Abstract
In whole adult mouse lung, full identification of airway nerves (or other cellular/subcellular objects) has not been possible due to patchy distribution and micron-scale size. Here we describe a method using tissue clearing to acquire the first complete image of three-dimensional (3D) innervation in the lung. We then created a method to pair analysis of nerve (or any other colabeled epitope) images with identification of 3D tissue compartments and airway morphometry by using fluorescent casting and morphometry software (which we designed and are making available as open-source). We then tested our method to quantify a sparse heterogeneous nerve population by examining visceral pleural nerves. Finally, we demonstrate the utility of our method in human tissue to image full thickness innervation in irregular 3D tissue compartments and to quantify sparse objects (intrinsic airway ganglia). Overall, this method can uniquely pair the advantages of whole tissue imaging and cellular/subcellular fluorescence microscopy.
Keywords: nerve, clearing, modeling, visceral pleura, morphometry
Clinical Relevance
Our tissue clearing with airway casting method can map novel whole lung nerve pathways such as previously undescribed visceral pleural nerve populations and their novel interconnectivity to airway/vascular compartments. An improvement in fully capturing and quantifying nerves across the whole lung is critical for mechanistic studies of normal physiology and disease neuroplasticity. This technique is not confined to nerves or confined to animal studies but is readily applicable to any antibody-based microscopy and, as we demonstrate, is readily applicable in human tissue.
Neuroplasticity, including changes in nerve structure and neurotransmitter content, occur in lung diseases, including asthma, cough, and chronic obstructive pulmonary disease (1–3). However, lung nerve subpopulations are sparsely and unevenly distributed, and this may explain inconsistent findings in attempting to characterize pulmonary neuroplasticity in disease using traditional histological methods (4–6). This sampling problem is compounded because the distribution of the disease itself is often patchy (7, 8). In asthma, for example, post mortem tissue specimens and in vivo ventilation data have demonstrated that the locations of pathology and poor ventilation are unpredictable and scattered across the lung (7, 8).
Our overall goal was to develop a method to study lung innervation that combines the advantages of whole lung imaging to capture the complete structure of an organ with the advantages of fluorescence microscopy to detect fine subcellular structure. Although lung nerves initially travel in conspicuous bundles, visualizing their individual processes at effector/receptor sites requires micron-scale optical resolution and a labeling method to increase signal contrast and visibility. This is not possible with current whole lung imaging approaches (e.g., micro-computed tomography and magnetic resonance imaging [MRI]) (9, 10). Whole lung imaging in animal models has a more limited range of labeling available and typically millimeter resolution with a higher resolution possible when examining a small portion of the lung (11–13). In contrast, fluorescence/brightfield microscopy has the capability to resolve individual neurites but has not previously imaged the whole lung due to optical/physical restraints. Design-based stereology and whole lung microdissection are the two current microscopy approaches that can apply fluorescence nerve images across the lungs to attempt capturing macro- and microanatomy (14, 15). Design-based stereology is a well-defined mathematical approach to reduce selection bias and deduce whole lung structure from rigorous randomized sampling. However, this approach is limited to common objects, such as muscle efferents, with sufficient sampling probability (14, 16).
We recently developed a new approach using whole lung microdissection to map a sparse epithelial nerve population with sampling probability too low for stereology (15). Although this technique can capture full airway epithelial nerve distribution, it is labor intensive (∼ 8 h to dissect one full mouse airway tree) and has the potential to disturb normal lung architecture. Imaging of nerves is also limited to a visible and surgically accessible tissue compartment (e.g., the epithelium). Here we introduce a method capable of overcoming the resolution and labeling limitations of whole lung imaging and the undersampling and tissue distortion of microscopic techniques to image and quantify nerve subpopulations at the individual neurite level across all tissues of the three-dimensional (3D) mouse lung. We also demonstrate the application of this method to full-thickness human airway tissues.
Analysis of whole lung microscopic images also presents challenges. Large 3D images must be subdivided on the basis of meaningful anatomy (e.g., vessels, ducts, ventricles, and lobes). These anatomic structures have nongeometric 3D shapes, so researchers would be faced with weeks of manual tracing to identify even gross tissue compartments. As a result, studies often condense their images to two dimensions (flattening) or crop their images, reducing sample size. This sacrifices data and increases sampling error (15).
In this paper we introduce a new technique that can produce whole lung 3D nerve images. Two major advantages of this approach are (1) quantification of sparse heterogeneous nerve populations and (2) identification of nerve populations in distinct tissue compartments. We demonstrate whole lung nerve imaging and identification of 3D tissue compartments of the mouse lung (airway, pleura, vascular, and airway branch generations) and then test our technique at detecting and quantifying sparse heterogeneous nerves by analyzing visceral pleural nerves. We also demonstrate our technique’s utility in human tissue by generating the first full-thickness images of tracheal innervation followed by identification of tissue compartment–specific nerves and quantification of sparse heterogeneous nerves (intrinsic ganglia).
Materials and Methods
Animal and Human Tissue
Tissue clearing, modeling, and computational analysis were demonstrated in intact murine airways from larynx down to both lungs still connected to the distal esophagus. Mouse airways were harvested en bloc from 6- to 10-week-old C57Bl/6 mice and separated from the heart.
We also demonstrated this method in full-thickness human tracheas and for visceral pleural nerves in a portion of the inferior left lung (medial surface). Organ donor tissues were provided by the Pacific Northwest Transplant Bank.
Tissue Immunohistochemistry and Clearing
Mouse tissues were fixed in Zamboni’s fixative overnight and then immunolabeled and cleared as described in the online supplement. Briefly, clearing is accomplished by dehydrating tissues in methanol and rehydrating them in a 1:2 vol/vol mixture of benzyl alcohol and benzyl benzoate. The refractive index of this mixture matches that of the tissue proteins, rendering the tissues completely transparent. Airway casts were generated by treatment with streptavidin covalently attached to a red fluorophore, taking advantage of binding to endogenous biotin expressed in the airway epithelium (17). Human trachea specimens were processed the same way for pan-neuronal staining but were also labeled for substance P with a rat antibody (clone NC1; BD Pharmigen, San Jose, CA) and goat anti-rat 555 secondary (A21434; Invitrogen, Carlsbad, CA).
Image Acquisition
All images were acquired using a confocal microscope with automated scanning provided by the OHSU Advanced Light Microscopy Core (Portland, OR) as detailed in the online supplement.
Modeling Airways with Fluorescent Casting
3D images of fluorescently casted airways were processed to make airway masks as detailed in the online supplement. Briefly, cast images were 95% automatically segmented, and then the largest two or three airway generations were semiautomatically identified using our custom software “Make 3D Mask” (G. Scott, https://sourceforge.net/projects/make3 dmask4ij/). Airway, visceral pleural, and vessel/other tissue compartments were identified using casting and our software as described in the online supplement.
Identification of Airway Regions and Quantification of Airway Morphometry and Nerve Density
We created a second computer program for FIJI (18) called “Airway Morph” (G. Scott, https://sourceforge.net/projects/airwaymorpholog/) to perform whole airway morphometry (e.g., length, cross-sectional diameter) across different branch generations as detailed in the online supplement.
3D Mapping of Visceral Pleural Nerves
To quantify visceral pleural nerve distribution in mice, nerves were modeled using computer tractography and measured in different lung regions as detailed in the online supplement.
Imaging and Quantifying Nerve Ganglia in Human Trachea
We demonstrated tissue clearing and full-thickness imaging to quantify sparse ganglia in full-thickness human trachea as described in the online supplement.
Statistical Analysis
A one-way ANOVA with Tukey’s post hoc test was used to compare visceral nerve length between different airway regions (see Figure 4) and to compare nerve density between tissue compartments (see Figure 3). A Pearson’s correlation was used to compare nerve density versus branch generation bins. A nonparametric Mann-Whitney two-tailed t test was used to compare ganglion size between adventitial versus submucosal human trachea layers. Data were analyzed using Prism 5 software (GraphPad Software, La Jolla, CA).
Figure 4.
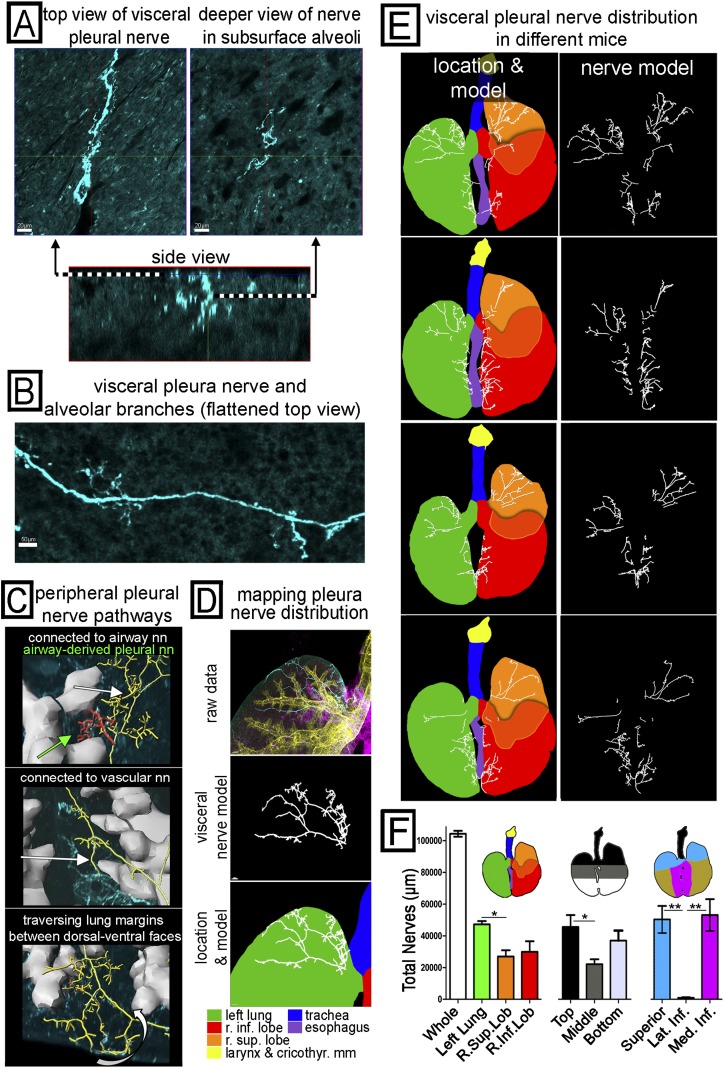
Mapping the pathways of peripheral, visceral pleural nerves and quantifying their overall distribution. (A) A 3D image sample of a visceral pleura nerve is shown in a top-down view of the pleural surface (upper left), a top-down view deeper in alveoli (upper right), and a side view of the same nerve (lower) oriented with the pleural surface on top. (B) A wider field of the pleural surface is shown in a flattened projection to demonstrate a visceral pleural nerve with branches (these usually penetrate alveoli). (C) Some of the novel peripheral pathways taken by visceral nerves are shown with airways (gray) and visceral nerves that have been identified and modeled (yellow, red). Visceral nerves are seen connecting to airway innervation (top, white arrow), connecting to vascular innervation (middle, white arrow), and traversing the lung margins (bottom, white arrow) connecting dorsal and ventral visceral nerve networks. We also found rare patches of visceral nerves (top, green arrow) derived from airway innervation but not connected to the main visceral nerve network. (D–F) Identification, modeling, and mapping of visceral nerve distribution across the whole airway. (D) Views of the superior left lung visceral pleural nerves in cyan from Figure 3A (top), the identified visceral pleural nerves (middle), and visceral pleural nerves overlaid on a lung diagram color coded by region (bottom). The color legend is shown. (E) Visceral pleural nerves shown with (left) and without (right) the color-coded lung diagram showing different regions in four different mouse lungs. (F) Aggregate nerve length based on three different schemes to subdivide the airways (lobe, top to bottom, and superior versus medial inferior versus lateral inferior) was quantified by the computer (n = 4). *P < 0.05; **P < 0.01. Visceral nerve pathways are shown in Videos E4–E6.
Figure 3.
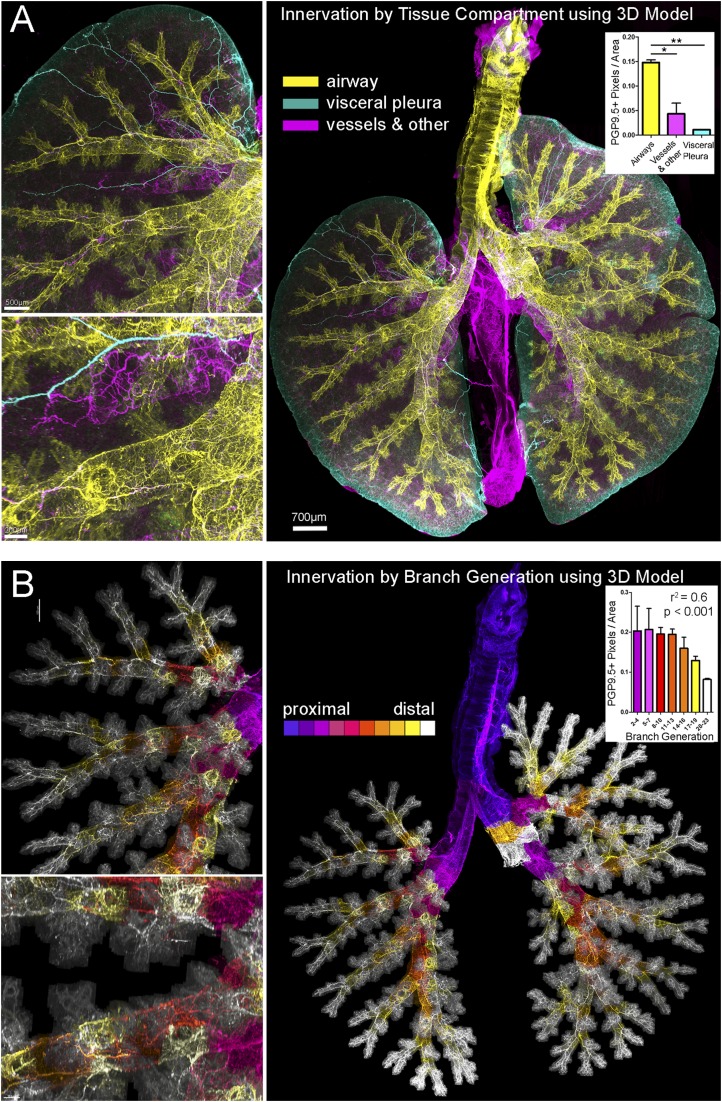
Combining 3D casting (as in Figure 2), computer modeling, and morphometric analysis with images of whole airway innervation. (A) Nerve images were color coded based on tissue compartment: airways are yellow; visceral pleural are cyan and vessel/other are magenta. The left superior lung is shown at increasing magnification (upper left, lower left) to demonstrate airway (yellow) and vascular (magenta) nerves running parallel to each other and visceral pleural nerves along the surface (cyan). The corresponding whole airway view (right) of innervation colored by tissue compartment (n = 2) was used to calculate nerve density in airway generations. *P < 0.05; **P < 0.01. (B) Nerve images were color coded based on airway generation identified in the cast 3D model from proximal (blue) to distal (white), and the density of nerves in each generation was calculated (inset). The Pearson’s correlation analysis is shown above the graphed data and was r2 = −0.7478 (P < 0.0001). The left superior lung is shown at the same magnified views (left) as in A to demonstrate density of nerves supplying distal airway generations. Airway generation figures and videos also show the two cut stems of the post-caval and middle lobes (white), which were removed before imaging. See Video E3 for innervation color coded by branch generation.
Results
Imaging Whole Airway 3D Innervation
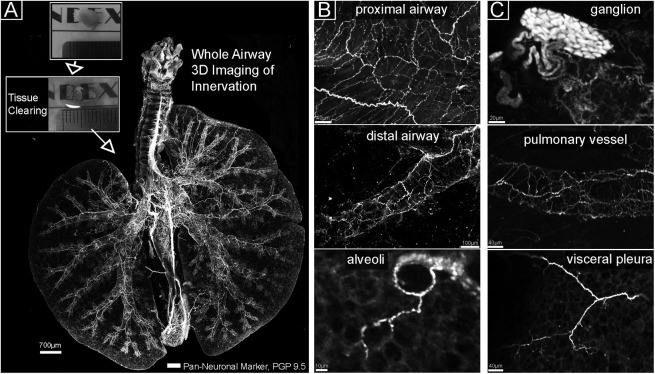
We developed a method to stain adult whole murine lungs (including the distal esophagus), optically clear the tissue, and image lung innervation in its entirety in a single imaging session using a computer-controlled laser scanning confocal microscopy. A flattened view of whole lung innervation is shown in Figure 1, and the 3D structure is best appreciated in Video E1 in the online supplement. Nerve morphology across the whole lung (Figure 1A) permitted simultaneous visualization of (1) airway and vascular innervation; (2) regional heterogeneity of neurons; and (3) detection of sparse objects such as ganglia (Figure 1C), alveolar nerve arborizations (Figure 1B), and visceral pleural nerves (Figure 1C, discussed below).
Figure 1.
Tissue clearing and whole airway imaging of nerves. (A) Flattened image of a whole mouse lung. Visible are all nerves stained with the pan-neuronal marker PGP 9.5 (white). The airway is oriented with the left lung on the left and superior/inferior right lung lobes on the right. The boxed areas show a murine lung lobe before (top) and after (bottom) optical clearing. (B, C) Magnified images from A show nerve populations in areas of the lung and a ganglion in the carina region.
Whole Airway 3D Morphometry
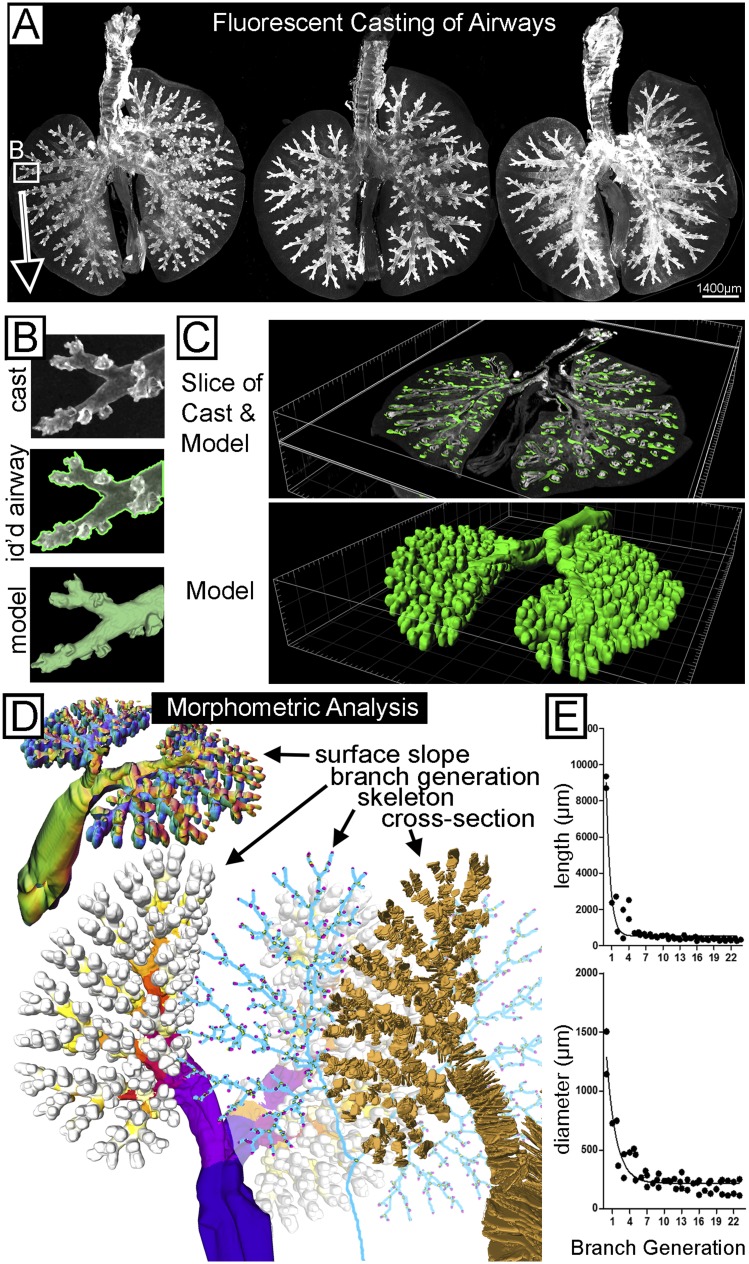
We developed a fluorescent casting method based on binding of fluorescently labeled streptavidin to endogenous biotin expressed in airway epithelium. We wrote analysis software to analyze 3D tissue compartments, a task that would otherwise require an impractical amount of manual tracing. Figure 2A shows three fluorescently cast and cleared mouse airways. Signal density was sufficient to automatically identify > 95% of the airways in a few minutes, including the distal airways that were hardest to manually delineate (Figure 2B). Signal gaps in the remaining largest three airway generations were corrected using our earlier software for 3D model generation from predefined seed points (15). We wrote new software (https://sourceforge.net/projects/airwaymorpholog/) to automatically analyze models from the casted airways shown in Figures 2C–2E. Figure 2D shows quantified morphometric values, including surface slope (top left), cross-sections (bottom right), airway skeleton (bottom middle), and branch generations (bottom left). These were used to output airway branch position, generation, length (Figure 2E and see Figure E1), cross-sectional diameter (Figures 2E and E1), volume, surface slope, and surface area. Our demonstration (n = 2) of calculating diameter and length closely followed that of previous studies with resin casts (19). Full structure of the airway cast and morphometric analysis are best seen in Video E2.
Figure 2.
Fluorescent casting and computer modeling of mouse airways. (A) Flattened images of three different representative mouse lungs. Airways were labeled with fluorophore-conjugated streptavidin, creating a fluorescent cast of the airways. (B) A magnified view (top) of distal airway cast from A. The computer identification of the airway wall (transparent green) is overlaid on the cast image (middle), and the resulting computer model tessellated from computer identification (bottom). (C) Oblique image slices (top) through a whole casted airway and overlaid three-dimensional (3D) model and (bottom) the whole 3D model. (D) Visual representations of local and global values calculated by our software from the airway model to automate morphometric analysis. The direction of local airway surface slope for analysis of shape and location was color coded by the computer and shown (upper left) in an oblique view of a coronal-cut airway model. Branch generation was color coded by the computer proximally to distally from blue to red to white (lower left). Airway generation figures and videos also show the two cut stems of the post-caval and middle lobes (marked white), which were removed before imaging. The skeletonized representation (blue, bottom middle) of the airway model made with Liu “skeleton” software was used for identification of branch-points (yellow) and end-points (pink). For calculating diameter, the computer made cross sections of the airway that look like successive disc shapes (lower right, gold). Every third cross section is shown for viewing purposes. (E) For demonstration purposes (n = 2), diameter and length were quantified over branch generations (a nonlinear decay fit is included).
Identifying Innervation of Distinct Tissue Compartments
An important advantage of this technique is that it allows us to combine casted airway modeling with nerve imaging in the same whole lung specimens to identify innervation in distinct tissue compartments. We initially distinguished three distinct gross nerve populations: airway, visceral pleural, and vessel/other. Figure 3A shows nerves in the airways (yellow) defined by the airway cast 3D model, nerves on the visceral pleural surface (cyan) defined by a mask of the lung surface, and the remaining lung with conspicuous vascular innervation (magenta). Airway nerves (yellow) consisted of large parallel bundles and a circumscribing plexus of nerves that followed the airway branching pattern. Vascular innervation (magenta) ran parallel to airway innervation but was less dense. Visceral pleural nerve distribution is described below. We then quantified overall nerve density in the three compartments shown in Figure 3B (one-way ANOVA with Tukey post test; P < 0.05).
To demonstrate automated parsing of airway nerve populations, we applied identification of airway generations (from the airway cast) to the nerve images. Figure 3B shows airway nerves color coded based on generation level from proximal to distal. The 3D structure of airway innervation in different branch generations is best seen in Video E3. We also demonstrated (n = 2) quantification of nerve density across airway generations and found that it decreased distally approximately 2.5-fold, similar to previous studies (20). The data are shown in Figure 3D and Figure E2 (r2 = 0.6; P < 0.0001).
Mapping Visceral Pleural Nerves with Whole Airway Nerve Imaging and Modeling
We also mapped an understudied population of sparse heterogeneous nerves on the visceral pleural surface (Figures 4A and 4B). Our technique fully captured these sparse nerves and allowed us to demonstrate novel visceral pleural nerve pathways and to accurately quantify their distribution. Identifying tissue compartments allowed us to detect novel connections between the visceral pleura and peripheral airways and vessels. We found that, although most visceral pleural nerves travel from medial locations to the periphery in the pleura, some pleural nerve arose from airway and vascular innervation and traveled through alveoli to the pleural surface. Many of these airway- and vessel-derived visceral pleural nerves connect with the main network of visceral pleural nerves. The 3D projection and connectivity of these different innervation patterns is best seen in Videos E5 and E6 and is shown at 1:15 in Video E4. We also found that some superior visceral nerves extend to the lateral margins of the lobe and around to the opposite dorsal/ventral side. An example of nerves traveling around the lung margins and others penetrating through parenchyma to connect dorsal and ventral visceral pleural surfaces is shown at the end of Video E6 and in Video E4, respectively. We also tracked the source of visceral pleural nerves and found that one group came from the hilum, but another population came from the esophagus. This second group projected from the esophagus onto the first 2 mm of medial surface of the inferior lungs in all mice studied (n = 4). The nerves passed through the mediastinum by traveling within the thin fascial plane connecting the distal esophagus to inferior lung lobes. A montage of brightfield images showing this connecting fascia is given in Figure E3. This projection pathway is best demonstrated in Video E4
The whole lung view also allowed us to more accurately quantify visceral pleural nerve distribution despite significant intra- and intersubject heterogeneity. We found one consistent location of a visceral pleural nerve (n = 4) along the pleura parallel to the large superior left/right pulmonary vessel, which is a useful landmark for distinguishing visceral pleural nerve populations coming from the lung hilum versus esophagus (described above). Figure 4C shows a visceral nerve parallel to the anterior segmental vessel of the left lung. Other nerve projections were seen in some mice and not in others, such as visceral pleural nerves traveling around the lung margins or between lobes (“interlobar”). Although the smaller-scale distribution of visceral pleural nerves varied between mice, we could quickly appreciate from whole lung views that the overall distribution was consistently confined to superior and inferior–medial lung surfaces and nearly absent from lateral inferior surfaces (Figure 4D). Using this whole lung observation, we produced criteria to split the lungs into a “superior” region bounded inferiorly by the superior pulmonary vessel (with a tracking visceral nerve described previously); a “medial inferior” region of the first 2 mm of medial left and right lung surface; and the “lateral inferior” region, consisting of the remaining lateral inferior lung. We then mapped visceral nerves by aggregate nerve length in these regions in comparison to two other common criteria for defining lung regions based on lung lobes or on vertical position relative to the carina and inferior margins (airway diagrams in Figure 4E). Our results show that criteria based on whole airway observation can predict the distribution (i.e., the presence and absence) of visceral nerves (Figures 4E, 4F, and E4). For this study, the postcaval (or cardiac) lobe and middle (or azygous) lobe were removed to make the left and right side the same thickness for microscope slide mounting, but in Figure E5 we demonstrate nerves on their visceral pleural surfaces.
Imaging Full-Thickness Innervation in Cleared Human Trachea
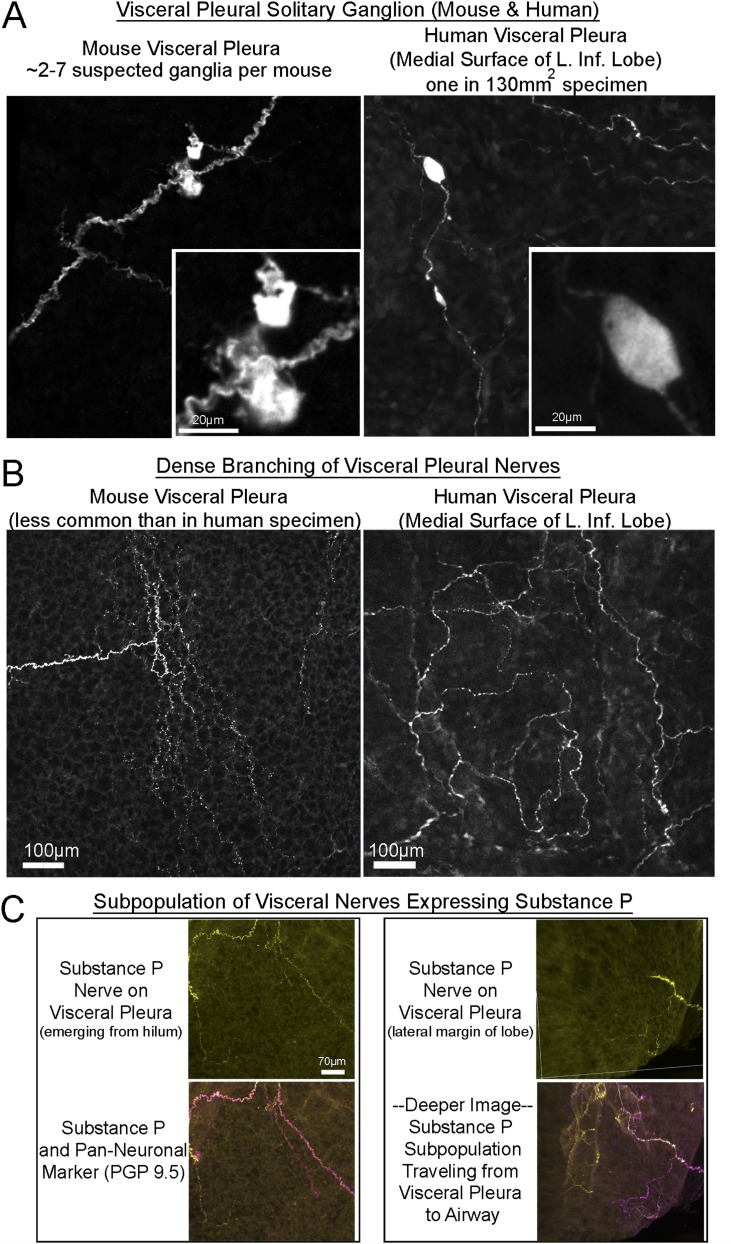
We also demonstrated the current technique for human studies (Figure 5A) and for detecting novel rare objects (e.g., solitary pleural ganglia and dense pleural patches) (Figure 5B) and pleural subpopulations (e.g., substance P nerves) (Figure 5C) thought to be important in inflammatory disease and cough (21), cleared tissue, computer modeling of tissue compartments, and quantification of sparse heterogeneous nerves (intrinsic ganglia in this case).
Figure 5.
Identifying new nerves on visceral pleural surface in mouse and human. (A, B) Images of nerves found in mouse lungs and the medial surface of the human left inferior lobe. (A) images showing novel visceral pleura ganglia stained for PGP 9.5 in mouse (left) and human (right) (see mouse ganglia in Video E4). (B) Areas of dense nerve branching are shown in mouse (left) and human (right) visceral pleura. (C) A subpopulation of visceral pleural nerves in mouse tissue that express substance P.
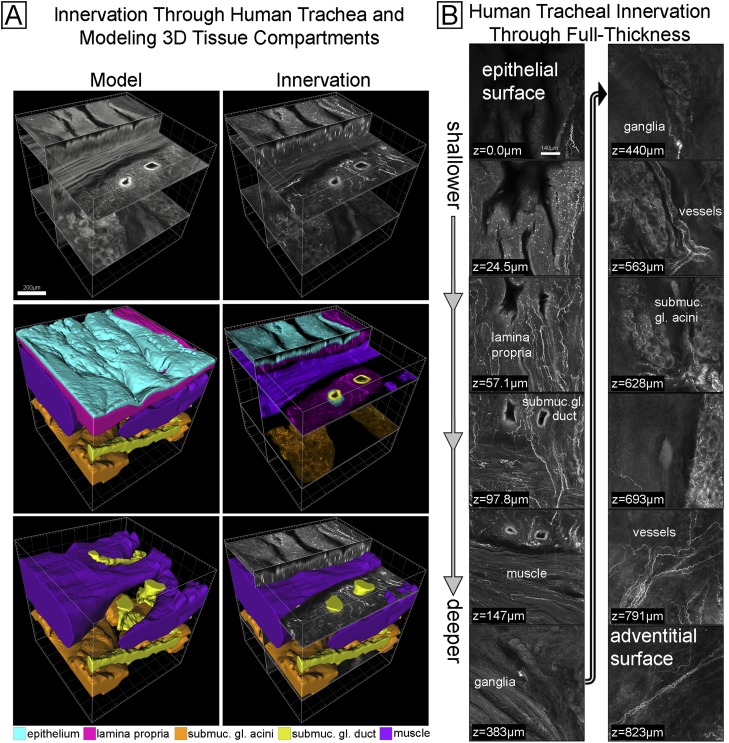
We used human trachea to demonstrate imaging past previous limits. We initially performed tissue clearing and nerve imaging in full-thickness pieces of human tracheas (n = 6) and could capture for the first time 3D innervation spanning the entire thickness from the epithelial surface to adventitial surface (Figure 6). Figure 6A shows an oblique view, and Figure 6B shows sequential cross-sectional views of human tracheal innervation at all depths. We used computer modeling to delineate smooth muscle, submucosal gland acini, submucosal gland ducts, the epithelium, and the lamina propria, all of which are complex structures that are impractical to trace manually. These delineations were made using tracheal autofluorescence emitted at a different wavelength to avoid the bias of seeing nerves. 3D models of each tissue compartment are shown in Figure 6A. We then combined tissue compartment models with nerve imaging as shown color coded in Figure 6A. The 3D structure of human tracheal tissue compartments and distribution of innervation is best seen in Video E7.
Figure 6.
Imaging innervation and modeling tissue compartments through full-thickness human trachea. (A) A full-thickness 3D image of innervation (upper right) and colored modeled tissue compartments (lower four images) made from red emission autofluorescence (upper left). The middle left and lower left images show the computer model of each tissue compartment. The right middle image shows the nerve image separated (by color) into distinct tissue compartments using the computer model. A color legend for the tissue compartments is included. (B) Select images of the data from A are shown from epithelium (top left) to the adventitia (bottom right). The presence of innervation in tissue compartments is labeled (e.g., lamina propria). See Video E7.
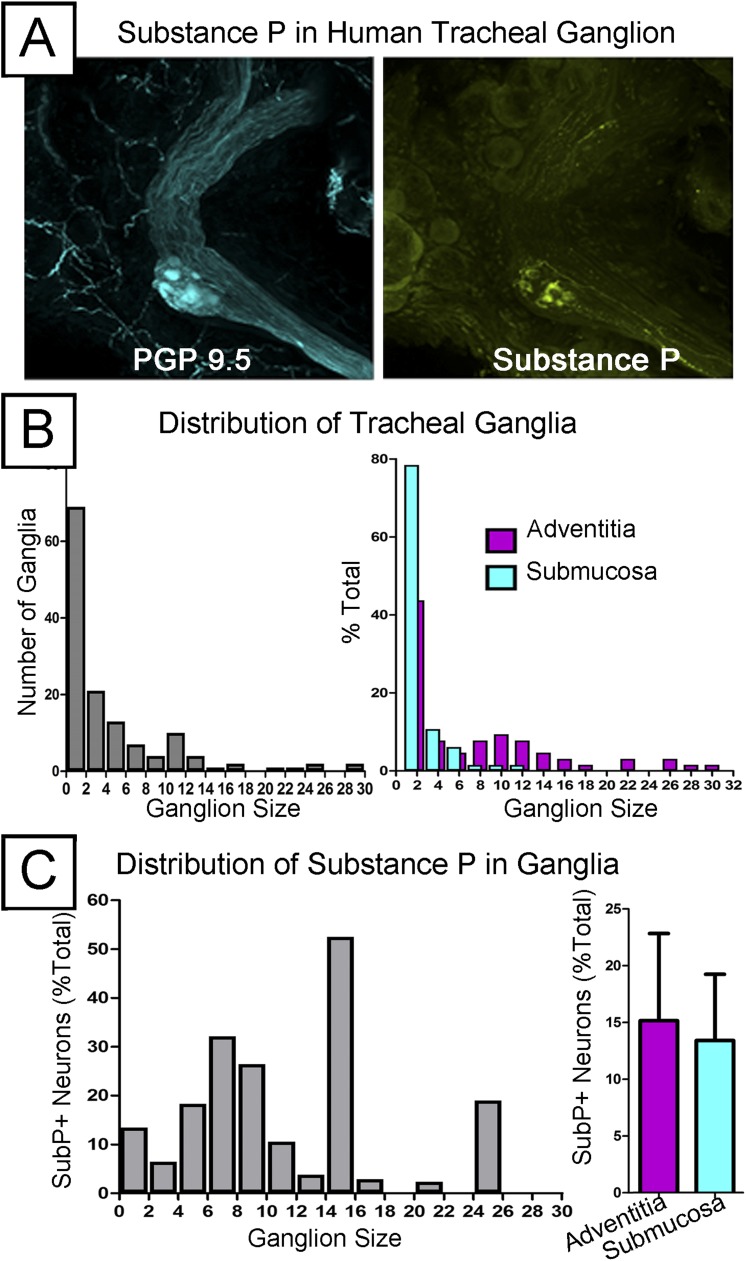
We next demonstrated the utility of this technique to detect and quantify sparsely distributed structures in human tissues. To do this, we quantified intrinsic airway ganglia, which vary in size from one neuron cell body to tens and are sparsely distributed in submucosal and adventitial layers. An overall demonstration of detecting sparse heterogeneous objects (including ganglia) in human trachea is shown in Video E8. In addition, we used substance P, a neuropeptide expressed in a subset of sensory nerves but uncommonly seen in intrinsic ganglia (discussed below), as a marker of an even rarer subset of intrinsic ganglia. Figure 7A shows an intrinsic tracheal ganglion adjacent to a nerve bundle (and nerves surrounding submucosal gland acini) and substance P expression in neuron cell bodies and connecting axons. This is best seen in Video E9. We then quantified the number and size distribution of intrinsic ganglia in human trachea segments (n = 6). Similar to findings in animal studies (22), we found that trachea contained more small intrinsic ganglia consisting of one or two neuron cell bodies than the other sizes (Figure 7B) and distinct size distributions of intrinsic ganglia in submucosal and adventitial layers with the adventitia having larger ganglia (Figure 7B). Finally, we found that substance P expression was present in a subset of ganglia but did not follow a distinct distribution in terms of ganglion size or layer (Figure 7C).
Figure 7.
Quantifying the distribution of human tracheal ganglia and the substance P subpopulation (n = 6). (A, left) Mid-sized intrinsic ganglia in human trachea stained with PGP 9.5. (A, right) Substance P was also labeled in these ganglia, where it stains neuron bodies and axons within the neighboring nerve bundle. (B, left) The relative abundance of different sized ganglia across the whole trachea and (right) their relative abundance when separated into submucosa and adventitia layers. (C) Quantification of the size distribution and overall layer separation in the substance P subpopulation.
Discussion
It is important to map both the macro- and microanatomy of lung nerves to understand their function and to capture disease-related changes (neuroplasticity). However, the ability to simultaneously capture large organ-wide and small neurite-sized scales has remained elusive, leading to a gap in our understanding, particularly of sparse or heterogeneous nerve populations. For example, innervation between distal tissue compartments remains understudied because it requires fine individual neurite-scale resolution ranging across the prohibitively large volume of the distal airways. In the current study, we addressed these challenges by developing a method to image whole lung innervation and identify tissue compartments and demonstrated its use in imaging and quantifying sparse heterogeneous nerve structures, including the visceral pleural nerves and human airway ganglia.
Whole Lung Imaging of Visceral Pleural Innervation
A series of studies in the 1920s and 1930s and one study in 2007 (23, 24) detected and studied visceral pleural nerves. These studies were laborious and limited to tissues targeted for section or dissection but provide an important first framework. They would capture visceral pleural nerves typically by flat-mounting pleural tissue after stripping lung or sectioning the lung and quantify pleural surface nerves in cross-section. In various species, the authors found visceral pleural nerves coming from the hilum that were unaffected by vagotomy and expressing various sensory receptors (24). In terms of distribution, these studies found visceral pleural nerves terminating in alveoli and distributed around medial and interlobar regions of the lung.
Our current method using whole lung imaging immediately gave us a comprehensive view of visceral pleural nerves and revealed novel distribution and interconnectivity. We found that pleural innervation was not confined to medial and interlobar spaces but was also abundant in the apical pleura of both lungs and extended along dorsal and ventral surfaces. We also detected new pleural nerve connectivity to vessels and airways, derivation from airways, wrapping around from dorsal to ventral surfaces. Our method also found novel connections between the distal esophagus and inferior lungs via the mediastinal fascia that may relate to the association between airway disease (e.g., asthma and cough) and esophageal disease (e.g., eosinophilic esophagitis, reflux) (25–30). The inability to image the whole lung and esophagus in prior studies likely led to undersampling, which explains differing conclusions about the distribution of visceral pleural nerves seen in these previous studies (23, 24, 31).
Identifying 3D Tissue Compartments and Airway Morphometry
Whole organ imaging and optical tissue clearing has a long history, particularly with xenopus and zebrafish research, and has recently regained popularity in mouse 3D brain and vascular mapping (32, 33). Newer studies using confocal microscopy have created striking large-scale 3D images, but these datasets come with considerable analysis hurdles. A primary hurdle is that tissue compartments have complex 3D structure that would take days to weeks to manually identify. In the current paper, after developing whole lung imaging, we addressed this issue using a novel fluorescent casting strategy and 3D image processing. We could then rapidly analyze distinct nerve populations in different tissue compartments for the first time and combine nerve analysis with airway morphometry. This is important because previous studies showed that identification of tissue compartments and analysis of airway structure (“airway morphometry”) was relevant to respiratory physiology and provided a useful internal frame of reference for describing locations in the lungs (19, 34). In the majority of previous studies, airways were identified by casting using a hardening reagent (e.g., resin, silicone) and analyzed by physical measurement (19). Casting methods could include distal airways and were accurate in that they produced airflow deposition models that matched in vivo measurements (19, 34). Because this method requires lengthy analysis and destroyed lung tissue, more recently airway morphometry has been demonstrated using microCT and MRI. For ease of use and higher resolution, some studies imaged lungs ex vivo that had been casted analogously to previous studies (35). By casting and acquiring high-contrast data, CT/MRI image processing similar to our custom software could semiautomatically identify the large airways (to the sixth generation). Subsequent studies of living mice did not label the airways and thus had to perform extensive manual analysis, including tracing airways slice-by-slice across 700 slices and manually quantifying diameters, length, and branch angles (10). The casting method in the current study, based on binding of labeled streptavidin to endogenous biotin in the airway epithelium, can be used with tissue clearing, whole organ laser-scanning microscopy, and colabeling other epitopes. As we demonstrated with innervation in different tissues (airways, pleura, vasculature, and third versus sixth branch generation), pairing morphometry with another epitope will allow future studies to measure a labeled structure and the 3D anatomic context in which it is located.
Full-Thickness Human Tracheal Innervation and Modeling Tissue Compartments
Large-scale distribution of microscopic nerves and sparse heterogeneous objects can be captured in human tissue with our technique. We obtained the first images of full-thickness trachea innervation and modeled full-thickness tissue compartments with complex 3D morphology. This allowed us to easily quantify a very sparsely and unevenly distributed set of structures—the instrinsic airway ganglia—which in previous studies required laborious tissue sampling (22, 36–38). We also pushed the limit of our technique by quantifying a rarer subset (15% total ganglia) that expresses substance P, which is implicated in lung disease and airway hyperresponsiveness (38, 39). This low sampling probability of substance P may explain contradictory findings of the presence of substance P in human intrinsic ganglia (36, 37). Our initial data about all intrinsic ganglia agrees with previous studies reporting that deeper (adventitial) ganglia contained more cell bodies than shallower (submucosal) ganglia (22). In terms of substance P, the images from the current study of human tracheas clearly demonstrate the presence of ganglionic substance P, which we could detect from our full-thickness images in a matter of minutes. Overall, we calculated a prevalence of substance P neurons in intrinsic human ganglia of 12.7%, which is similar to previous results in ferrets (38). We did not find substance P segregation based on tissue layer, which differs from previous studies in ferrets but may require more tissue given the high variability between our donors.
Limitations
One limitation of our technique is its invasiveness. An advantage of noninvasive whole-lung imaging (e.g., microCT, MRI) over our technique is the ability to perform noninvasive in vivo and longitudinal type studies in the same mouse (12). MRI has the additional advantage over microCT of not exposing mice to radiation, but MRI has lower ideal resolution in lung studies (9).
Another limitation of our technique is that it uses partially deflated and partially flattened lungs for imaging. Fully inflated lungs would need to be imaged piecemeal to physically fit in our slides, and their increased depth may decrease fluorescent signal from deeper tissues. Although the lungs are partially deflated, there is more than sufficient z-resolution and a full 1 to 2 mm of thickness to pick up the 3D structure throughout the lung, as demonstrated in Video E2 (15–60 s). The dehydration step of tissue clearing has also been shown to cause mild shrinkage of tissue (∼ 1–2%) (40). Like previous studies that have cleared tissue, future studies using this technique can compensate for shrinkage by processing their control group of specimens identically to their experimental group, including dehydration.
An advantage of our technique is that it does not use paraffin embedding, which causes greater tissue shrinkage that is not necessarily consistent between experimental groups (16, 41). We do not expect partial deflation or tissue clearing to greatly influence airway-to-bronchiolar morphometry when measured across the whole lung because our pilot data (Figures 2 and 1E) matched resin casting morphometry (19). Newer-generation clearing reagents are being developed to reduce tissue distortion that should be compatible with our approach (42).
Summary
We demonstrate a method that combines the advantages of organ-wide and cellular/subcellular imaging for an unprecedented view of the whole lung. Our method overcomes significant analysis hurdles and can automatically quantify a labeled structure (nerves in this case) in the context of its 3D tissue compartment. The approach can be readily used to study other sparse and heterogeneous structures, such as rare sites of microchimerism where fetal cells travel into the lung during pregnancy or the earliest micron-sized sites of tumor metastases (43, 44). Whole lung microscopic imaging has the potential to fill the gap between large-scale (e.g., CT, MRI) and small-scale (e.g., fluorescence microscopy) imaging data to provide a comprehensive analysis of cellular/subcellular structures throughout the lung.
Acknowledgments
Acknowledgments
The authors thank the members of the Pacific Northwest Transplant Bank for help procuring human tissue and the OHSU Advanced Light Microscopy Core for help with fluorescence imaging.
Footnotes
This work was supported from National Institutes of Health grants HL113023 (D.B.J.), AR061567 (D.B.J.), ES017592 (A.D.F.), and ES014601 (A.D.F.); by National Heart, Lung and Blood Institute Predoctoral F30 Fellowship 1F30HL106930–01 (G.D.S.); and by National Institutes of Health Training Grant T32HL83808 (G.D.S.).
Originally Published in Press as DOI: 10.1165/rcmb.2013-0284OC on January 28, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Muhlfeld C, Graulich T, Das SK, Krasteva G, Wessels L, Ruppert C, Kummer W, Hofler G.Tracheal innervation and alveolar epithelial type II cell lamellar bodies are reduced in the cancer cachectic mouse Am J Respir Crit Care Med 2012. [accessed 2013 Oct 1]. Available from: http://www.atsjournals.org/doi/abs/10.1164/ajrccm-conference.2012.185.1_MeetingAbstracts.A3552 [Google Scholar]
- 2.O’Connell F, Springall DR, Moradoghli-Haftvani A, Krausz T, Price D, Fuller RW, Polak JM, Pride NB. Abnormal intraepithelial airway nerves in persistent unexplained cough? Am J Respir Crit Care Med. 1995;152:2068–2075. doi: 10.1164/ajrccm.152.6.8520777. [DOI] [PubMed] [Google Scholar]
- 3.Scott GD, Fryer AD. Role of parasympathetic nerves and muscarinic receptors in allergy and asthma. Chem Immunol Allergy. 2012;98:48–69. doi: 10.1159/000336498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ollerenshaw SL, Jarvis D, Sullivan CE, Woolcock AJ. Substance P immunoreactive nerves in airways from asthmatics and nonasthmatics. Eur Respir J. 1991;4:673–682. [PubMed] [Google Scholar]
- 5.Howarth PH, Springall DR, Redington AE, Djukanovic R, Holgate ST, Polak JM. Neuropeptide-containing nerves in endobronchial biopsies from asthmatic and nonasthmatic subjects. Am J Respir Cell Mol Biol. 1995;13:288–296. doi: 10.1165/ajrcmb.13.3.7654385. [DOI] [PubMed] [Google Scholar]
- 6.Goldie RG, Fernandes LB, Rigsby PJ, Pudney CJ, Spalding LJ, O’Connor BJ, Page CP. Airway structure: a role for confocal microscopy? Pulm Pharmacol Ther. 1998;11:349–354. doi: 10.1006/pupt.1999.0161. [DOI] [PubMed] [Google Scholar]
- 7.Dunnill MS. The pathology of asthma, with special reference to changes in the bronchial mucosa. J Clin Pathol. 1960;13:27–33. doi: 10.1136/jcp.13.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lange EE, Altes TA, Patrie JT, Parmar J, Brookeman JR, Mugler JP, III, Platts-Mills TA. The variability of regional airflow obstruction within the lungs of patients with asthma: assessment with hyperpolarized helium-3 magnetic resonance imaging. J Allergy Clin Immunol. 2007;119:1072–1078. doi: 10.1016/j.jaci.2006.12.659. [DOI] [PubMed] [Google Scholar]
- 9.Turnbull DH, Mori S. MRI in mouse developmental biology. NMR Biomed. 2007;20:265–274. doi: 10.1002/nbm.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiesse J, Namati E, Sieren JC, Smith AR, Reinhardt JM, Hoffman EA, McLennan G. Lung structure phenotype variation in inbred mouse strains revealed through in vivo micro-CT imaging. J Appl Physiol (1985) 2010;109:1960–1968. doi: 10.1152/japplphysiol.01322.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavanaugh D, Johnson E, Price RE, Kurie J, Travis EL, Cody DD. In vivo respiratory-gated micro-CT imaging in small-animal oncology models. Mol Imaging. 2004;3:55–62. doi: 10.1162/15353500200403184. [DOI] [PubMed] [Google Scholar]
- 12.Badea CT, Athreya KK, Espinosa G, Clark D, Ghafoori AP, Li Y, Kirsch DG, Johnson GA, Annapragada A, Ghaghada KB. Computed tomography imaging of primary lung cancer in mice using a liposomal-iodinated contrast agent. PLoS ONE. 2012;7:e34496. doi: 10.1371/journal.pone.0034496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasilescu DM, Gao Z, Saha PK, Yin L, Wang G, Haefeli-Bleuer B, Ochs M, Weibel ER, Hoffman EA. Assessment of morphometry of pulmonary acini in mouse lungs by nondestructive imaging using multiscale microcomputed tomography. Proc Natl Acad Sci USA. 2012;109:17105–17110. doi: 10.1073/pnas.1215112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyde DM, Harkema JR, Tyler NK, Plopper CG. Design-based sampling and quantitation of the respiratory airways. Toxicol Pathol. 2006;34:286–295. doi: 10.1080/01926230600713509. [DOI] [PubMed] [Google Scholar]
- 15.Scott GD, Fryer AD, Jacoby DB. Quantifying nerve architecture in murine and human airways using three-dimensional computational mapping. Am J Respir Cell Mol Biol. 2013;48:10–16. doi: 10.1165/rcmb.2012-0290MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mühlfeld C, Papadakis T, Krasteva G, Nyengaard JR, Hahn U, Kummer W. An unbiased stereological method for efficiently quantifying the innervation of the heart and other organs based on total length estimations. J Appl Physiol (1985) 2010;108:1402–1409. doi: 10.1152/japplphysiol.01013.2009. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn C. Biotin stores in rodent lungs: localization to Clara and type II alveolar cells. Exp Lung Res. 1988;14:527–536. doi: 10.3109/01902148809087825. [DOI] [PubMed] [Google Scholar]
- 18.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madl P, Hofmann W, Oldham MJ, Asgharian B. Stochastic morphometric model of the BALB/c mouse lung. Anat Rec (Hoboken) 2010;293:1766–1775. doi: 10.1002/ar.21208. [DOI] [PubMed] [Google Scholar]
- 20.Larson SD, Schelegle ES, Hyde DM, Plopper CG. The three-dimensional distribution of nerves along the entire intrapulmonary airway tree of the adult rat and the anatomical relationship between nerves and neuroepithelial bodies. Am J Respir Cell Mol Biol. 2003;28:592–599. doi: 10.1165/rcmb.4889. [DOI] [PubMed] [Google Scholar]
- 21.Barnes PJ, Chung KF, Page CP. Inflammatory mediators of asthma: an update. Pharmacol Rev. 1998;50:515–596. [PubMed] [Google Scholar]
- 22.Baker DG, McDonald DM, Basbaum CB, Mitchell RA. The architecture of nerves and ganglia of the ferret trachea as revealed by acetylcholinesterase histochemistry. J Comp Neurol. 1986;246:513–526. doi: 10.1002/cne.902460408. [DOI] [PubMed] [Google Scholar]
- 23.Larsell O. The ganglia, plexuses, and nerve-terminations of the mammalian lung and pleura pulmonalis. J Comp Neurol. 1922;35:97–132. [Google Scholar]
- 24.Pintelon I, Brouns I, De Proost I, Van Meir F, Timmermans JP, Adriaensen D. Sensory receptors in the visceral pleura: neurochemical coding and live staining in whole mounts. Am J Respir Cell Mol Biol. 2007;36:541–551. doi: 10.1165/rcmb.2006-0256OC. [DOI] [PubMed] [Google Scholar]
- 25.Blake K, Teague WG. Gastroesophageal reflux disease and childhood asthma. Curr Opin Pulm Med. 2013;19:24–29. doi: 10.1097/MCP.0b013e32835b582b. [DOI] [PubMed] [Google Scholar]
- 26.Carr S, Watson W.Eosinophilic esophagitis. Allergy Asthma Clin Immunol20117SupplS8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harer KN, Enders FT, Lim KG, Alexander JA, Katzka DA. An allergic phenotype and the use of steroid inhalers predict eosinophilic oesophagitis in patients with asthma. Aliment Pharmacol Ther. 2013;37:107–113. doi: 10.1111/apt.12131. [DOI] [PubMed] [Google Scholar]
- 28.Canning BJ. Reflex regulation of airway smooth muscle tone. J Appl Physiol (1985) 2006;101:971–985. doi: 10.1152/japplphysiol.00313.2006. [DOI] [PubMed] [Google Scholar]
- 29.Joos GF, Germonpré PR, Pauwels RA. Neural mechanisms in asthma. Clin Exp Allergy. 2000;30:60–65. doi: 10.1046/j.1365-2222.2000.00100.x. [DOI] [PubMed] [Google Scholar]
- 30.Wu M, Majewski M, Wojtkiewicz J, Vanderwinden JM, Adriaensen D, Timmermans JP. Anatomical and neurochemical features of the extrinsic and intrinsic innervation of the striated muscle in the porcine esophagus: evidence for regional and species differences. Cell Tissue Res. 2003;311:289–297. doi: 10.1007/s00441-002-0696-7. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin AI. Nerves and nerve endings in the visceral pleura of the cat. J Physiol. 1933;80:101–104. doi: 10.1113/jphysiol.1933.sp003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokomizo T, Ng CE, Osato M, Dzierzak E. Three-dimensional imaging of whole midgestation murine embryos shows an intravascular localization for all hematopoietic clusters. Blood. 2011;117:6132–6134. doi: 10.1182/blood-2011-02-334037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldham MJ, Robinson RJ. Predicted tracheobronchial and pulmonary deposition in a murine asthma model. Anat Rec (Hoboken) 2007;290:1309–1314. doi: 10.1002/ar.20593. [DOI] [PubMed] [Google Scholar]
- 35.Chaturvedi A, Lee Z. Three-dimensional segmentation and skeletonization to build an airway tree data structure for small animals. Phys Med Biol. 2005;50:1405–1419. doi: 10.1088/0031-9155/50/7/005. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu T, Yamamoto M, Shimokata K, Nagura H. Distribution of substance P-immunoreactive and calcitonin gene-related peptide-immunoreactive nerves in normal human lungs. Int Arch Allergy Appl Immunol. 1991;95:23–28. doi: 10.1159/000235449. [DOI] [PubMed] [Google Scholar]
- 37.Laitinen LA, Laitinen A, Panula PA, Partanen M, Tervo K, Tervo T. Immunohistochemical demonstration of substance P in the lower respiratory tract of the rabbit and not of man. Thorax. 1983;38:531–536. doi: 10.1136/thx.38.7.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dey RD, Altemus JB, Rodd A, Mayer B, Said SI, Coburn RF. Neurochemical characterization of intrinsic neurons in ferret tracheal plexus. Am J Respir Cell Mol Biol. 1996;14:207–216. doi: 10.1165/ajrcmb.14.3.8845170. [DOI] [PubMed] [Google Scholar]
- 39.Wu ZX, Barker JS, Batchelor TP, Dey RD. Interleukin (IL)-1 regulates ozone-enhanced tracheal smooth muscle responsiveness by increasing substance P (SP) production in intrinsic airway neurons of ferret. Respir Physiol Neurobiol. 2008;164:300–311. doi: 10.1016/j.resp.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwadare T, Mori H, Ishiguro K, Takeishi M. Dimensional changes of tissues in the course of processing. J Microsc. 1984;136:323–327. doi: 10.1111/j.1365-2818.1984.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 41.Dorph-Petersen KA, Nyengaard JR, Gundersen HJ. Tissue shrinkage and unbiased stereological estimation of particle number and size. J Microsc. 2001;204:232–246. doi: 10.1046/j.1365-2818.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 42.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 43.Pritchard S, Wick HC, Slonim DK, Johnson KL, Bianchi DW. Comprehensive analysis of genes expressed by rare microchimeric fetal cells in the maternal mouse lung. Biol Reprod. 2012;87:42. doi: 10.1095/biolreprod.112.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis. 2009;26:357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]