Abstract
Human microsomal epoxide hydrolase (EPHX1) is active in the metabolism of many potentially carcinogenic or otherwise genotoxic epoxides, such as those derived from the oxidation of polyaromatic hydrocarbons. EPHX1 is polymorphic and encodes allelic variation at least two amino acid positions, Y113H and H139R. In a number of recent molecular epidemiological investigations, EPHX1 polymorphism has been suggested as a susceptibility factor for several human diseases. To better evaluate the functional contribution of EPHX1 genetic polymorphism, we characterized the enzymatic properties associated with each of the respective variant proteins. Enzymatic profiles were evaluated with cis-stilbene oxide (cSO) and benzo[a]pyrene-4,5-epoxide (BaPO), two prototypical substrates for the hydrolase. In one series of experiments, activities of recombinant EPHX1 proteins were analyzed subsequent to their expression using the pFastbac® baculovirus vector in Spodoptera frugiperda-9 (Sf9) insect cells, and purification by column chromatography. In parallel studies, EPHX1 activities were evaluated with human liver microsomes derived from individuals of known EPHX1 genotype. Using the purified protein preparations, rates of cSO and BaPO hydrolysis for the reference protein, Y113/H139, were approximately 2-fold greater than those measured with the other EPHX1 allelic variants. However, when activities were analyzed using human liver microsomal fractions, no major differences were evident in the reaction rates generated among preparations representing the different EPHX1 alleles. Collectively, these results suggest that the structural differences encoded by the Y113H and H139R variant alleles exert only modest impact on EPHX1-specific enzymatic activities in vivo.
Keywords: Epoxide hydrolase, EPHX1, Human, Genetic polymorphism, Metabolism
1. Introduction
Microsomal epoxide hydrolase (EPHX1; EC 3.3.2.3) is a smooth endoplasmic reticulum enzyme that is expressed relatively ubiquitously in most tissues and in many species [1,2]. Enzymatically, EPHX1 typically catalyzes the hydrolysis of epoxides to trans-dihydrodiols, and is responsible for the detoxification of a wide variety of suspected genotoxins [3]. In certain instances, the initial trans-dihydrodiol metabolites are further activated by subsequent P450 catalysis to form highly electrophilic and reactive dihydrodiol-epoxides that, in a stereoselective manner, form covalent adducts with DNA [4]. Thus, EPHX1 is important for its dual functional role in detoxication as well as bioactivation processes.
The gene and corresponding cDNA sequences encoding human EPHX1 have been characterized previously [5,6,2]. The translated EPHX1 protein is the product of a single gene [7,8], although alternatively spliced non-coding regions of exon 1 have been reported [9]. Previously, we established that the human EPHX1 protein is polymorphic, with amino acid substitutions at two positions, Y113H and H139R [5]. These data were confirmed using independent methods by other laboratories [10,11]. More recently, other EPHX1 single nucleotide polymorphisms (SNPs) were identified [12,13]. However, most of these latter SNPs represent polymorphisms either within non-coding regions of the transcriptional unit, or are synonomous and therefore do not alter the protein structure of EPHX1. A total of eight non-synonomous SNPs for EPHX1 are currently listed in the NCI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP), including the Y113H and H139R polymorphisms. The remaining six non-synonomous dbSNPs either have not yet been validated or were identified to date only within a single heterozygote individual. Thus, it appears likely that the Y113H and H139R SNPs remain as the most common human EPHX1 amino acid variants in the human population.
In addition to EPHX1, a large number of genetic polymorphisms have been cataloged for other bio-transformation enzymes. These include variants of both phase I and phase II metabolism pathways such as the cytochrome P450s and glutathione transferases, respectively [14]. The functional impact of these polymorphisms with respect to xenobiotic metabolism and associated toxicity can be quite variable. For example, CYP2D6 is highly polymorphic, with over 50 variant alleles identified in human populations. Individual CYP2D6 alleles result either the absence of functional changes or in any of a constellation of functional alterations that include amino acid changes, splicing defects, premature termination of translation, and frameshifts [15]. As a result, CYP2D6 enzymatic activity and idiosyncratic reactions to pharmacological substrates of CYP2D6 can vary greatly depending on genotype [16,15]. In addition to their impact in pharmacology, interindividual differences in cancer susceptibility also have been associated with genetic polymorphism within the biotransformation process [17,18].
Since the identification of EPHX1 polymorphisms, a large number of epidemiologic investigations have been conducted examining the association of cancer incidence and other disease endpoints with EPHX1 genotype. For example, McGlynn et al. [19] were among the first to report an apparent association between the incidence of hepatocellular carcinoma (HCC) and the EPHX1 H113 allele in a Chinese population. These results were intriguing, especially in light of the reported role of human EPHX1 in aflatoxin B1 metabolism [20], an important risk factor in the development of HCC. Although a similar association with EPHX1 H113 and HCC was reported separately [21], subsequent epidemiological studies examining this relationship further were unable to detect an association between EPHX1 genotype status and the frequency of HCC disease [22,23]. These include results from a more recent investigation by McGlynn et al. [24]. EPHX1 polymorphisms also have been studied with respect to several other disease endpoints and have been variously associated with colorectal polyp formation [25], lung cancer [26,27,28], orolaryngeal cancer [29], and sensitivity to 1,3-buadiene [30]. A review of selected molecular epidemiological investigations examining associations of EPHX1 genotype with cancer susceptibility has been published [17]. Elucidating the potential contribution of EPHX1 genotype as a risk factor in human disease, either alone or with combined interactions with other polymorphic loci, remains an important and active area of research investigation.
Although the existence of EPHX1 genetic polymorphism is firmly established, the relative enzymatic contribution of the commonly studied EPHX1 Y113H and H139R polymorphisms has been examined only to a limited extent. With the initial discovery of the respective EPHX1 SNPs, the resulting four allelic variants were evaluated for relative functional activities using benzo[a]pyrene-4,5-epoxide (BaPO) as substrate [5]. When normalized to inherent levels of immunoreactive EPHX1 protein, it was concluded that only minimal differences in enzymatic specific activities were apparent among the variants [5]. Another investigation similarly examined the enzymatic capacity of the respective EPHX1 variants but also failed to discern a correlation between EPHX1 polymorphism and enzymatic activity [31].
Given the potential important role that EPHX1 contributes to the chemical biotransformation process, and the suggested association of genetically encoded differences in EPHX1 protein structure with the incidence of certain diseases, it is important to more rigorously characterize the functional impact of established EPHX1 polymorphisms. In the present investigation, we re-evaluated the metabolic capability of human EPHX1 allelic variants with two well-characterized chemical substrates, cis-stilbene oxide (cSO) and benzo[a]pyrene-4,5-oxide. In these studies, we used purified EPHX1 allelic proteins from baculovirus-infected Spodoptera frugiperda-9 (Sf9) cells, a system not previously used for functional analysis of polymorphic EPHX1 variants, as well as microsomal preparations derived from high quality human livers of known EPHX1 genotype.
2. Materials and methods
2.1. Materials
The Bac-to-Bac™ Baculovirus Expression System and other supplies used in the construction of the bacmid, recombinant baculovirus, and Sf9 cells were obtained from Invitrogen Life Technologies, Carlsbad, CA. All reagents used in the purification of proteins were obtained from J.T. Baker, Phillipsburg, NJ. Protein purification columns were obtained from Amersham Pharmacia Biotech, Piscataway, NJ. [3H] cSO was a generous gift from Dr. Bruce Hammock, University of California, Davis. (+/−)-Benzo[a]pyrene-4,5-oxide was obtained from Midwest Research Institute, Kansas City, MO. (+/−)-Benzo[a]pyrene-4,5-dihydrodiol (BPDD) was a gift from Dr. Mont Juchau, University of Washington, Seattle. BaPO and BPDD were used in accordance with recommendations outlined in the National Cancer Institute handling guidelines and with approval from our institutional biosafety committee. Isooctane was obtained from Sigma Chemical Co., St. Louis, MO; HPLC grade methanol was obtained from Burdick and Jackson, Muskegon, MI. Access to human liver bank specimens was kindly provided by Dr. Kenneth Thummel, University of Washington.
2.2. Construction of recombinant bacmid and recombinant baculovirus
The coding region of the four EPHX1 alleles, previously cloned in the pSG5 expression vector [5], were amplified using PCR and primers that created artificial restriction sites (NotI FP, 5′-GAAGCGGCCGCAGTGCTTCTCCCTGTGCTG-3′; PstI RP, 5′-AGCCTGCAGGGCACTTGTGGGGGGAGGTGG-3′). The resulting fragments were gel purified, restricted and subcloned into the multiple cloning site of the pFastbac™ vector. Competent DH5α cells were transformed, plasmids were isolated, and the DNA sequence of each insert was confirmed by cycle sequencing with an ABI Model 377 sequencer (Applied Biosystems, Inc., Foster City, CA). Competent DH10Bac™ cells were transformed with the recombinant pFastbac plasmids, and bacmids were isolated following recombination of the insert into the bacmid DNA. Sf9 cells were transfected with the bacmid DNAs, and recombinant baculovirus stocks for each EPHX1 allele were obtained from these cultures.
2.3. Purification of microsomal epoxide hydrolase
Baculovirus stocks were used to infect Sf9 cells. Infection conditions were optimized for the viral titer, the multiplicity of infection, and the time duration for EPHX1 expression. The infected cells were grown for 5–6 days in 250 mL suspension cultures containing Grace’s Insect Medium supplemented with 10% fetal calf serum. At the end of the culture period cells were harvested, washed with phosphate-buffered saline, and the cell pellet was frozen until purification in 10 mM potassium phosphate buffer (pH 7.4) containing 1% Genapol C-100. The method of Lacourciere et al. [32] was used to purify EPHX1 protein from Sf9 cells, except that Tris–HCl buffer was replaced by potassium phosphate buffer. Purified proteins were stored in 10 mM potassium phosphate buffer (pH 7.4) at −80 °C. Protein fractions obtained during the protein purification procedure were monitored for EPHX1 content by western immunoblot analysis [5] and cSO hydrolysis [33].
2.4. Preparation of human liver microsomes
Microsomes were prepared [34] from livers of six human donors that were previously determined to possess homozygous EPHX1 genotypes [35]. The EPHX1 homozygous genotypes were: Y113/H139 (samples 127, 137), H113/H139 (103, 133), and Y113/R139 (104, 141). Microsomal protein concentrations were determined using the BCA Assay (Pierce, IL). The specific EPHX1 content in the human liver microsomal samples was determined by western immunoblot analysis [5]. Our institutional human subjects review committee approved the studies using human tissues.
2.5. Western immunoblot analysis
Human liver microsomes and purified epoxide hydrolase proteins were characterized initially by Western immunoblotting. Proteins were separated by SDS-PAGE on 10% acrylamide (ReadyGel®, Bio-Rad precast gels) and blotted onto PVDF membranes (Immobilon-P, Millipore Corp., Bedford, MA). The antibodies used, and the methods employed have been reported earlier [5]. Band intensities were quantified by scanning densitometry using Millipore BioImage software (Ann Arbor, MI). The intensities of the bands were reported as corrected OD, which refers to the average OD after background subtraction.
2.6. Enzyme assay and kinetics of cis-stilbene oxide hydrolysis
The hydration of cSO by purified EPHX1 enzymes or human liver microsomes was determined by the method of Gill et al. [33]. Briefly, [3H] cis-stilbene oxide was mixed with the enzyme preparation in a final volume of 100 μL containing 10 mM potassium phosphate buffer (pH 7.4) at 37 °C and reactions were conducted for 3 min. Initially, pooled human liver microsomal proteins (pooled from three livers) were used as the enzyme source to determine rate constants for the human enzyme, and to determine a suitable substrate concentration for further experiments with cSO. Typically, 0.25 μg of purified enzymes, or 5 μg of human liver microsomes, and cSO (56 μM) were used in the incubation mixture. Reactions were terminated by addition of isooctane (200 μL). The solution was mixed for 20 s and an aliquot (25 μL) of the aqueous phase was analyzed by liquid scintillation spectroscopy. Incubations without enzyme were performed in phosphate buffer to estimate non-enzymatic hydrolysis. Product formation was determined as a fraction of the total number of counts present in the incubation mixture.
2.7. Enzyme assay of benzo[a]pyrene-4,5-oxide hydrolysis
Hydration of BaPO was determined by the method of Omiecinski et al. [36], with modifications. Briefly, BaPO (0.66 or 240 μM) was incubated with the enzyme source, followed by HPLC product separation and fluorescence detection. Incubations, with purified enzyme or human liver microsomes, were carried out in a final volume of 150 μL in 10 mM potassium phosphate buffer (pH 7.4) at 37 °C for 15 min. Typically, 0.25–0.5 μg of purified enzymes, or 5 μg of human liver microsomes, were used in the incubation mixtures. The reactions were terminated by addition of 10 μL of perchloric acid, centrifuged, and 5–10 μL of acidified supernatant was injected directly onto the C18 HPLC column. Quantification was achieved by comparison of peak areas for enzymatically generated BPDD to a standard curve produced by spiking acidified incubation buffers with 0–5 nmoles of synthetic BPDD. All other procedures, involving HPLC separation, and fluorescence detection were similar to that reported previously [36].
2.8. Data analysis
Data were analyzed with Microsoft Excel software V 5.1 (Microsoft Corp., Redmond, WA) and the kinetic constants were calculated using K.cat software V 1.3.1 (BioMetallics Inc., Princeton, NJ). Curve fitting was performed assuming constant absolute error, and kinetic constants were calculated after analysis of data with velocity versus substrate concentration plots, Eadie-Hofstee plots, and Lineweaver-Burke plots. Statistical analysis of rate data was performed with Prism® v3.0 (Graphpad Software Inc., San Diego, CA). The rate data were tested for differences by performing a one-way ANOVA, with Tukey’s multiple comparison test. P values <0.05 were accepted as statistically significant.
3. Results
3.1. Purification of microsomal epoxide hydrolase proteins
The four allelic variants of the EPHX1 protein were expressed using baculovirus infection of Sf9 insect cells. Membrane fractions of the respective cell preparations were subjected to protein purification schemes as described in Section 2. Overall, the yield of EPHX1 from the infected cells was on the order of 0.5–2.0% of the total cellular protein. The integrity and specific EPHX1 content of the proteins purified to near homogeneity was assessed by immunoblotting analyses. Fig. 1 presents results from an immunoblot analysis of the respective purified proteins. A single immunoreactive band, corresponding to the expected molecular mass for EPHX1 (~49 kDa), was observed for each of the EPHX1 protein variants.
Fig. 1.
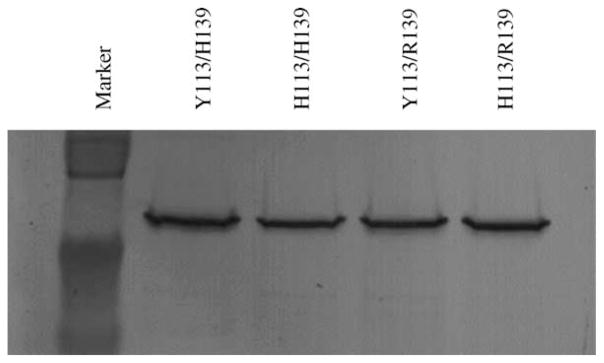
Western immunoblot of purified, baculovirus-derived EPHX1 proteins. Equal amounts (0.125 μg) of the EPHX1 allelic variants were loaded in the lanes and equivalence in levels among the purified proteins was verified by densitometry determination of band intensities.
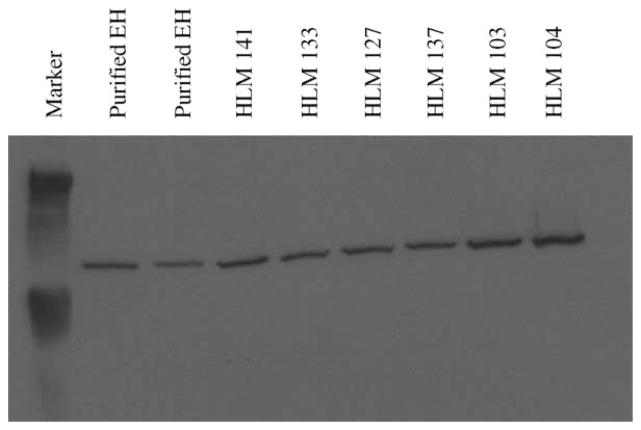
Microsomal samples prepared from human livers that had been stored at −80° also were assessed for EPHX1 activity. The results of Western immunoblot analysis conducted with these samples (5 μg protein/lane) are presented in Fig. 2. Computer densitometry was performed to determine the specific content of EPHX1 in each microsomal or purified protein preparation and the corresponding values were used to normalize subsequent reaction velocity calculations.
Fig. 2.
Western immunoblot analyses performed with human liver microsomes. Purified EPHX1 proteins (5 and 10 ng) were used as positive controls. Five micrograms of protein sample was loaded in each lane. Band intensities were determined by densitometry and the values generated were used to normalize the rate data with human liver microsomes (Table 2).
3.2. Enzymatic activity analyses
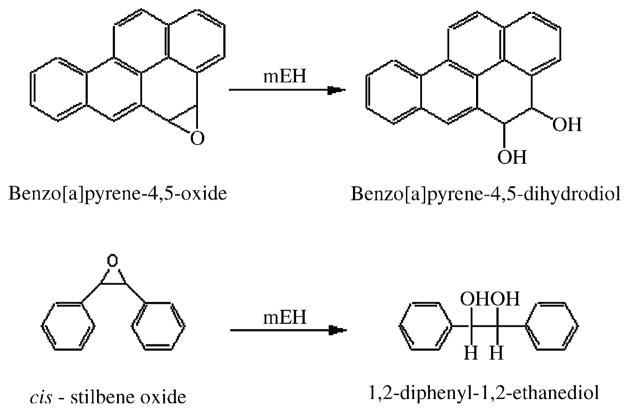
Metabolic activities were assessed using the prototypical EPHX1 substrates, BaPO and cSO. As depicted Fig. 3, BaPO is selectively metabolized to BPDD by EPHX1 [37], while cSO is selectively metabolized by EPHX1 to 1,2-diphenyl-1,2-ethanediol [33].
Fig. 3.
Metabolism of BaPO and cSO by microsomal epoxide hydrolase.
3.3. Hydrolysis of cis-stilbene oxide
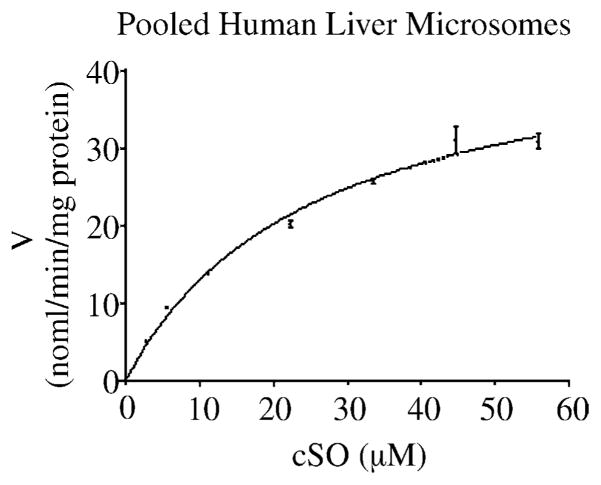
Hydrolysis of cSO was first evaluated using a pooled microsomal sample from three human livers. The microsomal sample catalyzed cSO hydrolysis with characteristic Michaelis–Menten kinetics, and average Km and Vmax values of 25 μM and 52 nmol/min/mg protein, respectively. These results are presented in Fig. 4.
Fig. 4.
V vs. S plot for pooled human liver microsomal proteins with cSO as substrate. Microsomes from three human livers were pooled and the reaction velocities were determined. All velocities were initial velocities. The data points represent mean values and the error bars represent standard deviations of the mean. The line depicts non-linear regression of the data points. The mean Km and Vmax values were 25 μM and 52 nmol/min/mg protein, respectively.
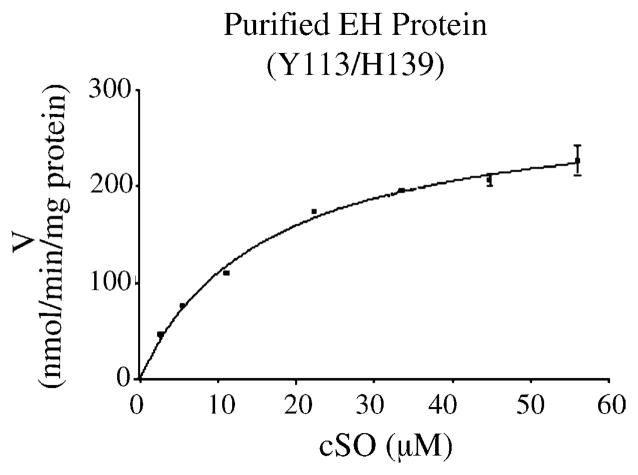
Using a saturating concentration of cSO substrate (56 μM), reaction velocities for the hydrolysis of cSO were determined for the purified EPHX1 protein preparations, and in liver microsomes obtained from individuals homozygous for three of the four known protein variants. To illustrate these results, Fig. 5 presents graphically the data obtained with the Y113/H139 reference protein. Tables 1 and 2 present the detailed results derived from the reaction rate studies. The reference enzyme preparations generated reaction rates that were greater than those obtained from the other allelic variants (Table 1). Although less than 2-fold, the differences were statistically significant (P < 0.01), as determined by ANOVA analysis. A similar trend in wild type versus variant EPHX1 activities also was obtained with the human liver microsomes (Table 2), although the differences in the latter case were not statistically significant (P > 0.05). Per unit of protein, the purified enzyme preparations yielded reaction rates that were approximately 4–5-fold higher than those obtained using the liver microsomal fractions.
Fig. 5.
V vs. S plot illustrating results obtained with cSO as substrate and a purified sample of Y113/H139 EPHX1 reference protein. See Tables 1 and 2 for detailed reaction rate analyses.
Table 1.
Metabolism of EPHX1 substrates by purified epoxide hydrolase
| Protein | cSO (nmol/min/mg protein) | BaPO (0.66 μM) (nmol/min/mg protein) | BaPO (240 μM) (nmol/min/mg protein) |
|---|---|---|---|
| Y/H | 286 ± 10a | 8.2 ± 1.2a | 57 ± 3.8 |
| H/H | 158 ± 13.5 | 4.4 ± 0.7 | 45 ± 7 |
| Y/R | 178 ± 18.3 | 5.2 ± 0.6 | 50 ± 5.3 |
| H/R | 146 ± 9.8 | 5.1 ± 0.9 | 37 ± 2.2a |
All values are mean ± S.D. or reaction rates (n = 3).
Significantly different from other allelic variants (P <0.01).
Table 2.
Metabolism of EPHX1 substrates by human liver microsomes
| Protein | Allele | cSO (nmol/min/mg protein) | BaPO (0.66 μM) (nmol/min/mg protein) | BaPO (240 μM) (nmol/min/mg protein) |
|---|---|---|---|---|
| HLM 127 | Y/H | 67.7 ± 7.9 | 1.00 ± 0.04 | 9.24 ± 0.59 |
| HLM 137 | Y/H | 52.5 ± 3.3 | 0.89 ± 0.08 | 8.19 ± 0.36 |
| HLM 133 | H/H | 34.4 ± 3.2 | 0.98 ± 0.006 | 7.68 ± 0.7 |
| HLM 103 | H/H | 33.6 ± 1.3 | 0.88 ± 0.04 | 8.57 ± 0.67 |
| HLM 141 | Y/R | 41.5 ± 2.8 | 0.82 ± 0.04 | 8.14 ± 1.39 |
| HLM 104 | Y/R | 39.1 ± 1 | 0.77 ± 0.06 | 9.53 ± 0.67 |
All values are mean ± S.D. or reaction rates (n = 3); HLM: human liver microsomes.
3.4. Hydrolysis of benzo[a]pyrene-4,5-oxide
The hydrolysis of BaPO to BPDD was similarly evaluated with the purified EPHX1 proteins. The reaction rates were measured at two BaPO substrate concentrations, 0.66 and 240 μM, levels of substrate used in previous investigations and representative of limiting and saturating substrate concentrations, respectively [35]. These results are presented in Table 1. At the lower substrate concentration the reference allele exhibited a significantly higher (2-fold) reaction rate compared to the EPHX1 protein variants (P < 0.01). However, relative to each other, the mEH variants themselves demonstrated no apparent differences in reaction rates, as determined by a one-way ANOVA (P > 0.05). The higher substrate concentration enabled approximately 7–10-fold increases in reaction rates compared to the low substrate concentration. The reaction rates noted for the H/R allele in particular were significantly lower (P < 0.01) than the other three EPHX1 allelic variants, although these differences were less than 2-fold in magnitude. The activities of remaining EPHX1 variants did not differ significantly (P > 0.05) from reference protein at the saturating level of BaPO substrate.
Human liver microsomes, homozygous for three of the four allelic EPHX1 variants, were also tested with BaPO using the same two different substrate concentrations (Table 2). In the human liver microsomes studies, rates of BaPO hydrolysis were normalized to EPHX1 content in the microsomal samples, as determined by densitometric analysis (Fig. 2). Analysis of the reaction rates at 0.66 μM by one-way ANOVA indicated no statistically significant difference (P > 0.05) among the various microsome sample preparations. Although analysis of the reaction rates at 240 μM by a one-way ANOVA revealed an apparent difference in the reaction rates (P < 0.05) among the six microsomal samples, when analyses were conducted using a paired comparison test, no significant difference among the microsomal samples (P > 0.05) was detected. Similar to the results obtained with the purified Sf9-derived preparations, 9–12-fold increases in reaction rates were observed in the microsomal samples subsequent to increasing the BaPO substrate concentration from 0.66 to 240 μM. The purified EPHX1 preparations demonstrated an approximately 4–6-fold increase in BaPO hydrolysis rates relative to the human liver microsome samples. However, the overall trends exhibited in measured BaPO hydrolysis rates between the two systems were similar.
4. Discussion
Microsomal epoxide hydrolase plays a pivotal role in the generation of bay-region diol-epoxides of the carcinogenic polyaromatic hydrocarbons, such as benzo[a]pyrene [38]. The critical nature of EPHX1 bioactivation in polyaromatic hydrocarbon-induced carcinogenesis was demonstrated in EPHX1 null mice, which were completely resistant to the tumorigenic effects of dimethylbenz[a]anthracene in a complete carcinogenesis assay [39]. However, EPHX1 plays a protective role in other chemically mediated toxicities. For example, expression of EPHX1 protects against styrene 7,8-oxide induced genotoxicity in mammalian cells [40].
In the current investigation we sought to more definitively assess the functional relationships of EPHX1 allelic status. In part, we exploited baculovirus infected insect cells engineered to over produce the respective EPHX1 allelic variants. Enzymatic activities were analyzed using EPHX1 enzyme derived either from purified, baculovirus-expressed EPHX1 protein or from human liver microsomes prepared from tissues of known EPHX1 genotype. Two prototypical EPHX1 substrate probes were employed in the analyses, cSO and BaPO. For cSO, the reaction rates for EPHX1-mediated hydrolysis were determined and compared among the different allelic variants. In the case of BaPO, enzymatic activities were measured at two substrate concentrations, representing saturating and near-Km levels of substrate, respectively. By performing enzymatic assays with purified EPHX1 enzymes, variables such as uniformity of expression and potential influences from competing reaction pathways in cell lines were more easily controlled. The assays conducted with human liver microsomal preparations that were homozygous for three of the four EPHX1 allelic protein variants enabled comparison of activity measurements within lipid environments more closely resembling those occurring in vivo (the remaining protein variant, H113/R139, was not assayed because the available liver bank samples did not include any individuals of this genotype).
For cSO, hydrolysis rates (Tables 1 and 2) were obtained using a substrate concentration of 56 μM, subsequent to the determination of enzymatic rate constants obtained with pooled human liver microsome fractions (Fig. 4). Experiments conducted with both purified enzymes and with microsomal preparations indicated a modest (<2-fold) difference in cSO metabolism among the four EPHX1 different allelic variants, with the reference Y113/H139 genotype exhibiting the highest levels of activity. Although the derived cSO rate constants were approximately 4-fold higher than those reported by Kitteringham et al. [31], our results were otherwise consistent with those previous studies in that no strong association was observed between EPHX1 allelic status and corresponding rates of cSO hydrolysis.
The results obtained from the BaPO experiments demonstrated similar trends to those obtained with cSO. Using purified enzymes, relative activities demonstrated for BaPO hydrolysis between the respective EPHX1 allelic variants, again with the Y113/H139 allele exhibiting the highest activity (Table 1). In contrast, the results of assays conducted with human liver microsome samples demonstrated no statistically significant difference in BaPO hydrolysis among the tested variants (Table 2). The current results, obtained with BaPO and human liver microsomes, also were consistent with data reported previously from assays using S9 fractions obtained from human livers [35].
A potential source of variability in estimating the in vivo activity of EPHX1 is enzyme stability. The phenomenon of varying stability of polymorphic variants has been documented with other biotransformation proteins, including the soluble form of human epoxide hydrolase, EPHX2 [41], and the N-acetyltransferases [42]. For example, specific amino acid substitutions in N-acetyltransferase, NAT2, have been shown to alter the stability of the resulting protein, thereby influencing the metabolic capacity of the enzyme [42]. The thiopurine S-methyltransferases (TPMT) are also polymorphic, and patients with TPMT deficiency are at a high risk of hematopoietic toxicity [43]. In studies conducted to assess the functional consequences of TPMT polymorphism, lower TPMT content in some patients is the consequence of enhanced degradation of certain allelic variants of the enzyme [44]. Analyses of translational rates and protein half-lives for the respective EPHX1 protein variants has been investigated previously in our laboratory using transient transfection techniques in COS-1 cells [45]. Although not statistically significant, the H113/H139 EPHX1 variant tended to exhibit a shorter protein half-life than the remaining variants. The results from the studies reported here suggest only modest differences in the measured rates of specific activity among the respective EPHX1 variants, however, inherent allelic differences in protein stability also may lead to altered rates of clearance of epoxide species in vivo. The relationship between EPHX1 protein stability and epoxide metabolism in vivo remains unclear, although it is likely that potential differences in enzyme stability, together with polymorphisms in the 5′-flanking region affecting transcriptional rates [46], may combine to modify metabolism and clearance of epoxides. In addition, results from several studies suggest that interactions between specific polymorphic loci, such as between EHPX1 and the cytochrome P450s [28], and/or between EPHX1 and the glutathione-S-epoxide transferases [29], will likely determine unique genotypes that are most highly susceptible to chemically-initiated diseases. Further studies conducted on large population samples will required to thoroughly address these gene–gene interaction determinants [18].
Given the broad substrate specificity inherent in EPHX1, it is intriguing to ask the additional question as to whether the model EPHX1 substrates used here, i.e., cSO and BaPO, serve as adequate surrogates for predicting EPHX1 activities among the large array of environmental epoxides that may otherwise be enzymatic substrates. An early study compared EPHX1 reaction rates among 11 substrates using microsome preparations obtained from nine different human livers [47]. The substrates included several polyaromatic hydrocarbon epoxides, together with styrene 7,8-oxide and octane 1,2-oxide. Across the entire panel of nine livers, the results demonstrated high correlations (r = 0.87 to r = 0.99) of EPHX1 activity for each substrate when compared against EPHX1 activity for each of the other substrates [47], indicating that enzymatic activities obtained with any one are predictive for others. However, in a more recent study, EPHX1 hydrolytic activities were evaluated across three substrates: cis-stilbene oxide, carbamazepine-10,11-epoxide, and naphthalene oxide, within a panel of microsome samples obtained from 15 human livers [31]. These investigators provided evidence supporting the idea of substrate-specific variation for EPHX1. Therefore, although the data presented in the current investigation suggest no major specific activity alterations associated with the 113/139 EPHX1 allelic substitutions; it remains possible that individual EPHX1 variants may possess unique substrate preferences and associated activity differences.
Furthermore, other provocative biological roles for EPHX1 have been proposed, including function as a bile acid transporter [48], participation in vitamin K1 oxide reductase activity [49], and a role in endogenous metabolism of steroid epoxides [50]. It remains to be determined whether EPHX1 polymorphism may functionally influence these processes.
Acknowledgments
The authors are grateful to Dr. Kenneth Thummel for providing human liver tissues and to Dr. Bruce Hammock for radiolabeled substrates. This work was supported by funding from the NIH; ES04978 (C.J.O.), ES07033, and GM32165 (A.E.R.).
Abbreviations
- BaPO
benzo[a]pyrene-4,5-epoxide
- BPDD
benzo[a]pyrene-4,5-dihydrodiol
- cSO
cis-stilbene oxide
- EPHX1
microsomal epoxide hydrolase
- HCC
hepatocellular carcinoma
- HLM
human liver microsomes
- Sf9
Spodoptera frugiperda-9
- TPMT
thiopurine S-methyl transferases
References
- 1.Coller JK, Fritz P, Zanger UM, Siegle I, Eichelbaum M, Kroemer HK, Murdter TE. Distribution of microsomal epoxide hydrolase in humans: an immunohistochemical study in normal tissues, and benign and malignant tumours. Histochem J. 2001;33:329–336. doi: 10.1023/a:1012414806166. [DOI] [PubMed] [Google Scholar]
- 2.Beetham JK, Grant D, Arand M, Garbarino J, Kiyosue T, Pinot F, Oesch F, Belknap WR, Shinozaki K, Hammock BD. Gene evolution of epoxide hydrolases and recommended nomenclature. DNA Cell Biol. 1995;14:61–71. doi: 10.1089/dna.1995.14.61. [DOI] [PubMed] [Google Scholar]
- 3.Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 4.Shou M, Gonzalez FJ, Gelboin HV. Stereoselective epoxidation and hydration at the K-region of polycyclic aromatic hydrocarbons by cDNA-expressed cytochromes P4501A1, 1A2, and epoxide hydrolase. Biochemistry. 1996;35:15807–15813. doi: 10.1021/bi962042z. [DOI] [PubMed] [Google Scholar]
- 5.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994;3:421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassett C, Robinson KB, Beck NB, Omiecinski CJ. The human microsomal epoxide hydrolase gene (EPHX1): complete nucleotide sequence and structural characterization. Genomics. 1994;23:433–442. doi: 10.1006/geno.1994.1520. [DOI] [PubMed] [Google Scholar]
- 7.Hartsfield JK, Jr, Sutcliffe MJ, Everett ET, Hassett C, Omiecinski CJ, Saari JA. Assignment1 of microsomal epoxide hydrolase (EPHX1) to human chromosome 1q42.1 by in situ hybridization. Cytogenet Cell Genet. 1998;83:44–45. doi: 10.1159/000015164. [DOI] [PubMed] [Google Scholar]
- 8.Skoda RC, Demierre A, McBride OW, Gonzalez FJ, Meyer UA. Human microsomal xenobiotic epoxide hydrolase. Complementary DNA sequence, complementary DNA-directed expression in COS-1 cells, and chromosomal localization. J Biol Chem. 1988;263:1549–1554. [PubMed] [Google Scholar]
- 9.Gaedigk A, Leeder JS, Grant DM. Tissue-specific expression and alternative splicing of human microsomal epoxide hydrolase. DNA Cell Biol. 1997;16:1257–1266. doi: 10.1089/dna.1997.16.1257. [DOI] [PubMed] [Google Scholar]
- 10.Gaedigk A, Spielberg SP, Grant DM. Characterization of the microsomal epoxide hydrolase gene in patients with anticonvulsant adverse drug reactions. Pharmacogenetics. 1994;4:142–153. doi: 10.1097/00008571-199406000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Green VJ, Pirmohamed M, Kitteringham NR, Gaedigk A, Grant DM, Boxer M, Burchell B, Park BK. Genetic analysis of microsomal epoxide hydrolase in patients with carbamazepine hypersensitivity. Biochem Pharmacol. 1995;50:1353–1359. doi: 10.1016/0006-2952(95)02009-8. [DOI] [PubMed] [Google Scholar]
- 12.Saito S, Iida A, Sekine A, Eguchi C, Miura Y, Nakamura Y. Seventy genetic variations in human microsomal and soluble epoxide hydrolase genes (EPHX1 and EPHX2) in the Japanese population. J Hum Genet. 2001;46:325–329. doi: 10.1007/s100380170067. [DOI] [PubMed] [Google Scholar]
- 13.Maekawa K, Itoda M, Hanioka N, Saito Y, Murayama N, Nakajima O, Soyama A, Ishida S, Ozawa S, Ando M, Sawada J. Non-synonymous single nucleotide alterations in the microsomal epoxide hydrolase gene and their functional effects. Xenobiotica (Lond) 2003;33:277–287. doi: 10.1080/0049825021000061615. [DOI] [PubMed] [Google Scholar]
- 14.Wormhoudt LW, Commandeur JN, Vermeulen NP. Genetic polymorphisms of human N-acetyltransferase, cytochrome P450, glutathione-S-transferase, and epoxide hydrolase enzymes: relevance to xenobiotic metabolism and toxicity. Crit Rev Toxicol. 1999;29:59–124. doi: 10.1080/10408449991349186. [DOI] [PubMed] [Google Scholar]
- 15.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P4502D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 16.Cascorbi I. Pharmacogenetics of cytochrome P4502D6: genetic background and clinical implication. Eur J Clin Invest. 2003;33(Suppl 2):17–22. doi: 10.1046/j.1365-2362.33.s2.3.x. [DOI] [PubMed] [Google Scholar]
- 17.Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione-S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]
- 18.Yoshimura K, Hanaoka T, Ohnami S, Kohno T, Liu Y, Yoshida T, Sakamoto H, Tsugane S. Allele frequencies of single nucleotide polymorphisms (SNPs) in 40 candidate genes for gene-environment studies on cancer: data from population-based Japanese random samples. J Hum Genet. 2003;48:654–658. doi: 10.1007/s10038-003-0096-1. [DOI] [PubMed] [Google Scholar]
- 19.McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoebonnie A, Oforiadjei D, Chen GC, London WT, Shen FM, Buetow KH. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci USA. 1995;92:2384–2387. doi: 10.1073/pnas.92.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly EJ, Erickson KE, Sengstag C, Eaton DL. Expression of human microsomal epoxide hydrolase in saccharomyces cerevisiae reveals a functional role in aflatoxin B(1) detoxification. Toxicol Sci. 2002;65:35–42. doi: 10.1093/toxsci/65.1.35. [DOI] [PubMed] [Google Scholar]
- 21.Tiemersma EW, Omer RE, Bunschoten A, van’t Veer P, Kok FJ, Idris MO, Kadaru AM, Fedail SS, Kampman E. Role of genetic polymorphism of glutathione-S-transferase T1 and microsomal epoxide hydrolase in aflatoxin-associated hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prevent. 2001;10:785–791. [PubMed] [Google Scholar]
- 22.Wild CP, Yin F, Turner PC, Chemin I, Chapot B, Mendy M, Whittle H, Kirk GD, Hall AJ. Environmental and genetic determinants of aflatoxin-albumin adducts in the Gambia. Int J Cancer. 2000;86:1–7. doi: 10.1002/(sici)1097-0215(20000401)86:1<1::aid-ijc1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Wong NA, Rae F, Bathgate A, Smith CA, Harrison DJ. Polymorphisms of the gene for microsomal epoxide hydrolase and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a Caucasian population. Toxicol Lett. 2000;115:17–22. doi: 10.1016/s0378-4274(00)00166-1. [DOI] [PubMed] [Google Scholar]
- 24.McGlynn KA, Hunter K, LeVoyer T, Roush J, Wise P, Michielli RA, Shen FM, Evans AA, London WT, Buetow KH. Susceptibility to aflatoxin B1-related primary hepatocellular carcinoma in mice and humans. Cancer Res. 2003;63:4594–4594. [PubMed] [Google Scholar]
- 25.Ulrich CM, Bigler J, Whitton JA, Bostick R, Fosdick L, Potter JD. Epoxide hydrolase Tyr113His polymorphism is associated with elevated risk of colorectal polyps in the presence of smoking and high meat intake. Cancer Epidemiol Biomarkers Prevent. 2001;10:875–882. [PubMed] [Google Scholar]
- 26.Gsur A, Zidek T, Schnattinger K, Feik E, Haidinger G, Hollaus P, Mohn-Staudner A, Armbruster C, Madersbacher S, Schatzl G, Trieb K, Vutuc C, Micksche M. Association of microsomal epoxide hydrolase polymorphisms and lung cancer risk. Br J Cancer. 2003;89:702–706. doi: 10.1038/sj.bjc.6601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee WJ, Brennan P, Boffetta P, London SJ, Benhamou S, Rannug A, To-Figueras J, Ingelman-Sundberg M, Shields P, Gaspari L, Taioli C. Microsomal epoxide hydrolase polymorphisms and lung cancer risk: a quantitative review. Biomarkers. 2002;7:230–241. doi: 10.1080/13547500210121882. [DOI] [PubMed] [Google Scholar]
- 28.Lin P, Wang SL, Wang HJ, Chen KW, Lee HS, Tsai KJ, Chen CY, Lee H. Association of CYP1A1 and microsomal epoxide hydrolase polymorphisms with lung squamous cell carcinoma. Br J Cancer. 2000;82:852–857. doi: 10.1054/bjoc.1999.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JY, Schantz SP, Lazarus P. Epoxide hydrolase genotype and orolaryngeal cancer risk: interaction with GS™1 genotype. Oral Oncol. 2003;39:483–490. doi: 10.1016/s1368-8375(03)00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Rahman SZ, Ammenheuser MM, Ward JB., Jr Human sensitivity to 1,3-butadiene: role of microsomal epoxide hydrolase polymorphisms. Carcinogenesis. 2001;22:415–423. doi: 10.1093/carcin/22.3.415. [DOI] [PubMed] [Google Scholar]
- 31.Kitteringham NR, Davis C, Howard N, Pirmohamed M, Park BK. Interindividual and interspecies variation in hepatic microsomal epoxide hydrolase activity: studies with cis-stilbene oxide, carbamazepine 10, 11-epoxide and naphthalene. J Pharmacol Exp Ther. 1996;278:1018–1027. [PubMed] [Google Scholar]
- 32.Lacourciere GM, Vakharia VN, Tan CP, Morris DI, Edwards GH, Moos M, Armstrong RN. Interaction of hepatic microsomal epoxide hydrolase derived from a recombinant baculovirus expression system with an azarene oxide and an aziridine substrate analogue. Biochemistry. 1993;32:2610–2616. doi: 10.1021/bi00061a019. [DOI] [PubMed] [Google Scholar]
- 33.Gill SS, Ota K, Hammock BD. Radiometric assays for mammalian epoxide hydrolases and glutathione-S-transferases. Anal Biochem. 1983;131:273–282. doi: 10.1016/0003-2697(83)90166-5. [DOI] [PubMed] [Google Scholar]
- 34.Haugen DA, Coon MJ. properties of electrophoretically homogeneous phenobarbital-inducible and beta-naphthoflavone-inducible forms of liver microsomal cytochrome P-450. J Biol Chem. 1976;251:7929–7939. [PubMed] [Google Scholar]
- 35.Hassett C, Lin J, Carty CL, Laurenzana EM, Omiecinski CJ. Human hepatic microsomal epoxide hydrolase: comparative analysis of polymorphic expression. Arch Biochem Bio-phys. 1997;337:275–283. doi: 10.1006/abbi.1996.9794. [DOI] [PubMed] [Google Scholar]
- 36.Omiecinski CJ, Aicher L, Holubkov R, Checkoway H. Human peripheral lymphocytes as indicators of microsomal epoxide hydrolase activity in liver and lung. Pharmacogenetics. 1993;3:150–158. doi: 10.1097/00008571-199306000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Jerina DM, Dansette PM, Lu AYH, Levin W. Hepatic microsomal epoxide hydrase: a sensitive radiometric assay for hydration of arene oxides of carcinogenic aromatic hydrocarbons. Mol Pharmacol. 1977;13:342–351. [PubMed] [Google Scholar]
- 38.Sims P, Grover PL, Swaisland A, Pal K, Hewer A. Metabolic activation of benzo(a)pyrene proceeds by a diol-epoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- 39.Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. Targeted disruption of the microsomal epoxide hydrolase gene. microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- 40.Herrero ME, Arand M, Hengstler JG, Oesch F. Recombinant expression of human microsomal epoxide hydrolase protects V79 Chinese hamster cells from styrene oxide-but not from ethylene oxide-induced DNA strand breaks. Environ Mol Mutagen. 1997;30:429–439. doi: 10.1002/(sici)1098-2280(1997)30:4<429::aid-em8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 41.Sandberg M, Hassett C, Adman ET, Meijer J, Omiecinski CJ. Identification and functional characterization of human soluble epoxide hydrolase genetic polymorphisms. J Biol Chem. 2000;275:28873–28881. doi: 10.1074/jbc.M001153200. [DOI] [PubMed] [Google Scholar]
- 42.Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular-genetics of human polymorphic N-acetyltransferase - enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric Nat2 allozymes. Hum Mol Genet. 1994;3:729–734. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- 43.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, Pui CH, Evans WE. Mercaptop-urine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 44.Tai HL, Fessing MY, Bonten EJ, Yanishevsky Y, d’Azzo A, Krynetski EY, Evans WE. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics. 1999;9:641–650. doi: 10.1097/01213011-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 45.Laurenzana EM, Hassett C, Omiecinski CJ. Post-transcriptional regulation of human microsomal epoxide hydrolase. Pharmacogenetics. 1998;8:157–167. doi: 10.1097/00008571-199804000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Raaka S, Hassett C, Omiencinski CJ. Human microsomal epoxide hydrolase: 5′-flanking region genetic polymorphisms. Carcinogenesis. 1998;19:387–393. doi: 10.1093/carcin/19.3.387. [DOI] [PubMed] [Google Scholar]
- 47.Kapitulnik J, Levin W, Morecki R, Dansette PM, Jerina DM, Conney AH. Hydration of arene and alkene oxides by epoxide hydrase in human liver microsomes. Clin Pharmacol Ther. 1977;21:158–165. doi: 10.1002/cpt1977212158. [DOI] [PubMed] [Google Scholar]
- 48.von Dippe P, Amoui M, Stellwagen RH, Levy D. The functional expression of sodium-dependent bile acid transport in Madin-Darby canine kidney cells transfected with the cDNA for microsomal epoxide hydrolase. J Biol Chem. 1996;271:18176–18180. doi: 10.1074/jbc.271.30.18176. [DOI] [PubMed] [Google Scholar]
- 49.Guenthner TM, Cai D, Wallin R. Co-purification of microsomal epoxide hydrolase with the warfarin-sensitive vitamin K1 oxide reductase of the vitamin K cycle. Biochem Pharmacol. 1998;55:169–175. doi: 10.1016/s0006-2952(97)00431-0. [DOI] [PubMed] [Google Scholar]
- 50.Fandrich F, Degiuli B, Vogel Bindel U, Arand M, Oesch F. Induction of rat liver microsomal epoxide hydrolase by its endogenous substrate 16 alpha, 17 alpha-epoxyestra-1,3,5-trien-3-ol. Xenobiotica (Lond) 1995;25:239–244. doi: 10.3109/00498259509061848. [DOI] [PubMed] [Google Scholar]