Abstract
Histone acetylation plays important roles in gene regulation. However, the functions of individual histone acetyltransferases (HATs) in specific developmental transcription programs are not well defined. To define the functions of Gcn5, a prototypical HAT, during mouse development, we have created a series of mutant Gcn5 alleles. Our previous work revealed that deletion of Gcn5 leads to embryonic death soon after gastrulation. Embryos homozygous for point mutations in the catalytic center of Gcn5 survive longer, but die soon after E16.0 and exhibit defects in cranial neural tube closure. Embryos bearing a hypomorphic Gcn5flox(neo) allele also exhibit neural closure defects and die at or soon after birth. We report here that Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ embryos exhibit anterior homeotic transformations in lower thoracic and lumbar vertebrae. These defects are accompanied by a shift in the anterior expression boundary of Hoxc8 and Hoxc9. These data provide the first evidence that Gcn5 contributes to Hox gene regulation and is required for normal anteroposterior patterning of the mouse skeleton.
Keywords: acetyltransferase, chromatin, histone, Hox gene
Introduction
Homeotic genes regulate segment identity in developing embryos. In Drosophila, anteroposterior patterning is determined by two clusters of homeotic genes, the Antennapedia complex (AnT-C) and Bithorax complex (Bx-C), which were both derived from an ancestral homeotic cluster, HOM-C (McGinnis & Krumlauf 1992; Duboule & Morata 1994). Mammalian homeotic genes are homologous to AnT-C and are classified into four linkage groups, HoxA, HoxB, HoxC, and HoxD (Holland & Garcia-Fernandez 1996). Hox gene expression along the anteroposterior (AP) axis of embryos correlates with the physical order of these genes within the Hox clusters. Expression of genes within the AnT-C and Bx-C clusters is initiated by transiently expressed maternal and segmentation gene products such as bicoid and hunchback (Ingham 1988). Once initiated, spatial and quantitative patterns of Hox gene expression are maintained by Trithorax group (TrxG) and Polycomb group (PcG) proteins, which are required for Hox gene activation and repression, respectively (Singh 1994; Paro 1995; Simon & Tamkun 2002).
Interestingly, Drosophila Trx, along with its yeast (Set1) and mammalian (hSet1, MLL, and MLL2) homologues, functions as a histone H3 lysine 4 (K4)-specific methyltransferase (Briggs et al. 2001; Beisel et al. 2002; Milne et al. 2002; Klymenko & Muller 2004; Glaser et al. 2006; Klymenko et al. 2006). Conversely, the PcG group protein Ezh2 functions as an H3 K27-specific methyltransferase (Cao et al. 2002; Czermin et al. 2002). Methylation of histones affects gene expression by regulating the binding of transcription factors, silencing proteins, or additional chromatin modifying complexes to gene promoters. In particular, methylation of K9 or K27 in H3 is associated with heterochromatin formation and silencing of chromosomal domains (Nakayama et al. 2001; Peters et al. 2001; Kuzmichev et al. 2002; Plath et al. 2003; Bernstein et al. 2005). H3 K4 methylation is excluded from such domains and instead is enriched in active genes (Strahl et al. 1999; Bernstein et al. 2002; Santos-Rosa et al. 2002; Bernstein et al. 2005). Modulation of histone methylation levels and patterns by TrxG and PcG proteins provides a molecular mechanism for the regulation of Hox genes across large chromosomal loci.
Methylation of H3 at K4 is often tied to increased acetylation of H3 and other histones (Bernstein et al. 2005). In fact, H3 K4 methylation may directly recruit histone acetyltransferase (HAT) complexes. A component of yeast SAGA and SLIK HAT complexes, Chd1, binds to methyl K4 in H3 (Pray-Grant et al. 2005). Mammalian Chd1 also binds specifically to this histone modification (Sims et al. 2005). Moreover, the MLL methyltransferase complex associates with Mof, an H4-specific acetyltransferase (Dou et al. 2005). Given the importance of TrxG proteins in maintaining Hox gene expression, these findings suggest that acetylation will also be important to Hox gene regulation. Indeed, several lines of evidence link histone acetylation to modulation of the spatial expression of Hox genes. During early development, both histone H3 K4 methylation and H3 K9 acetylation mark transcriptionally active and ‘poised’ chromatin regions within the HoxB cluster (Chambeyron & Bickmore 2004). At the Hoxb8 locus, for example, H3 K9 acetylation is much higher in Hoxb8-expressing regions of the embryo than in regions where Hoxb8 is repressed (Fujimura et al. 2006). Also, sequential activation of genes during neurogenesis within the mouse Hoxd4 locus is linked to sequential changes in histone modifications, including methylation of H3 K4 methylation followed by H3 and H4 acetylation (Rastegar et al. 2004). Consistent with this notion, Zebrafish Moz and Drosophila CBP have been reported to maintain the expression of specific Hox genes (Petruk et al. 2001; Miller et al. 2004). However, to date no HAT has been linked to anteroposterior skeletal patterning in mice.
Only a few mouse mutant studies have been carried out to determine the roles of specific HATs during mammalian development (Yao et al. 1998; Xu et al. 2000; Yamauchi et al. 2000; Lin & Dent 2006). Our lab previously reported that deletion of Gcn5, the catalytic subunit of mammalian SAGA-like complexes (e.g. STAGA and TFTC) (Brand et al. 1999; Martinez et al. 2001) causes embryonic lethality by E11.0 (Xu et al. 2000). Gcn5 null embryos have normal morphology until E7.5 but exhibit high levels of apoptosis, which leads to loss of mesodermal lineages. We subsequently found that embryos homozygous for point mutations that destroy the catalytic activity of Gcn5 survive longer but exhibit cranial neural tube defects (Bu et al. 2007). Thus Gcn5 has HAT independent functions required for cell survival and early embryo development, but Gcn5 HAT activity is required for neural tube closure. We recently reported the creation of a conditional Gcn5flox allele, and a hypomorphic Gcn5flox(neo) allele (Lin et al. 2008). Embryos homozyogous for the hypomorphic allele, as well as Gcn5flox(neo)/Δ embryos, survive until E18.5, but many of these mice exhibit exencephaly stemming from neural tube closure defects. We report here that these Gcn5 mutant mice also exhibit rib and vertebral malformations consistent with anterior transformations of specific thoracic and lumbar skeletal segments, defects previously observed in Hoxc8 and Hoxc9 mutant mice (Le Mouellic et al. 1992; Suemori et al. 1995). These studies provide the first evidence that proper expression of Gcn5 is important for spatial regulation of specific Hox genes and for normal skeletal patterning.
Materials and methods
Skeletal preparations
Pups or E18.5 embryos were eviscerated and fixed in 95% ethanol overnight. They were then stained in alcian blue solution (0.015% alcian blue 8GX in a 1 : 4 mixture of acetic acid and 95% ethanol) overnight, soaked in 2% KOH for 24 h, stained in 1% KOH, 0.005% alizarin red overnight, and finally cleared in 1% KOH/20% glycerol for about 2 days (Nagy et al. 2003).
Histology
E18.5 embryos were dissected and rinsed in phosphate-buffered saline (PBS) and fixed in PBS containing 4% paraformaldehyde for 24 h. Embryos were rinsed twice in PBS for 30 min each, dehydrated by serial washes in 70% (v/v) ethanol, 80% ethanol, 95% ethanol and 100% ethanol and then embedded in paraffin by standard methods (Nagy et al. 2003). Serial sections (10 μm) were prepared and stained with hematoxylin and eosin (Nagy et al. 2003).
Whole mount in situ hybridizations
E9.5–E12.5 mouse embryos were dissected and fixed overnight with 4% paraformaldehyde. The yolk sac was used to prepare genomic DNA for genotyping. Whole mount RNA in situ hybridization was carried out as described (Nagy et al. 2003). Hoxc8 and Hoxc9 probes were kindly provided by Dr Armin Schumacher.
Staging of mouse embryos
Mouse embryos were staged according to the number of somites. Mouse embryos with 35–39 somites are approximately E10.5. Mouse embryos with 45–47 are approximately E11.5. The embryos harvested at 15 days postcoitum were approximately E15.5. The embryos harvested at 18 days postcoitum were approximately E18.5.
Results
Anteroposterior skeletal transformations in Gcn5flox(neo)/Δ and Gcn5flox(neo)/flox(neo) mutants
Our previous studies demonstrated that deletion of Gcn5 in the mouse causes loss of mesodermal lineages, including paraxial mesoderm and chordamesoderm (Xu et al. 2000). However, paraxial mesoderm and somites formed in Gcn5flox(neo)/Δ and Gcn5flox(neo)/flox(neo) embryos, where Gcn5 expression is reduced but is still detectable (Lin et al. 2008). In wild-type animals, somites further differentiate into the sclerotome, myotome and dermatome, which in turn give rise to specific organs such as skeleton, skeletal muscles, and skin, respectively. No obvious defects in muscle or skin development were observed in Gcn5flox(neo)/Δ or Gcn5flox(neo)/flox(neo) embryos (Lin et al. 2008), but Gcn5 mutants exhibited several abnormalities in skeletal shape and patterning.
Analyses of the vertebral columns of Gcn5flox(neo)/Δ or Gcn5flox(neo)/flox(neo) embryos at E18.5 and pups at P0 indicated that specific lumbar segments of the skeleton were transformed into more anterior thoracic segments (Fig. 1 and Table 1). In most (12 out of 15) Gcn5flox(neo)/+ embryos, the presacral vertebrae included the classic wild-type pattern of seven cervical, 13 thoracic and six lumbar vertebrae (C7/T13/L6) (Fig. 1A,B). Some (3 of 15) Gcn5flox(neo)/+ embryos exhibited a reduced number of lumbar vertebrae (C7/T13/L5) (Table 1). This level of variability has been reported in wild-type mice (Kessel & Gruss 1991; Le Mouellic et al. 1992). Similarly, the majority of Gcn5+/Δ mice had seven cervical, 13 thoracic and six lumbar vertebrae (C7/T13/L6) (Fig. 1C); only two Gcn5+/Δ mice had five lumbar vertebrae (C7/T13/L5), indicating that mice bearing at least one wild type allele of Gcn5 develop normally. In contrast, 45% (10 of 22) of Gcn5flox(neo)/flox(neo) E18.5 embryos exhibited a transformation of the first lumbar vertebra (L1) into a thoracic vertebra (C7/T14/L5; Fig. 1D; Table 1) with a 14th pair of ribs emanating from the normal position of L1. Strikingly, all Gcn5flox(neo)/Δ mice examined exhibited this transformation (Fig. 1E and Table 1). Both Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ mutants consistently had 26 presacral vertebrae, further indicating that the development of a 14th pair of ribs arose from transformation of L1 to T14, not from development of an extra thoracic vertebra. This transformation did not occur in Gcn5flox/flox mice (Table 1), further indicating that it arose from the presence of the neomycin cassette in intron 2 and lowered the expression of Gcn5 (Lin et al. 2008).
Fig. 1.
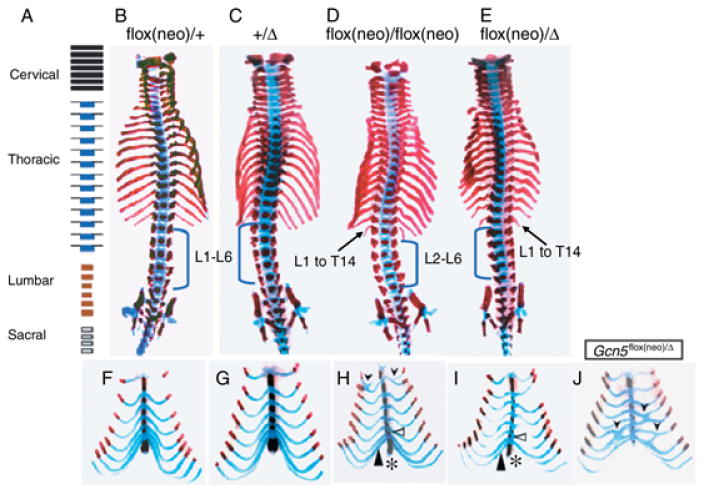
Anterior homeotic skeletal transformations in Gcn5 mutants. (A) Diagram showing normal distribution of cervical, thoracic, lumbar and sacral vertebrae. (B–E) Ventral views of Alizarin Red S/Alcian Blue stained skeletal preparations of E18.5 embryos. The blue brackets mark the lumbar vertebra. Arrows indicate transformation of L1 to a 14th thoracic vertebrae in the Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ embryos. (F–I) Ventral views of ribs and sternum of E18.5 embryos. Black arrowheads in (I and J) point to rib fusions, and the open triangles in (H) and (I) point to extra sternabrae. The filled triangles in (H) and (I) point to the eighth rib (T8) attached to the sternum, which may occur either bilaterally (majority) or unilaterally. The asterisks indicate malformed xiphoid processes.
Table 1. Summary of skeletal phenotypes.
| Morphology | Gcn5 genotype | |||||
|---|---|---|---|---|---|---|
| +/+ | flox(neo)/+ | +/Δ | flox/flox | flox(neo)/flox (neo) | flox(neo)/Δ | |
| Number of ribs attached to sternum | ||||||
| 7 | 10 | 15 | 11 | 19 | 3 | 0 |
| 8 | 0 | 0 | 0 | 3 | 19 | 20 |
| Number of ribs | ||||||
| 13 | 10 | 15 | 11 | 22 | 12 | 0 |
| 14 | 0 | 0 | 0 | 0 | 10 | 20 |
| Location of transitional vertebra | ||||||
| 10 | 10 | 15 | 11 | 22 | 5 | 0 |
| 12 | 0 | 0 | 0 | 0 | 16 | 18 |
| 13 | 0 | 0 | 0 | 0 | 1 | 2 |
| Anapophysis | ||||||
| L2 | 9 | 14 | 11 | 20 | 12 | 0 |
| L3 | 1 | 1 | 0 | 2 | 9 | 17 |
| L4 | 0 | 0 | 0 | 0 | 1 | 3 |
| Total | 10 | 15 | 11 | 22 | 22 | 20 |
Another homeotic transformation was found in the area of the sternum in both the Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ mutants at E18.5 and P0 (Fig. 1F–J). Six ossified segments make up the sternum of wild-type mice, including the manubrium at the top, four sternebrae, and finally the xiphoid process. The first six pairs of ribs are attached to the sternum, separating it into sternebrae. The seventh pair of ribs is normally attached to the sternum at the same site as the sixth pair of ribs (Fig. 1F,G). The eighth pair of ribs is not attached to the sternum in wild-type animals, as was observed in Gcn5flox(neo)/+ and Gcn5+/Δ heterozygotes (Fig. 1F,G). In all Gcn5flox(neo)/Δ and some Gcn5flox(neo)/flox(neo) mutants, an extra sternebrae developed between the fourth sternebrae and the xiphoid process (open arrowheads, Fig. 1H,I; Table 1). Also the eighth pair of ribs was attached to the sternum at the same site as the seventh pair of ribs in the Gcn5flox(neo)/Δ mutants (filled arrowhead, Fig. 1I) and the size of xiphoid process was smaller in Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ embryos than in their wild-type or heterozygous littermates (asterisk in Fig. 1H,I; Table 1). The penetrance of this malformation was lower in Gcn5flox(neo)/flox(neo) homozygous mice (19 of 22; 86%) than in Gcn5flox(neo)/Δ mice (20 of 20; 100%). No Gcn5flox(neo)/+ or Gcn5+/Δ heterozygous mice and only three of 22 Gcn5flox/flox mice exhibited this malformation.
A fraction of Gcn5flox(neo)/flox(neo) mice (six of 22) and Gcn5flox(neo)/Δ mice (11 of 20) exhibited rib fusions, and some had fusions at multiple sites (Fig. 1H,J and data not shown). As a result, the sternum in some mice had one or several fewer sternebrae even though additional sternebrae may have developed between the fourth sternebrae and xiphoid process upon transformation of L1 to T14. Some rib fusions occurred at the site of attachment with the sternum in the Gcn5flox(neo)/Δ and Gcn5flox(neo)/flox(neo) mice (Fig. 1I and data not shown). In other mutants, ribs fused to form a common ventral rib, which then attached to the sternum (Fig. 1H). Rib fusions occurred both symmetrically and asymmetrically in Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ mutants (Fig. 1IH, and data not shown). For example, in the embryo shown in Figure 1(H), the eighth rib on one side is attached to the sternum, whereas the eighth rib on the other side extends freely and is not attached. Similar anterior skeletal transformations were observed in Gcn5flox(neo)/flox(neo) mice that survived beyond birth, indicating that these malformations were not linked to perinatal lethality.
Gcn5flox(neo)/flox(neo) and Gcn5flox(neo)/Δ mice also exhibited transformations in the ventral vertebral column. In wild-type (not shown), Gcn5flox(neo)/+ or Gcn5+/Δ mice (Fig. 2A,B; Table 1), the spinous processes were found anterior to the 10th thoracic vertebra and extended caudally. A high percentage (15 of 22; 70%) of Gcn5flox(neo)/flox(neo) mutants exhibited T12 to T10 transformations, as evidenced by the rounded morphology of T12 in the mutant embryos (Fig. 2C; Table 1). An even higher penetrance of T12 to T10 transformation (18 out of 20; 90%) was observed in Gcn5flox(neo)/Δ mutants (Fig. 2D; Table 1). The other two Gcn5flox(neo)/Δ mutants examined showed T13 to T10 transformations, so 100% of these embryos exhibited anterior transformations in the ventral thoracic skeleton.
Fig. 2.
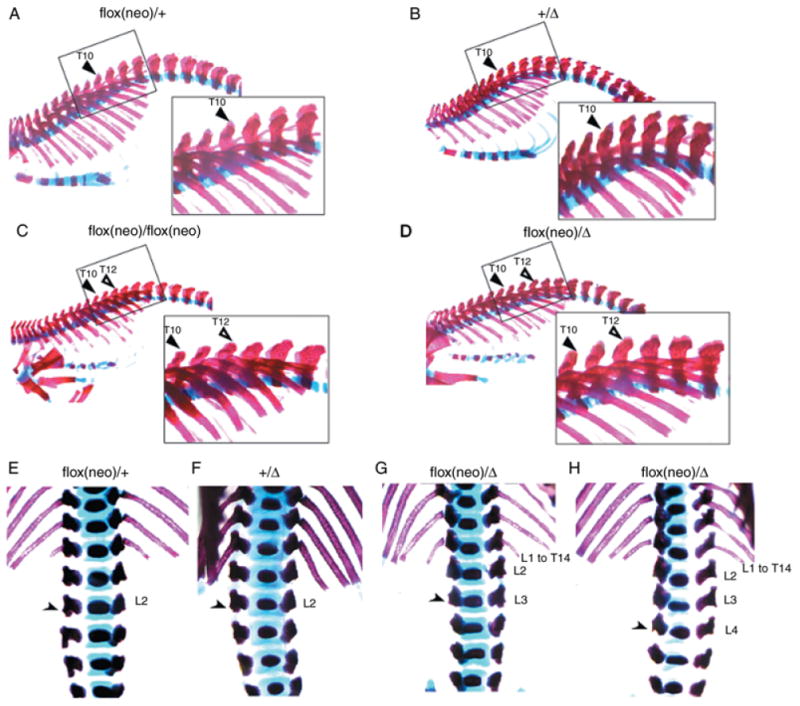
Anterior transformations of thoracic and lumbar vertebrae in Gcn5 mutants. Lateral views of vertebral columns stained with Alizarin Red S/Alcian Blue. In A–D, the filled triangle indicates the 10th thoracic vertebra (T10) and the open triangle indicates the 12th thoracic vertebra (T12). In E–H, the arrowheads point to anapophyses. L1–L4, lumbar vertebra.
Another anterior transformation was observed in the upper lumbar region of Gcn5flox(neo)/Δ E18.5 embryos. Normally, L2 is the last vertebra to bear small lateral processes of the pedicles, called anapophysis, as was observed in Gcn5flox(neo)/+ and Gcn5+/Δ embryos (Fig. 2E,F). In 85% (17 of 20) of Gcn5flox(neo)/Δ mutants, anapophysis was found on L3 (Fig. 2G; Table 1), indicating that conversion of L3 to L2 accompanied conversion of L1 to T14 in these embryos. In the other 15% of these mutants (three out of 20), anapophysis was found on the L4 vertebrae (Fig. 2H; Table 1). These transformations were observed at a lower frequency in Gcn5flox(neo)/flox(neo) embryos; L3 to L2 transformations were observed in 40% (nine out of 22) of Gcn5flox(neo)/flox(neo) embryos, and only one of 22 Gcn5flox(neo)/flox(neo) embryos exhibited anapophysis on L4.
Altogether, these data indicate that lowered Gcn5 function leads to anterior transformation of a region extending from L3 to T8.
Defective Hox gene regulation in Gcn5 hypomorphic mutants
Hox genes encode critical regulators of anteroposterior patterning in the mouse. The homeotic transformation phenotypes observed above in Gcn5flox(neo)/Δ mutants are similar to phenotypes described for mutations in Hoxb9, Hoxc8 and Hoxc9 (Le Mouellic et al. 1992; Suemori et al. 1995; Chen & Capecchi 1997). Since Gcn5 is a histone acetyltransferase and histone H3 acetylation is enriched at Hox gene loci (Petruk et al. 2001; Chambeyron & Bickmore 2004; Miller et al. 2004; Fujimura et al. 2006), we reasoned that Gcn5 might regulate Hox gene expression. Abnormal levels or spatial patterns of Hox gene expression in the Gcn5 mutants might then lead to the homeotic transformations observed above.
To test this possibility, we analyzed the expression of Hoxc8 and Hoxc9 in E10.5–11.5 Gcn5+/+ and Gcn5flox(neo)/Δ embryos with equal numbers of somites. The anterior expression boundary of Hoxc8 was shifted posteriorly by one somite at both time points in Gcn5flox(neo)/Δ mutants (Fig. 3). Similarly, the anterior boundary of Hoxc9 was shifted posteriorly by one somite at E10.5 (Fig. 4). For each analysis, at least three embryos of each genotype were examined, and all embryos gave consistent results (data not shown). In contrast, levels and spatial distribution of Hoxb9 and Hoxa9 expression were unaffected in the Gcn5 mutants (data not shown). Collectively, these data suggest that Gcn5 plays a role in mouse skeletal patterning through spatial regulation (directly or indirectly) of specific Hox genes.
Fig. 3.
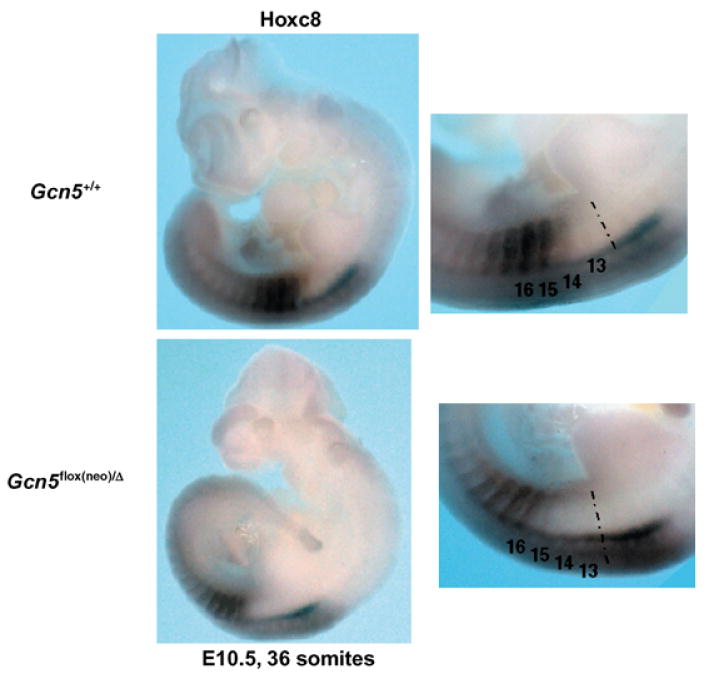
Hoxc8 expression is altered in Gcn5 mutants. Whole mount in situ hybridization to monitor spatial distribution of Hoxc8 expression in Gcn5+/+ and Gcn5flox(neo)/Δ embryos at E10.5 embryos with 36 somites. Panels on the right show views from panels on the left at increased magnification.
Fig. 4.
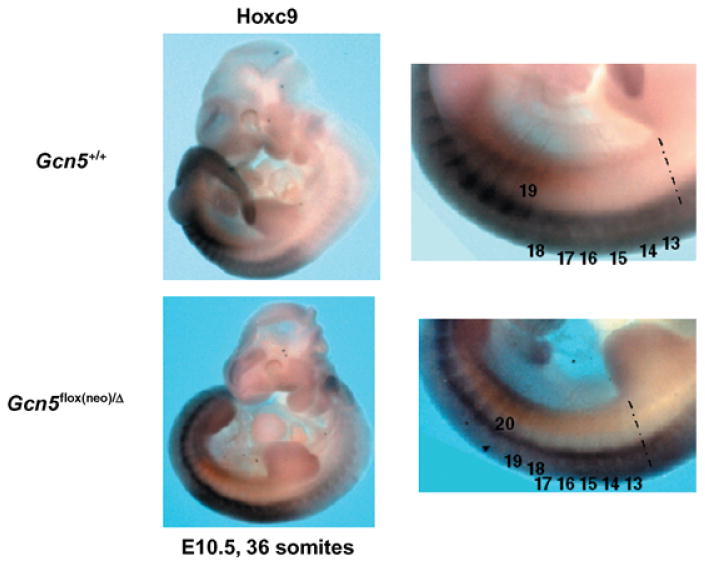
Hoxc9 expression is altered in Gcn5 mutants. Whole mount in situ hybridization to monitor spatial distribution of Hoxc9 expression in Gcn5+/+ and Gcn5flox(neo)/Δ embryos at E10.5 embryos with 36 somites. Panels on the right show views from panels on the left at increased magnification.
Discussion
Our previous studies demonstrated that Gcn5 is essential for mouse embryo development and survival beyond E11.0 and that Gcn5 HAT activity and expression is required for cranial neural tube closure (Xu et al. 2000; Bu et al. 2007; Lin et al. 2007; Lin et al. 2008). Our studies of a hypomorphic allele of Gcn5 reported here demonstrate that proper expression of Gcn5 is also required for normal axial skeletal patterning. The increased survival of these mice relative to that of Gcn5 null or Gcn5hat/hat mutants reflects the lowered expression of Gcn5 in mice bearing the Gcn5flox(neo) allele (Lin et al. 2008) versus its absence or lack of catalytic activity in our other mutants (Xu et al. 2000; Bu et al. 2007).
Skeletal patterning is controlled by an orchestration of Hox gene expression along the anteroposterior axis of the developing embryo. The rostral-caudal expression boundaries of individual Hox genes along with overlapping, combinatorial patterns of Hox gene expression govern proper development of the spine, ribs, and limbs (Favier & Dolle 1997). Our hypomorphic mutant Gcn5 embryos exhibit several abnormalities observed in mice bearing mutations in Hox genes that act at different levels along the anterior-posterior axis of the developing mouse (Fig. 5). For example, as in our Gcn5 mutants, L1 to T14 transformations were observed in Hoxc8 and Hoxc9 mutant mice (Le Mouellic et al. 1992; Suemori et al. 1995). Rib fusions occurred in Hoxb9 and Hoxc9 single knockout mice and Hoxa9/Hoxb9 double mutants (Le Mouellic et al. 1992; Suemori et al. 1995; Chen & Capecchi 1997). Axial skeletal transformations from T8 to T7 were observed in Hoxc8 and Hoxc9 mutant embryos. Lack of the ventral curvature of the vertebral column occurred in Hoxb1-Hoxb9 locus deletion mutants (Medina-Martinez et al. 2000). Gcn5 is known to function as a transcriptional coactivator (Roth et al. 2001), and our observation that Hoxc8 and Hoxc9 expression is altered in Gcn5flox(neo)/Δ embryos suggests that Gcn5 might directly regulate the spatial distribution of these and other Hox genes. Since the Hox genes encode transcription factors, it is also possible that Gcn5 functions as a coactivator for downstream targets of Hox proteins. Gcn5, then, could mediate a feed-forward loop, first activating Hox gene expression and then activating downstream Hox targets.
Fig. 5.
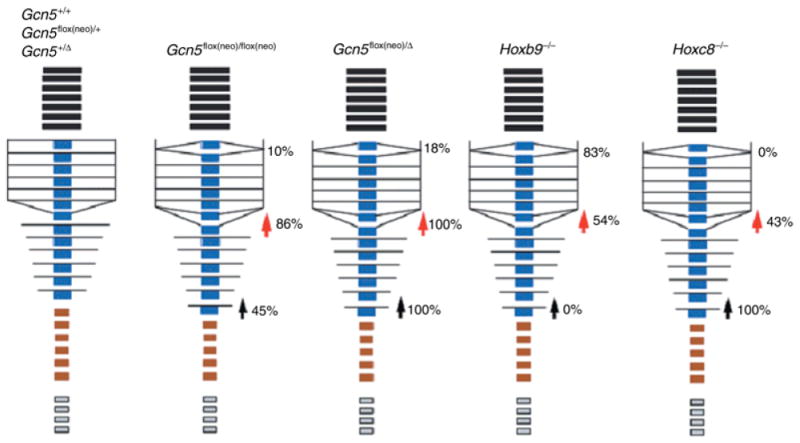
Comparison of anterior vertebral transformations in Gcn5, Hoxb9, and Hoxc8 mutants. Black boxes represent cervical vertebrae; blue boxes represent thoracic vertebrae; brown boxes represent lumbar vertebrae; gray boxes represent caudal vertebrae. Rib fusions are indicated by altered positions of lines in the thoracic area. The red arrowheads indicate T8 to T7 transformations. Black arrowheads indicate L1 to T14 transformations. Percentages indicate the penetrance of both bilateral and unilateral malformations.
If Gcn5 plays a direct role in activating Hox genes, it likely cooperates with TrxG proteins known to regulate these genes, including Set1 and MLL homologues in the mouse. These proteins, like Gcn5, modify histone H3. Set1 and MLL proteins methylate K4 in H3, whereas Gcn5 acetylates multiple downstream lysines (e.g. K9, K14, K18, and K23). Several studies indicate that methylation of H3 K4 is physically and functionally linked to H3 acetylation (Schneider et al. 2004; Kim et al. 2005; Pray-Grant et al. 2005; Sims et al. 2005; Barski et al. 2007; Govind et al. 2007). Complete loss of MLL functions leads to embryonic lethality, with defects in neural, skeletal, hematopoietic, and craniofacial development (Yu et al. 1995; Hess et al. 1997; Yu et al. 1998). Interestingly, embryos carrying a hypomorphic allele of MLL that lack the catalytic SET domain have some features similar to those of embryos carrying our Gcn5 hypomorphic allele, including rib fusions, sternal defects, and decreased expression of Hoxc8 and Hoxc9 (Terranova et al. 2006). However, these MLL mutant embryos exhibited posterior as well as anterior axial transformations, and the anterior boundaries of Hoxc8, Hoxc9, and Hoxd4 were unchanged in these mutants. These data suggest that MLL and Gcn5 may share some functions in axial skeletal patterning but that they may also have some distinct functions in Hox gene regulation. Mutations in other chromatin modifiers such as Bmi-1 and Mel 18 affect expression of different subsets of Hox genes (Akasaka et al. 1996; van der Lugt et al. 1996). The orchestration of Hox gene expression for proper embryo patterning, then, may reflect an upstream orchestration in the regulation and functions of multiple histone modifying enzymes.
Acknowledgments
This work was supported by a grant from the NIH (GM067718) to S.Y.R.D. We thank members of the Dent lab and members of the Michelle Barton lab for helpful discussions. We thank Zhilian Xia for technical assistance and Dr Armin Schumacher (Baylor College of Medicine) for the gift of Hoxc8 and Hoxc9 probes for whole mount in situ analyses. DNA sequencing was carried out by the DNA Analysis Core facility, and Gcn5 mutant mice were generated with help of the core Genetically Engineered Mouse Facility at UTMDACC (both supported by NCI CA16672).
Footnotes
Author contributions: W. L. participated in the design of the study, characterized the function of Gcn5 alleles, analyzed the skeletal phenotypes and collected embryonic samples, and drafted the manuscript. C. H. C. carried out whole mount RNA in situ hybridization. R. R. B. aided in the evaluation of the data. S. Y. R. D. coordinated the study, provided key reviews of the results and manuscript content, and obtained funding for this study. All authors read and approved the final manuscript
References
- Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18 285–18 289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev Biol. 1997;181:186–196. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3, K4 methyltransferase MLL1 and the H4, K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Favier B, Dolle P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- Fujimura Y, Isono K, Vidal M, et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development. 2006;133:2371–2381. doi: 10.1242/dev.02405. [DOI] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J. Hox genes and chordate evolution. Dev Biol. 1996;173:382–395. doi: 10.1006/dbio.1996.0034. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mouellic H, Lallemand Y, Brulet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992;69:251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- Lin W, Dent SY. Functions of histone-modifying enzymes in development. Curr Opin Genet Dev. 2006;16:137–142. doi: 10.1016/j.gde.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. Developmental potential of Gcn5 (−/−) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- Lin W, Zhang Z, Srajer G, et al. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn. 2008;237:928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Bradley A, Ramirez-Solis R. A large targeted deletion of Hoxb1-Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev Biol. 2000;222:71–83. doi: 10.1006/dbio.2000.9683. [DOI] [PubMed] [Google Scholar]
- Miller CT, Maves L, Kimmel CB. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–2461. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer RR. Manipulating the Mouse Embryo; a Laboratory Manual. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York, USA: 2003. [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Paro R. Propagating memory of transcriptional states. Trends Genet. 1995;11:295–297. doi: 10.1016/s0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rastegar M, Kobrossy L, Kovacs EN, Rambaldi I, Featherstone M. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol Cell Biol. 2004;24:8090–8103. doi: 10.1128/MCB.24.18.8090-8103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via Its tandem chromodomains. J Biol Chem. 2005;280:41 789–41 792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PB. Molecular mechanisms of cellular determination: their relation to chromatin structure and parental imprinting. J Cell Sci. 1994;107:2653–2668. doi: 10.1242/jcs.107.10.2653. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14 967–14 972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suemori H, Takahashi N, Noguchi S. Hoxc-9 mutant mice show anterior transformation of the vertebrae and malformation of the sternum and ribs. Mech Dev. 1995;51:265–273. doi: 10.1016/0925-4773(95)00371-1. [DOI] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard Y, Wakamiya M, Behringer RR, Roth SY. Loss of GCN5 leads to increased apoptosis and mesodermal defects during mouse development. Nat Gen. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, et al. Distinct but overlapping roles of the histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA. 2000;97:11 303–11 306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10 632–10 636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]


