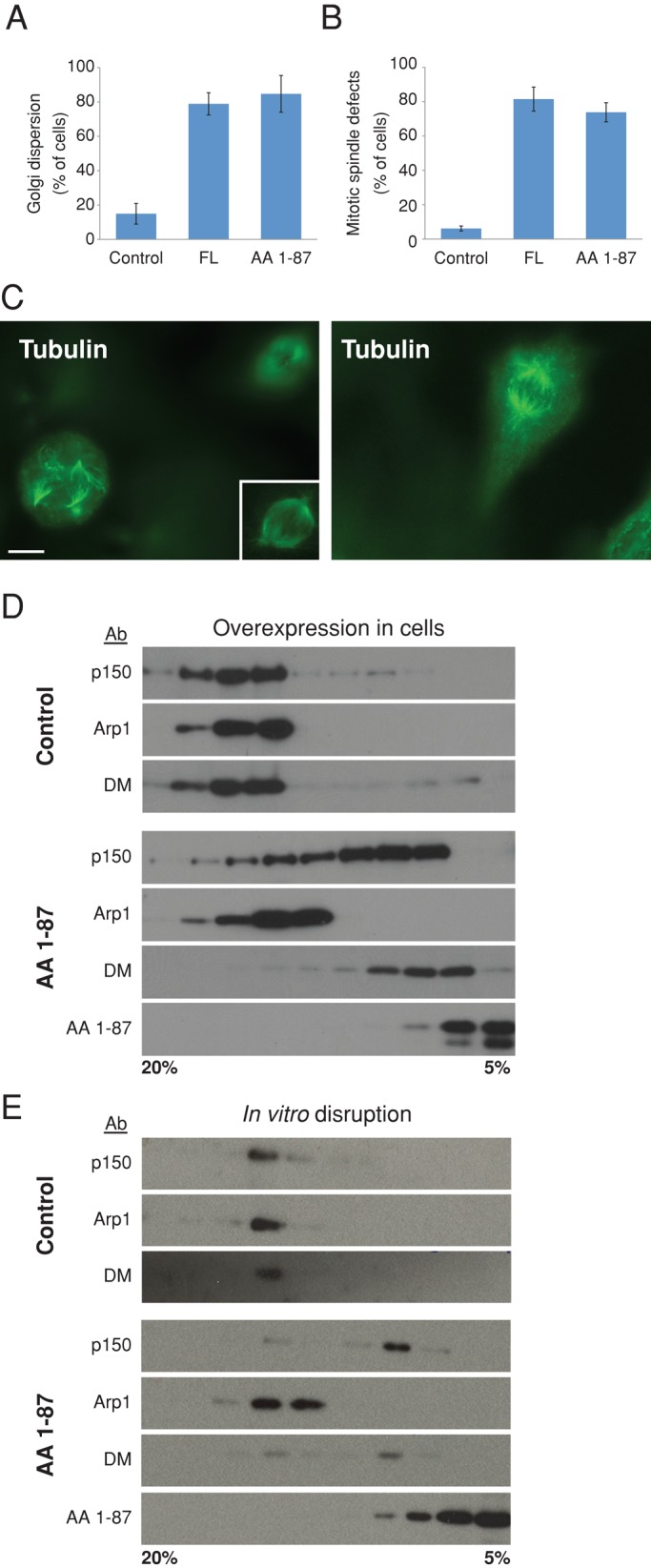
Effect of dynamitin AA 1–87 on dynactin structure and function in vitro and in vivo. (A, B) cDNAs encoding myc-tagged proteins were introduced into Cos-7 cells ∼24 h before fixation and staining for giantin to visualize the Golgi complex or tubulin to visualize mitotic spindles. Overexpressing cell populations were scored for the presence of a dispersed Golgi complex (A) or deranged spindle (B; mean ± SD; three experiments; 700 < n < 800 cells per condition for Golgi and n = 400 per condition for spindles). The results shown here were obtained using myc-tagged proteins, but similar results were obtained using mCherry-tagged dynamitin species. Myc-tagged dynactin p62 AA 370–467 and monomeric red fluorescent protein (mRFP) were used as controls in A, and CMV-β gal was used as a control in B. (C) Left, representative image of a cell expressing myc-tagged AA 1–87, stained for tubulin. The nonexpressing cell at the upper right is in a different focal plane, and the inset shows this cell's spindle in focus. (See Supplemental Figure S1 for a merged image showing myc staining.) Right, A control cell expressing CMV-β gal, stained for tubulin. Scale bar, 5 μm. (D) cDNA encoding TAP-tagged AA 1–87 (or buffer as a control) was electroporated into Cos-7 cells. After 48 h, detergent lysates were subjected to velocity sedimentation into a 5–20% sucrose gradient. Gradient fractions were analyzed by immunoblotting to detect the dynactin subunit p150Glued, Arp1, or dynamitin (DM). Dynamitin AA 1–87 was detected using an antibody to TAP. Similar results were obtained with myc- or mCherry-tagged AA 1–87. (E) Purified bovine dynactin (10 μg) was mixed with 100× molar excess of recombinant (6X-His) dynamitin AA 1–87 and subjected to velocity sedimentation as in D. AA 1–87 was detected using an antibody to the Xpress tag.
The dynamitin N-terminus binds the Arp filament but does not interact with shoulder-sidearm subunits
To better understand the mechanism by which the dynamitin N-terminus triggers dynactin disassembly, we used TAP to identify binding partners. Cos-7 cells were transfected with TAP-tagged AA 1–87 or a control protein and allowed to express the proteins for 48 h, and then cytosols were prepared and subjected to affinity purification on streptavidin and calmodulin beads (see Materials and Methods). Components of the Arp1 filament (p62, Arp1/actin, Arp11, CapZ α and β, p27, and p25) were the predominant species on silver-stained SDS gels of the TAP pull down (Figure 3A). The identity of these proteins was confirmed by immunoblotting (Figure 3C, lane 3) and mass spectrometry (Supplemental Table S1). The TAP-tagged dynamitin N-terminus showed no evidence of interaction with p150Glued, dynamitin, or p24, verifying that it binds the Arp filament directly and not through the shoulder/arm complex. Similar results were obtained in an experiment in which we immobilized purified hexahistidine (6X-His)-tagged dynamitin AA 1–87 or a control protein on beads and then added purified bovine dynactin. Once again, the dynamitin N-terminal fragment could pull down Arp1 and its end-binding proteins (CapZ, Arp11, p62, p27, and p25) but not other dynactin subunits (Figure 3B).
FIGURE 3:
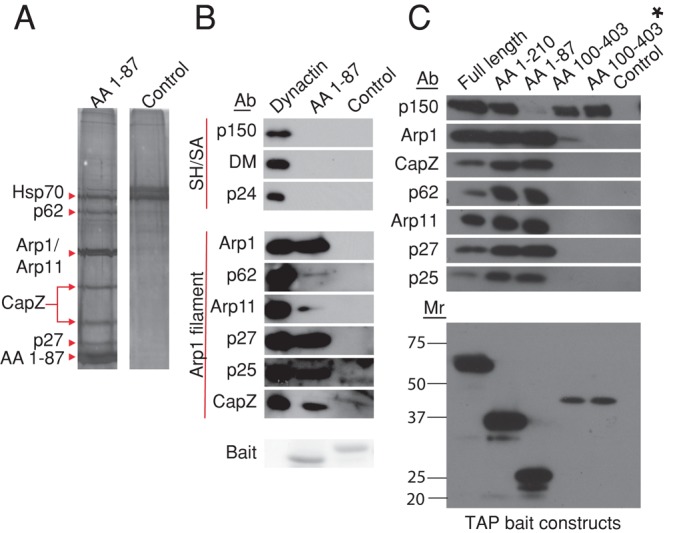
Isolation of binding partners for dynamitin N- and C-terminal fragments. (A) cDNAs encoding TAP-tagged AA 1–87 or a control (TAP-Mef2a, provided by Stratagene) were electroporated into Cos-7 cells, then lysates were prepared and subjected to tandem affinity purification on streptavidin and calmodulin resins as described in Materials and Methods. The final eluates were evaluated on a silver-stained SDS–polyacrylamide gel. Dynactin subunits (and Hsp70, the only other major component of the AA 1–87 eluate) are indicated with arrowheads. (B) Beads bearing purified 6X-His-AA 1–87 or a control (6X-His-TrbB) were mixed with purified bovine dynactin, and proteins were eluted using imidazole (see Materials and Methods). Samples were analyzed by immunoblotting. Arp1 filament and shoulder/arm (Sh/SA) components are indicated. p62 and Arp11 are very minor components that can behave erratically on blots, explaining the relatively weak signal. The “bait” protein present in each sample (6X-His-AA 1–87 or 6X-His TrbB) was detected by Ponceau S staining of the PVDF membrane. (C) Top, lysates prepared from cells expressing TAP-tagged dynamitin fragments or an untransfected control were incubated with streptavidin beads. Binding partners were eluted and analyzed by immunoblotting for the proteins indicated. For the sample labeled AA 100–403*, the lysate overexpressing TAP AA 100–403 was supplemented with recombinant 6X-His-AA 1–87 to release the shoulder/arm complex from the Arp1 filament before addition of beads. Bottom, TAP-tagged “bait” proteins present in the bead eluates were detected by immunoblotting (for calmodulin-binding protein).
Dynamitin AA 100–403 binds the shoulder/arm complex but does not cause dynactin disruption
In our previous structure–function analysis of dynamitin we focused on three α-helical coiled-coil motifs in AA ≈ 100–403 that are necessary for dynamitin self-association, p24 binding, and stable association with the Arp filament (Maier et al., 2008) and proposed that one or more was necessary for disruption. A later report indicated that AA 100–403, a fragment that contains all these motifs, did not cause organelle or spindle derangement when overexpressed (Jacquot et al., 2010). We, too, found that overexpression of TAP-tagged dynamitin AA 100–403 did not trigger release of p150Glued or endogenous dynamitin from the Arp filament and further observed that AA 100–403 did not cosediment with Arp1 when cytosols were subjected to sucrose gradient centrifugation (Supplemental Figure S2), suggesting that this fragment cannot bind the dynamitin protomers that are incorporated into dynactin. Together with our foregoing results, these findings suggest that the interaction of the dynamitin N-terminus with the Arp filament contributes significantly to disruption. The major function of the C-terminus may be to stabilize the dynactin shoulder/arm complex.
To verify that the AA 100–403 fragment was properly folded and able to interact with the shoulder/arm component, p150Glued, we used TAP to pull down AA100–403 plus any associated proteins (Figure 3C, lane 4, AA 100–403). TAP-AA 100–403 recruited p150Glued, indicating that it was active for binding, but it also pulled down a minor amount of Arp1, making it difficult to rigorously exclude the possibility of Arp-filament binding. We realized that a trace amount of overexpressed TAP AA 100–403 might have been incorporated into the dynactin shoulder/arm complex during biosynthesis, which would lead to a small amount of Arp1 being present in the TAP pull down. To rigorously demonstrate that binding of p150Glued to AA 100–403 was independent of Arp1, we added purified AA 1–87 to the cytosol to release the shoulder/arm complex from the Arp filament. When the resulting sample was incubated with immobilized TAP-AA 100–403, p150Glued was once again pulled down, but Arp1 was not (Figure 3C, lane 5, AA 100–403*).
Dynamitin AA 1–87 binds directly to Arp1 but not to other dynactin components
These affinity purification experiments clearly showed that dynamitin AA 1–87 binds dynactin's cargo-binding domain but did not identify which subunit (i.e., Arp1, CapZ, p27, p25, Arp11, or p62) was involved. Each dynactin molecule contains four dynamitin protomers (Eckley et al., 1999), two of which remain tightly associated with the Arp filament after dynamitin-mediated disruption (Melkonian et al., 2007). Arp1 is the only component of the filament that is present in two or more copies, making it an obvious candidate for a binding partner. Unfortunately, binding studies with purified Arp1 are extraordinarily difficult. Arp1 is completely insoluble when expressed in bacteria or insect cells, and so a recombinant form is not available, and cultured animal cells do not contain any free Arp1 protomers even when the protein is overexpressed. To get around these problems, we used native Arp1 isolated from purified bovine dynactin (as in Bingham and Schroer, 1999; see Materials and Methods) for binding assays. Purified Arp1 was found to bind purified 6X-His-AA 1–87 that had been immobilized on beads, but it did not bind a 6X-His–tagged control protein, indicating that binding was specific (Figure 4A). Conventional G-actin processed in parallel showed no evidence of binding to AA 1–87 (Figure 4B). Purified 6X-His-AA 1–87 did not copellet with polymerized F-actin (Figure 4C), further verifying the specificity of its interaction with Arp1. Together these findings demonstrate that AA 1–87 interacts directly and specifically with Arp1.
FIGURE 4:
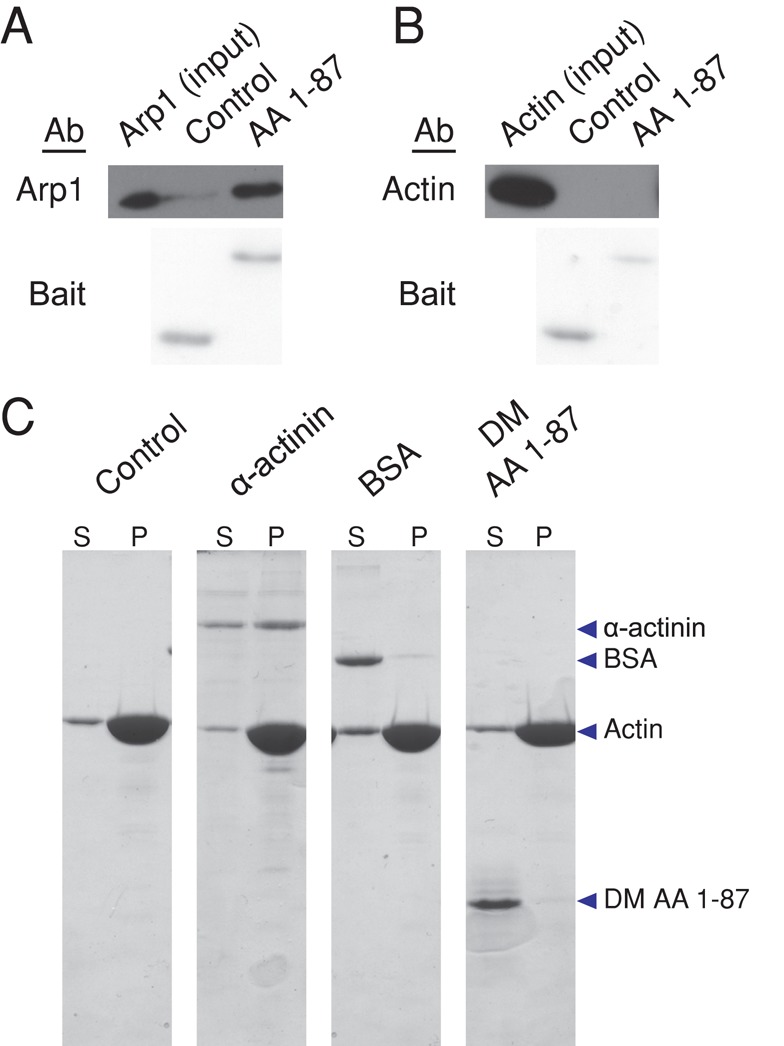
Analysis of dynamitin AA 1–87 binding to Arp1 and actin. (A, B) Arp1 isolated from bovine dynactin by KI treatment and gel filtration (A; Bingham and Schroer, 1999) or G-actin (B) was dialyzed into G-buffer for 1 h and then mixed with Talon beads bearing purified 6X-His-AA 1–87 or a control (6X-His-Fis1ΔTM). Samples were then analyzed by immunoblotting to detect Arp1 (2% of input and 2.5% of beads) or actin (2% of input and beads). Bait proteins were detected on the PVDF membrane by Ponceau S staining. (C) F-actin (see Materials and Methods) was incubated with buffer alone, α-actinin, BSA, or purified 6X-His-AA 1-87 (DM AA 1–87) for 30 min at room temperature. F-actin and bound proteins were then pelleted, and equal proportions of the supernatants (S) and pellets (P) were evaluated by SDS–PAGE, followed by Coomassie blue staining.
Dynactin's Arp filament is “capped” at both ends by protein complexes that prevent subunit addition and loss. The end nearest the shoulder-sidearm carries the conventional actin-capping protein, CapZ/CP (Schafer et al., 1994), whereas the opposite end terminates in the complex of Arp11, p62, p27, and p25 (Eckley et al., 1999). Although dynamitin AA 1–87 binds Arp1 directly, these proteins might participate in binding as well. To determine whether other Arp-filament components contributed to interactions with dynamitin AA 1–87, we embarked on a series of binding studies using cell extracts that had been depleted of dynactin components via RNAi.
The actin capping protein, CapZ, is located at the “barbed” end of the Arp filament in the immediate vicinity of the dynactin shoulder (Schafer et al., 1994; Imai, Narita, Maeda, and Schroer, unpublished data), making it an obvious potential binding partner for dynamitin. Previous work indicated that CapZ can be depleted from cells using RNAi (Mejillano et al., 2004), and we, too, found that cells transfected with CapZ small interfering RNAs (siRNAs) for 48 h largely lacked CapZ. However, when we analyzed the depleted cell lysates more carefully using sucrose gradient sedimentation, we detected a small pool of CapZ that sedimented at 20S (Supplemental Figure S3). This represents CapZ that is incorporated into dynactin and thus apparently resistant to depletion owing to dynactin's long half-life (Brown et al., 2005). The fact that CapZ cannot be depleted from the dynactin-associated pool prevented us from using RNAi to eliminate CapZ as a candidate. Instead, we examined binding of free cytosolic CapZ to 6X-His-AA 1–87. Free CapZ dimers were separated from the dynactin-associated, 20S pool using sucrose gradient sedimentation, and then the CapZ-containing fractions were incubated with 6X-His-AA 1–87 adsorbed to beads. CapZ showed no evidence of interaction with AA 1–87 (Figure 5B), but it did bind a known binding partner (mCAH3; Fujiwara et al., 2010; Supplemental Figure S4). We conclude that dynamitin AA 1–87 does not bind CapZ.
FIGURE 5:
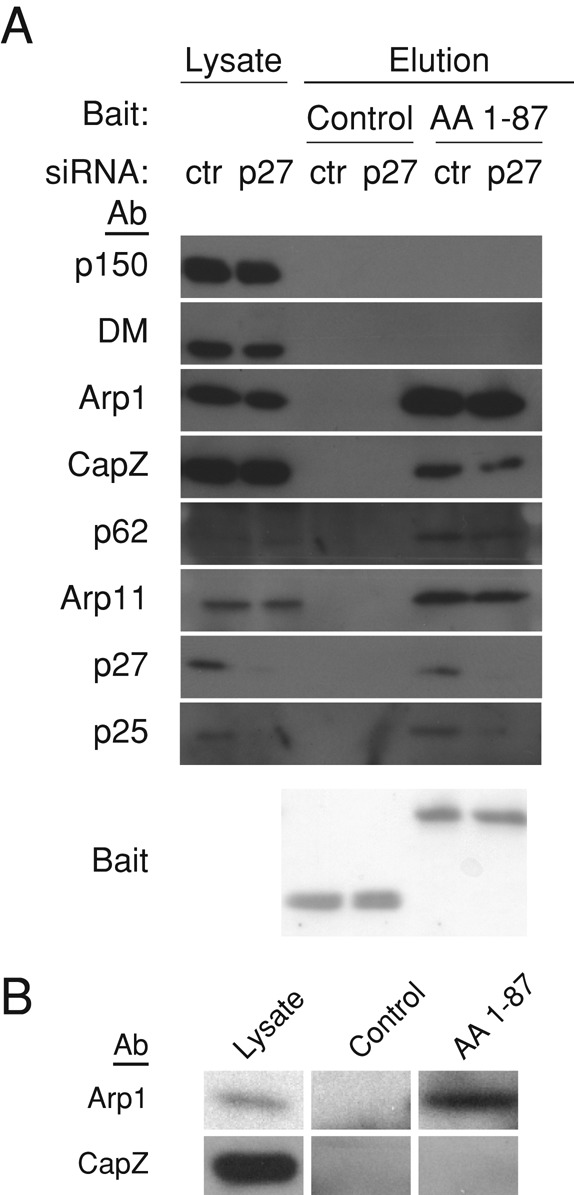
Analysis of the contributions of p27/p25 and CapZ to dynamitin–Arp1 binding. (A) Lysates of Cos-7 cells electroporated with p27 or control (ctr) siRNAs were incubated with 6X-His-AA 1–87 or a control (Fis1∆TM) before addition of Talon beads. Dynactin subunits in the eluates were detected by immunoblotting (1% of lysate and 5% of eluates). Bait proteins were detected by Ponceau S staining of the PVDF membrane. (B) Pools of cytosolic proteins (“lysate” lane) containing dynactin (top: 20S fraction) or free CapZ (bottom; 4–5S fraction) were separated by sucrose gradient sedimentation and then mixed with beads bearing 6X-His-AA 1-87 or a control (6X His-TrbB). The 4–5S fractions (bottom left) and the bead eluates (bottom middle and right) were immunoblotted for CapZ. Binding of Arp1 (in the 20S fraction; top left) to control or AA 1–87 beads was assayed in parallel (top middle and right).
We showed previously that the dynactin components p27 and p25 can be selectively and coordinately depleted from cells using RNAi (Yeh et al., 2012; Figure 5A). p150Glued, dynamitin and Arp1 still cosediment as a 20S particle that contains all the remaining dynactin subunits. Similarly, Arp1 can still be pulled down with p150Glued when p25 is deleted from Aspergillus (Zhang et al., 2011). These data are strong evidence that the interaction of dynamitin with Arp1 binding does not require p27 or p25. To verify that loss of p27 and p25 had no effect on the ability of AA 1–87 to bind the Arp filament, lysates prepared from cells depleted of p27 and p25 were mixed with 6X-His-AA 1–87, and then AA 1–87 and any associated proteins were pulled out using beads. The Arp filaments in the depleted lysate bound the resin equally well as undepleted controls (Figure 5A), providing clear demonstration that p27 and p25 are not required for dynamitin binding to the Arp1 filament.
Yeast dynamitin (Jnm1p) has been shown to interact with Arp11 (Arp10p; Clark and Rose, 2005) in a two-hybrid assay, but this interaction has not been reported in other species. No free pool of Arp11 or its binding partner p62 can be detected in cultured cells, and both proteins are insoluble when expressed in bacteria, so we could not perform direct binding assays on either using purified proteins. Unfortunately, it is not possible to use RNAi in experiments like those used to test the role of p27/p25 because depletion of either Arp11 or p62 also causes loss of Arp1 (Yeh et al., 2012). Despite these challenges, we were able to obtain anecdotal evidence indicating that p62 and Arp11 do not bind the dynamitin N-terminus. The purified Arp1 used in the direct binding experiment (Figure 4A) contains trace amounts of Arp11 and p62, CapZ, and p27/p25 (Bingham and Schroer, 1999; Eckley et al., 1999). Although Arp1 reproducibly bound dynamitin N-terminus immobilized on beads in these experiments, none of the other proteins (p62, Arp11, CapZ, p27, or p25) did and were instead always found in the column flowthrough (unpublished data).
DISCUSSION
The organization of subunits in the dynactin molecule has been generally defined (Schafer et al., 1994; Eckley et al., 1999; Imai et al., 2006; Imai, Narita, Maeda, and Schroer, unpublished data), but a high-resolution structure is not available. We show here that dynamitin occupies a key position by serving as a bipartite binding platform that anchors the dynein-binding p150Glued subunit to the Arp1 filament. The present biochemical demonstration that dynamitin binds Arp1 directly and specifically corroborates previous reports of close associations between these proteins in yeast (Clark and Rose, 2005; Amaro et al., 2008) and solves the long-standing question of how the shoulder-sidearm is tethered to the Arp1 filament.
Several lines of evidence suggest that the N-terminal portion of dynamitin is structurally distinct from the rest of the polypeptide. Sequence analysis predicts AA 1 to ∼100 to be largely unstructured except for a short α-helix (AA 55–62; predicted by DomPred; Marsden et al., 2002). In keeping with this prediction, the dynamitin N-terminus is highly labile to proteolysis (Tanimoto, Maeda, Imai, and Maeda, unpublished data; Maier et al., 2008). The rest of the sequence, by contrast, is predicted to be folded into a series of α-helices that we showed support homo- and hetero-oligomerization (Maier et al., 2008). The N-terminus is required for orderly homo-oligomerization in vitro (Maier et al., 2008) but does not contribute to interactions with the shoulder component, p24 (Tanimoto, Maeda, Imai, Maeda, Ketcham, and Schroer, unpublished observations). The present work reveals an important new role: Arp1 binding.
Many questions remain regarding the dynamitin/Arp1 interaction. It must be of sufficient affinity and stability to allow p150Glued to remain bound to the Arp1 filament even under tension, but soluble N-terminus can readily displace the entire shoulder/arm complex. The dynamitin N-terminus has also been reported to bind calmodulin (Yue et al., 2000). The ability of the same peptide to participate in multiple interactions may reflect its intrinsic disorder.
The dynamitin N-terminus contains a number of conserved charged residues, most of which are acidic, and this intrinsic negative charge may be accentuated by phosphorylation. The Arp1 primary sequence predicts a basic surface, suggesting that the interaction with dynamitin is electrostatic. Alanine-scanning mutagenesis of yeast Arp1 identified a cluster of charged amino acids near the C-terminus (K369, D371, E374, and D375; Clark and Rose, 2005) that appeared to be important for dynamitin and p150Glued binding. However, similar residues are present in actin, which we show does not bind dynamitin, suggesting that this may not be the binding site for dynamitin on Arp1.
We previously proposed that free dynamitin triggers subunit release by destabilizing contacts among dynamitin, p24, and p150Glued (Melkonian et al., 2007; Maier et al., 2008). The propensity of full-length dynamitin to oligomerize allows two exogenous protomers to be exchanged for two endogenous protomers (cartoon in Figure 6). Dynamitin AA 1–210, which lacks two of the three known oligomerization motifs, caused subunit release without remaining associated with the Arp filament (Maier et al., 2008), indicating that the subunit release does not require stable dynamitin–dynamitin binding. Our present findings verify that disruption does not depend upon dynamitin–dynamitin binding. The dynamitin N-terminus can trigger release of all four endogenous dynamitin protomers when it is overexpressed in vivo (Figure 2D; cartoon in Figure 6), whereas full-length dynamitin and AA 1–210 release only two protomers when mixed with purified dynactin in vitro (Melkonian et al., 2007; Maier et al., 2008). The simplest proposal that reconciles these findings is that disruption can occur via two different mechanisms. One (our earlier model) involves dynamitin–dynamitin binding and shoulder remodeling mediated by motifs in AA 100–403. The other (as defined here) involves direct binding of the dynamitin N-terminus to Arp1, leading to displacement of the entire shoulder/arm complex.
FIGURE 6:
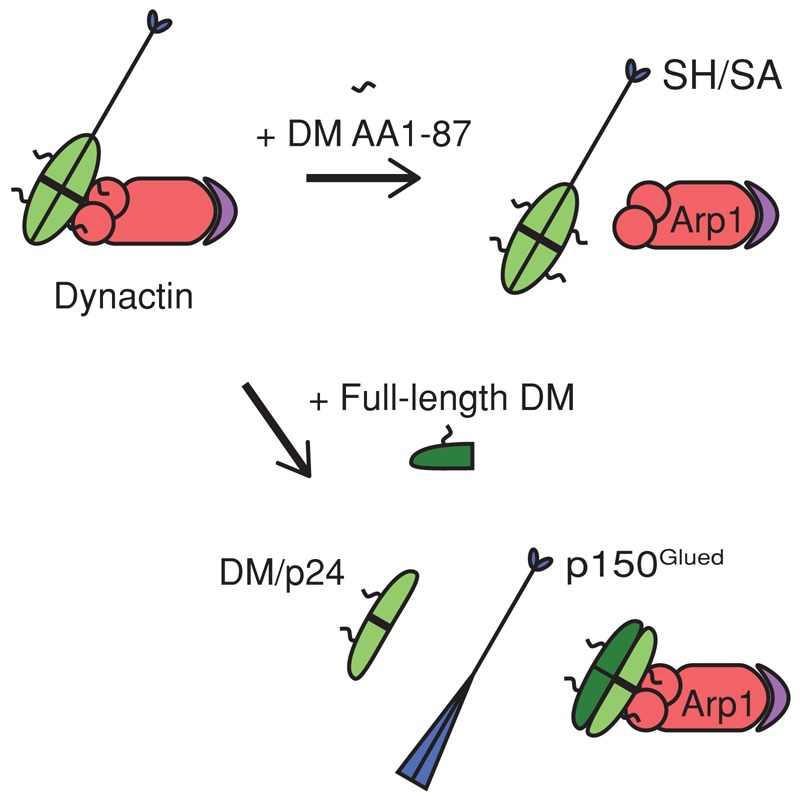
Proposed model of dynamitin interactions with other dynactin components. The unstructured dynamitin N-terminus (AA 1–87; black squiggle) binds directly to Arp1 (red) to anchor the shoulder and arm (SH/SA) structure to the Arp filament. The remainder of dynamitin (light green) is engaged in interactions with other dynamitin protomers and with p24 (thick black line) Top, excess AA 1–87 (DM 1–87) displaces the entire shoulder and arm structure from the Arp filament (purple depicts the pointed-end complex: p62, Arp11, p27, p25). Bottom, exogenous full-length dynamitin (DM, dark green) triggers subunit release by interacting with other shoulder/arm components and remodeling their interactions. This leads to displacement of a dynamitin/p24 (2:1) complex (light green; the thick black line bisecting the oval represents one p24 protomer) and the p150Glued dimer (blue). Two exogenous dynamitin protomers are exchanged for two endogenous protomers (Melkonian et al., 2007).
Previous work on dynamitin focused on its role in dynactin disintegration. The discovery that it binds Arp1 directly suggests that it may also play a role in dynactin assembly. Dynactin's Arp filament is highly uniform in length, yet the mechanism by which length is specified is unknown. Polymerization of purified Arp1 yields filaments ∼50 nm in length (Bingham and Schroer, 1999), which are longer than the ∼35-nm filaments found in dynactin, so an extrinsic mechanism is assumed to exist to limit assembly The unequal stoichiometries of dynamitin and Arp1 in dynactin (4 vs. ≥5; Imai, Narita, Maeda, and Schroer, unpublished data) precludes a simple one-to-one assembly model in which dynamitin templates a four-protomer Arp1 polymer. The fact that full-length dynamitin can displace only two of the four dynamitin protomers and leaves the other two bound to the Arp filament suggests that the two pairs of dynamitin protomers experience different environments, with the N-termini of two being closely bound to Arp1, and two being exposed on the periphery. Engagement of the N-termini of only one pair of dynamitins with Arp1 may allow for shoulder anchoring while leaving the remaining two dynamitin N-termini free to associate with extrinsic proteins such as calmodulin (Yue et al., 2000). We propose that the shoulder/arm complex binds an Arp1 oligomer (a dimer or trimer, the two stable forms of isolated Arp1; Bingham and Schroer, 1999) via direct interactions with two dynamitin protomers. This assembly intermediate must also incorporate CapZ (and possibly actin) via an undefined mechanism. A second “half-dynactin” may form via association of Arp oligomer(s) with the pointed-end subunits Arp11 and p62, both of which are also able to bind Arp1 (Garces et al., 1999; Karki et al., 2000; Eckley and Schroer, 2003). These two half-molecules could join by Arp1/Arp1 annealing (Bingham and Schroer, 1999), leading to formation of the holocomplex, a structure whose overall integrity may involve interactions between shoulder/arm and pointed-end components.
MATERIALS AND METHODS
Constructs
Full-length chicken dynamitin–green fluorescent protein (GFP; as described in Quintyne et al., 1999) was used as the template for PCR-based subcloning of the constructs described in this article (see Table 1). Commercial vectors pRSET-A (Invitrogen, Carlsbad, CA) and pNTAP-C (Interplay Mammalian TAP system; Stratagene, La Jolla, CA) were used for making His-DM AA 1–87 and the TAP-tagged constructs respectively, as listed in Table 1. Myc-tagged constructs were made using the parent vector pCMV5-myc2 (Liu and Yin, 1998). Myc-p62 AA 370–467 was subcloned from GFP-p62 (Quintyne et al., 1999) and used as a negative control in this study.
Table 1:
Constructs used.
| Construct | Vector | Up RE | Down RE | Primers | |
|---|---|---|---|---|---|
| His-DM AA 1–87 | pRSET A | BamHI | EcoRI | Forward | GATTATGGATCCGCCGACCCCAAATAC |
| Reverse | ATAATCGAATTCTCACTCCCCAGACTCATAG | ||||
| Myc-DM | pCMV5 myc2 | EcoRI | BamHI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | TATTAGGGATCCTCACTGCAGCCGCTTGATG | ||||
| Myc-DM AA 1–87 | pCMV5 myc2 | EcoRI | BamHI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | TATTAGGGATCCTCACTCCCCAGACTCATAG | ||||
| Myc-DM AA 1–78 | pCMV5 myc2 | EcoRI | BamHI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | TATTAGGGATCCTCAGCTCTTGCTGATGCG | ||||
| Myc-p62 AA 370–467 | pCMV5 myc2 | EcoRI | BamHI | Forward | TCTATGAATTCTGAAAGAGCTCATCTTAGCCGGC |
| Reverse | AATGCTGGATCCTTAAGGAAGAAGAGGGC | ||||
| TAP DM | pNTAP-C | EcoRI | XhoI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | ATAATCCTCGAGTCACTGCAGCCGCTTG | ||||
| TAP DM AA 1–87 | pNTAP-C | EcoRI | XhoI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | ATAATCCTCGAGTCATCACTCCCCAGACTC | ||||
| TAP DM AA 1–78 | pNTAP-C | EcoRI | XhoI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | ATAATCCTCGAGTCAGCTCTTGCTGATG | ||||
| TAP DM AA 1–210 | pNTAP-C | EcoRI | XhoI | Forward | GATTATGAATTCAGATGGCCGACCCCAAATAC |
| Reverse | ATAATCCTCGAGTCAGAAGGTGAGCGCGTC | ||||
| TAP DM AA 100–403 | pNTAP-C | EcoRI | XhoI | Forward | GATTATGAATTCAGCCCCAGCAGCGGTACCAG |
| Reverse | ATAATCCTCGAGTCAGAAGGTGAGCGCGTC | ||||
The vectors, restriction sites, and primers used in making each construct are shown. RE, restriction sites.
Antibodies
p150Glued and dynamitin: monoclonal antibody (mAb) product numbers 610473 and 611002, respectively, from BD Transduction Laboratories, San Jose, CA. Arp1 and p62: mAb 45A and 62B (Schafer et al., 1994). CapZ α: mAb 5B12 (Schafer et al., 1996). Arp11: affinity-purified rabbit antibody raised against the C-terminal of Arp11 (a gift from M. Way, Cancer Research United Kingdom, London, UK). p27: mAb 27A (Melkonian et al., 2007). p25: DCTN5 polyclonal antibody (10182-1-AP; ProteinTech Group, Chicago, IL). p24: affinity-purified rabbit polyclonal antibody R5700 (Pfister et al., 1998).
Giantin: G1/133 (Linstedt and Hauri, 1993). TGN46 (AbD Serotec). Tubulin: mAb DM1A (T9026; Sigma-Aldrich, St. Louis, MO) or rat mAb (YL1/2; AbD Serotec, Oxford, UK). TAP-tagged proteins: anti–calmodulin-binding protein epitope tag (07-482; Millipore, Billerica, MA); Xpress epitope in 6X-His–tagged proteins (R91025, Invitrogen); Myc epitope (ab9106; AbCam, Cambridge, UK).
Recombinant protein purification
Plasmids encoding 6X-His–tagged recombinant proteins were transformed into BL21-CodonPlus (DE3)-RIL cells, and colonies were grown overnight in a 5-ml culture. One liter of Luria broth supplemented with 100 μg/ml ampicillin and 35 μg/ml chloramphenicol was inoculated with the overnight culture, grown to OD 0.4–0.6, and then induced with 0.1 mM isopropyl-β-d-thiogalactoside overnight at 37°C before harvesting. Bacteria were pelleted and resuspended in 25 ml of binding buffer (50 mM sodium phosphate, 300 mM NaCl, 5 mM imadazole, pH 7.0) and lysed using a high-pressure homogenizer (Avestin EmusiFlex C3). The lysate was centrifuged at 15,000 × g for 20 min, and 5 ml of the supernatant was loaded onto a 0.5-ml Talon Spin-column (Clontech, Mountain view, CA). The column was washed 10 times with wash buffer (50 mM sodium phosphate, 300 mM NaCl, 25 mM imidazole, pH 7.0) and eluted with 1–2 ml of 150 mM imidazole in 50 mM sodium phosphate and 300 mM NaCl, pH 7.0. In some experiments, 250 mM imidazole was used instead. The p150Glued fragment CC1 was purified as in King et al. (2003). Dynamitin AA 1–210 (wt and L118P variant) and AA 1–81 were purified as in Maier et al. (2008).
Circular dichroism spectroscopy
All proteins were in 50 mM sodium phosphate, 150 mM NaCl, pH 7.0. CC1 was 0.13 mg/ml, AA 1–87 was 0.28 mg/ml, AA 1–78 was 0.18 mg/ml, AA 1–210 was 0.072 mg/ml, and AA 1–210 L118P was 0.342 mg/ml. Spectra were recorded in a 0.1-cm quartz cuvette at 25°C with a Jasco J-710 spectropolarimeter. The scans were collected from 195 to 260 nm using four consecutive accumulations, the average of which gave the final CD spectra. The continuing scans were done at scanning speed 50 nm/min with 4 s per point response time under standard sensitivity settings. The data pitch was 0.2 nm, and the bandwidth for samples was 1 nm. Each spectrum was subtracted by a buffer spectrum and further normalized by amino acid number and concentration. Eventually, measurements were reported in deg cm2 dmol–1. The α-helical content was estimated from fa = ([θ222] + 2340)/(−30,300) (Chen and Yang, 1971), where θ222 represents the mean residue ellipticity at 222 nm and fa is the fraction of α-helical content.
Cell culture, transfection, and RNAi
Cos-7 cells were grown in high-glucose DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO), 1% l-glutamine, and penicillin/streptomycin (Life Technologies) and 1% MEM nonessential amino acids (Life Technologies) at 37°C with 5% CO2. For most experiments using transient transfection, cells were grown to 70–90% confluency and harvested with 0.25% trypsin-EDTA (Life Technologies), and then 5 × 106 cells were resuspended in 0.5 ml Opti-MEM (Life Technologies) and electroporated with 10 μg of DNA at 240 V using an Electro Cell Manipulator 600 (BTX, Holliston, MA). Cells were grown on coverslips in six-well dishes overnight before being processed for immunofluorescence or up to 48 h in 10-cm dishes before being harvested for TAP or velocity sedimentation analysis. For analysis of mitotic cells, 4 × 105 cells were seeded/35-mm well in a six-well plate on a coverslip (0.2% gelatin coated) 24 h before transfection. Lipofectamine 2000 (Invitrogen) reagent, 5 μl, was incubated with 125 μl of Opti-MEM (Life Technologies) for 5 min and then mixed with 1 μg of plasmid DNA and 125 μl of Opti-MEM for 20 min at room temperature. The mixture was added dropwise to the well. Media were changed to remove the transfection reagent 3 h later, and coverslips were processed for immunofluorescence 24 h after that. For p27 knockdown experiments, siRNA oligos (CCACCUAAAGAAGACUAUGdTdG) at a final concentration of 133 nM were electroporated into 0.5 ml of Cos-7 cells, and cells were harvested 48–72 h later. For CapZ knockdown experiments, siRNA oligos targeting human CapZ β (GGAUUACCUUUUGUGUGACdTdT) at a final concentration of 133 nM (3.33 μl of 20 μM siRNA) were electroporated into 0.5 ml of Cos-7 cells, and cells were harvested 48–96 h later as indicated in the legend to Supplemental Figure S3. See Supplemental Materials and Methods for details of the CapZ siRNA experiments.
Immunofluorescence
Cells were rinsed with phosphate-buffered saline (PBS) and then fixed in −20°C methanol for 5 min or 4% formaldehyde in PBS at 37°C for 15 min. Formaldehyde-fixed cells were quenched with 50 mM ammonium chloride in PBS for 5 min and then permeabilized with 0.1% Triton X-100 in PBS/1% bovine serum albumin (BSA) for 15 min. All coverslips were incubated in blocking buffer (1% BSA in PBS) for 15 min before antibody addition. Primary antibodies were diluted in blocking buffer and added to cells for 1 h at room temperature in a humidified chamber, rinsed three times with blocking buffer, and then incubated with secondary antibody for 30 min. Before mounting, coverslips were rinsed three final times with blocking buffer. 4′,6-Diamidino-2-phenylindole (1 μg/ml) was included in the penultimate rinse. Cover slips were briefly rinsed in Milli-Q water (Millipore) and mounted using Fluoromount (Sigma-Aldrich) and dried overnight in the dark before imaging.
Immunofluorescence microscopy was performed on an Axiovert 100 LM microscope (Carl Zeiss, Jena, Germany). Images were captured using a CoolSnap digital monochrome camera and processed using SlideBook and Photoshop (Adobe). To quantify Golgi disruption, a minimum of 100–400 transfected cells were scored per condition per experiment in three independent experiments. To measure mitotic index, at least 1000 transfected cells were scored per condition per experiment in three independent experiments. To quantify spindle disruption, at least 100 transfected mitotic cells were scored per condition per experiment in three independent experiments.
Dynactin disruption and sucrose gradient analysis
For disruption experiments using purified proteins, 10 μg (9 × 10−6 mole) of purified bovine dynactin was mixed with a 25× molar excess (9 × 10−4 mole; see Maier et al., 2008) of the different recombinant dynamitin constructs in a total volume of 0.1–0.2 ml of sedimentation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol [DTT]) and then incubated on ice for 30 min. In some cases, 9 × 10−4 mole of the purified recombinant dynamitin fragments was added to 500 μg of Cos-7 detergent lysates (prepared as described later). After incubation, the samples were sedimented into a 5–20% sucrose gradient in a Beckman SW55 Ti rotor (Beckman Coulter, Brea, CA) at 30,500 rpm for 16 h at 4°C. Fractions (0.5 ml) were collected, and dynactin subunits were detected using SDS–PAGE, followed by immunoblotting. For analysis of dynactin integrity in cells overexpressing different dynamitin constructs, cells were harvested 18–48 h posttransfection, lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton X-100) for 30 min at 4°C, and then centrifuged at 44,700 rpm in a Beckman SW55Ti rotor for 30 min. The supernatants were collected, and 500 μg to 1 mg of protein in a volume of 0.2 ml was sedimented into a 5–20% sucrose gradient and analyzed as described.
Tandem affinity purification
TAP-tagged dynamitin constructs (Interplay Mammalian TAP system) were electroporated into Cos-7 cells for 48 h before harvesting. TAP controls were either TAP-Mef2a (Stratagene) or the empty TAP parent vector. Cells were lysed with lysis buffer and subjected to TAP on streptavidin and calmodulin beads according to the manufacturer's instructions, except that 1 mg of lysate was used rather than lysate prepared from 1 × 107 cells. As indicated in some experiments, only the first step of the purification (streptavidin-bead binding) was performed. See Supplemental Materials and Methods for mass spectrometry of samples isolated by TAP.
Talon bead binding
For binding to Talon beads (Clontech), purified proteins were dialyzed overnight into binding buffer (50 mM sodium phosphate, 300 mM NaCl, 5 mM imidazole, pH 7.0). In most bead-binding experiments, proteins were freshly purified and dialyzed. In some cases, proteins were first flash-frozen in liquid nitrogen and stored at −80°C, then precleared by centrifugation (14,000 rpm for 30 min, Eppendorf 5417C) to remove any aggregates. Proteins were not frozen more than once. Each binding reaction contained 7.3 nmol of protein (e.g., 100 μg of 6X-His-AA 1–87) and 12.5 μl of beads. 6X-His-TrbB (a conjugal transfer protein in Escherichia coli; Hemmis et al., 2011) or 6X-His-Fis1ΔTM (Picton et al., 2009) proteins were used as negative control. To assay binding to dynactin, 7.3 nmol (100 μg) of 6X-His-AA 1–87 in binding buffer was bound to 12.5 μl of Talon beads for 30 min and then 10 μg purified bovine dynactin (Quintyne et al., 1999) in binding buffer was added and the mixture rotated for 30 min at 4°C. Beads were washed three times with binding buffer and proteins eluted with elution buffer (150 mM imidazole, 300 mM NaCl, 50 mM sodium phosphate, pH 7.0). For experiments involving siRNA-treated cells, 100 μg of 6X-His-AA 1–87 was incubated for 30 min at 4°C with 500 μg of cell lysis (prepared using lysis buffer; see earlier description). Talon beads were added, the mixture incubated another 30 min, and the beads were washed and eluted as described. For CapZ binding, proteins in a detergent lysate made from untreated Cos-7 cells were sedimented into a 5–20% sucrose gradient, and fractions containing free CapZ (4–5S) or dynactin (19–20S) were used.
Arp1 isolation and binding assays
Arp1 was isolated from bovine dynactin using gel filtration in the presence of 0.7 M KI as described previously (Bingham and Schroer, 1999; Eckley et al., 1999) and then cryostored in 50% sucrose. Samples were thawed, passed through a 0.22-μm syringe-tip polyvinylidene fluoride (PVDF) filter, and subjected to a second round of chromatography on a Superose12 column in column buffer (2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT) containing 0.7 M KI to remove sucrose and any oligomers. Fractions containing Arp1 monomers (and trace amounts of p62, Arp11, actin, CapZ α/β, p27, and p25; Bingham and Schroer, 1999) were pooled, dialyzed into column buffer without KI for 1 h, and used immediately in binding assays.
Actin copelleting assay
Human platelet actin (Cytoskeleton, Denver, CO) was resuspended in G-buffer (5 mM Tris-HCl, pH 8.0 and 0.2 mM CaCl2) at 1 mg/ml, precleared by centrifuging at 150,000 × g in a SW55 Ti rotor for 90 min at 4°C, and then polymerized by addition of F-buffer (50 mM KCl, 2 mM MgCl2 and 1 mM ATP), followed by incubation for 4 h at room temperature. 6X-His-tagged-AA 1–87 was dialyzed into 10 mM Tris-Cl, pH 7.5, and 20 mM NaCl. A 40-μg amount of 6X-His-AA 1–87 (15.1 μM), α-actinin (2 μM), or BSA (2 μM) was added to 160 μg of F-actin and incubated for 30 min at room temperature. The mixtures were centrifuged at 150,000 × g in a SW55Ti rotor for 90 min at room temperature, and the supernatant was carefully separated from the pellet. Equal proportions of the supernatants and pellets were analyzed by SDS–PAGE, followed by Coomassie blue staining.
Supplementary Material
Acknowledgments
We are grateful to the following for providing reagents for this work: John Cooper for CapZ siRNA oligos, D. Mark Eckley for bovine Arp1, John A. Hammer, III for the CapZ binding protein mCAH3, Casey Hemmis for 6X-His-TrbB (control), Bingnan Kang for TAP parent vectors, Stephen M. King for the p24 Ab, Adam Linstedt for the giantin antibody (G1/133), Maria Matos for pCMV5-myc2, Mollie Meffert for mCherry-dynamitin and mCherry vector, Lora Picton for 6X-His-Fis1ΔTM (control), Arne Schon for his assistance with the CD, Fengyi Wan for the TAP-Mef-2a plasmid, and Michael Way for the affinity-purified Arp11 Ab. Our thanks also go to John Cooper, Michael Matunis, Kathy Wilson, and Yixian Zheng for valuable advice during the course of study. We thank the Schroer lab members for their advice and comments on the manuscript. This work was supported by National Institutes of Health grants to T.A.S. (RO1 GM44589) and J.Y. (P41 RR011823).
Abbreviations used:
- AA
amino acids
- CP
actin-capping protein
- DM
dynamitin
- 6xHis
hexahistidine
- KI
potassium iodide
- mRFP
monomeric red fluorescent protein
- RNAi
RNA interference
- SH/SA
shoulder/arm
- siRNA
small interfering RNA
- TAP
tandem-affinity purification
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0842) on May 14, 2014.
*Present address: Department of Genome Sciences, University of Washington, Seattle, WA 98195.
REFERENCES
- Amaro IA, Costanzo M, Boone C, Huffaker TC. The Saccharomyces cerevisiae homolog of p24 is essential for maintaining the association of p150Glued with the dynactin complex. Genetics. 2008;178:703–709. doi: 10.1534/genetics.107.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham JB, Schroer TA. Self-regulated polymerization of the actin-related protein. Arp1 Curr Biol. 1999;9:223–226. doi: 10.1016/s0960-9822(99)80095-5. [DOI] [PubMed] [Google Scholar]
- Brown CL, Maier KC, Stauber T, Ginkel LM, Wordeman L, Vernos I, Schroer TA. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 2005;6:1114–1124. doi: 10.1111/j.1600-0854.2005.00347.x. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintanance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Yang JT. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem Biophys Res Commun. 1971;44:1285–1291. doi: 10.1016/s0006-291x(71)80225-5. [DOI] [PubMed] [Google Scholar]
- Cheong FK. The Structural Role of Dynamitin in Dynactin. PhD Thesis. Baltimore, MD: Johns Hopkins University; 2010. [Google Scholar]
- Cheong FK, Schroer TA. The role of dynactin in dynein-mediated motility. In: King SM, editor. Dyneins: Structure, Biology and Disease. San Diego, CA: Academic Press; 2011. pp. 505–521. [Google Scholar]
- Clark SW, Rose MD. Alanine scanning of Arp1 delineates a putative binding site for Jnm1/dynamitin and Nip100/p150Glued. Mol Biol Cell. 2005;16:3999–4012. doi: 10.1091/mbc.E05-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of 50kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 filament pointed end. J Cell Biol. 1999;147:307–319. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Schroer TA. Interactions between the evolutionarily conserved, actin-related protein, Arp11, protein, actin and Arp1. Mol Biol Cell. 2003;14:2645–2654. doi: 10.1091/mbc.E03-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Remmert K, Hammer JA., 3rd Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem. 2010;285:2707–2720. doi: 10.1074/jbc.M109.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces JA, Clark IB, Meyer DI, Vallee RB. Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr Biol. 1999;9:1497–1500. doi: 10.1016/s0960-9822(00)80122-0. [DOI] [PubMed] [Google Scholar]
- Hemmis CW, Berkmen M, Eser M, Schildbach JF. TrbB from conjugative plasmid F is a structurally distinct disulfide isomerase that requires DsbD for redox state maintenance. J Bacteriol. 2011;193:4588–4597. doi: 10.1128/JB.00351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Narita A, Schroer TA, Maeda Y. Two-dimensional averaged images of the dynactin complex revealed by single particle analysis. J Mol Biol. 2006;359:833–839. doi: 10.1016/j.jmb.2006.03.071. [DOI] [PubMed] [Google Scholar]
- Jacquot G, Maidou-Peindara P, Benichou S. Molecular and functional basis for the scaffolding role of the p50/dynamitin subunit of the microtubule-associated dynactin complex. J Biol Chem. 2010;285:23019–23031. doi: 10.1074/jbc.M110.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Tokito MK, Holzbaur EL. A dynactin subunit with a highly conserved cysteine-rich motif interacts directly with Arp1. J Biol Chem. 2000;275:4834–4839. doi: 10.1074/jbc.275.7.4834. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Yin HL. Identification of the binding partners for flightless I, A novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J Biol Chem. 1998;273:7920–7927. doi: 10.1074/jbc.273.14.7920. [DOI] [PubMed] [Google Scholar]
- Maier KC, Godfrey JE, Echeverri CJ, Cheong FK, Schroer TA. Dynamitin mutagenesis reveals protein-protein interactions important for dynactin structure. Traffic. 2008;9:481–491. doi: 10.1111/j.1600-0854.2008.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden RL, McGuffin LJ, Jones DT. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 2002;11:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Melkonian KA, Maier KC, Godfrey JE, Rodgers M, Schroer TA. Mechanism of dynamitin-mediated disruption of dynactin. J Biol Chem. 2007;282:19355–19364. doi: 10.1074/jbc.M700003200. [DOI] [PubMed] [Google Scholar]
- Pfister KK, Benashski SE, Dillman JF, III, Patel-King RS, King SM. Identification and molecular characterization of the p24 dynactin light chain. Cell Motil Cytoskeleton. 1998;41:154–167. doi: 10.1002/(SICI)1097-0169(1998)41:2<154::AID-CM6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Picton LK, Casares S, Monahan AC, Majumdar A, Hill RB. Evidence for conformational heterogeneity of fission protein Fis1 from Saccharomyces cerevisiae. Biochemistry. 2009;48:6598–6609. doi: 10.1021/bi802221h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at fibroblast centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siglin AE, Sun S, Moore JK, Tan S, Poenie M, Lear JD, Polenova T, Cooper JA, Williams JC. Dynein and dynactin leverage their bivalent character to form a high-affinity interaction. PLoS One. 2013;8:e59453. doi: 10.1371/journal.pone.0059453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc Natl Acad Sci USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittman T, Hyman A. Recombinant p50/dynamitin as a tool to examine the role of dynactin in intracellular processes. Methods Cell Biol. 1999;61:137–143. doi: 10.1016/s0091-679x(08)61978-0. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L, Lu S, Garces J, Jin T, Li J. Protein kinase C-regulated dynamitin-macrophage-enriched myristoylated alanine-rice C kinase substrate interaction is involved in macrophage cell spreading. J Biol Chem. 2000;275:23948–23956. doi: 10.1074/jbc.M001845200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Penalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


