Abstract
Microsomal epoxide hydrolase (EPHX1) catalyzes hydration reactions that determine the cellular disposition of reactive epoxide derivatives. Whereas the previously defined EPHX1 exon 1 sequence (E1) is derived from a promoter proximal to exon 2 of the EPHX1 coding region, in this investigation, we identified an alternative EPHX1 exon 1 sequence, E1-b, originating from a gene promoter localized ~18.5 kb upstream of exon 2. Northern hybridizations demonstrated that the E1-b variant is widely expressed and that the E1-b promoter functions as the primary driver of EPHX1 expression in human tissues. In contrast, the E1 promoter directs expression only in the liver. To examine the basis for liver-specific usage of the E1 promoter, we identified several potential cis-regulatory elements that included GATA (−110/−105) and hepatocyte nuclear factor 3 (HNF3) (−96/−88) motifs. GATA-4 was the principal GATA family member interacting with its respective motif, whereas both HNF3α and HNF3β were capable of interacting with the HNF3 element. GATA-4 and HNF3α/HNF3β DNA binding complexes were enriched in hepatic cells. Site-directed mutagenesis and transactivation analyses of the E1 promoter revealed that GATA-4 is probably a principal factor that regulates liver-specific expression of the E1 variant, with HNF3α and HNF3β acting to negatively regulate GATA-4 function in hepatic cells.
Human microsomal epoxide hydrolase (mEH; EC 3.3.2.3) is an important biotransformation enzyme that functions to detoxify deleterious chemical epoxide intermediates generated by phase I oxidation reactions (Fretland and Omiecinski, 2000). The enzyme also participates in bioactivation reactions, notably by contributing to the formation of highly reactive and carcinogenic bay-region diol-epoxides generated by polyaromatic hydrocarbon metabolism (Sayer et al., 1985; Miyata et al., 1999). In addition to its role in xenobiotic metabolism, mEH has been implicated as a participant in endogenous steroid metabolism (Fandrich et al., 1995), as a bile acid transporter (Alves et al., 1993), and in the vitamin K reductase complex (Guenthner et al., 1998). A wide variation of interindividual mEH activities have been reported in human tissues (Mertes et al., 1985; Omiecinski et al., 1993; Hassett et al., 1997).
Human mEH is encoded by a single gene (EPHX1) located on chromosome 1q42.1 (Skoda et al., 1988; Hartsfield et al., 1998). The primary coding sequences of human, rat, and rabbit mEH gene are highly conserved, with similarities exceeding 75% (Hassett et al., 1989). Polymorphisms in the human EPHX1 coding for amino acid substitutions in the respective proteins have been described previously (Hassett et al., 1994a), and various reports have associated genetic polymorphism in EPHX1 with increased risk of lung cancer, hepatocellular carcinoma, and other disease syndromes (McGlynn et al., 1995; Ulrich et al., 2001; Lee et al., 2002). In addition to genetically encoded structural differences in bio-transforming enzymes, interindividual differences in transcriptional controls and in tissue-specific regulation are likely contributors to disease risk resulting from chemical exposures.
Comparison of proximal 5′-flanking region sequences of mEH gene across several vertebrate species reveals few apparent similarities, suggesting that the regulation of mEH gene expression in different species may involve distinct regulatory mechanisms. In this regard, it is of interest that the mouse and rat mEH genes seem readily inducible by several xenobiotic chemicals (Hardwick et al., 1983; Honscha et al., 1991; Cho and Kim, 1998), whereas in primary human hepatocytes, EPHX1 is only modestly responsive to prototypical inducing agents (Hassett et al., 1997).
It has become clear that the use of alternative promoters is an important source of generating protein and regulatory diversity (Landry et al., 2003). To evaluate alternative regulatory mechanisms, a previous study identified eight putative splice-variant forms of EPHX1 mRNA with different exon 1 sequences (Gaedigk et al., 1997) that differed from the classic exon 1 (E1) structure characterized earlier (Skoda et al., 1988; Hassett et al., 1994b). In the present investigation, we localized the positions of these putative E1-variant EPHX1 exons 1 with in silico analyses to distances ranging from ~15 to >46 kb 5′ of exon E1. In our analyses, we discovered that the identities of the previously identified EPHX1 alternative exons 1 were compromised by sequence similarities of E1 exons to the gene encoding the signal recognition particle 9-kDa protein (SRP9) (NM_003133) that resides upstream of EPHX1 on chromosome 1 (Hsu et al., 1995).
To assess the putative alternative EPHX1 exon 1 structures further, we applied a more stringent 5′-rapid amplification of cDNA ends (5′-RACE) protocol and examined mRNA transcript sequences derived from several human tissues. We discovered that an alternative promoter, termed the E1-b promoter, localized ~18.5 kb 5′ upstream from the coding region of EPHX1, is used preferentially to drive expression of EPHX1 mRNA transcripts in most cell types. The previously identified promoter, E1, which is directly proximal to the EPHX1 coding region (Skoda et al., 1988; Hassett et al., 1994b), is responsible for driving EPHX1 expression specifically in the liver, both in adult and fetal stages. Because the regulation of the EPHX1 E1 promoter seems biologically important for both developmental and mature liver function, we sought to better characterize the mechanistic features associated with the liver-specific expression profile of the EPHX1 E1 promoter. Our results clarify the context of EPHX1 alternative-promoter usage and alternative exon 1 structures and delineate potential regulatory features that define the basis of tissue-specific expression for EPHX1.
Materials and Methods
Cell Culture
The hepatoma cell lines HepG2 and Huh7 were grown in minimal essential medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, 0.1 mM nonessential amino acids, and 1.5 g/l sodium bicarbonate. A transformed human embryonic kidney cell line, 293A (Qbiogene Inc., Carlsbad, CA), and the HeLa human cervix adenocarcinoma cell line were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. All cell lines were grown at 37°C under 5% CO2.
5′-RACE Analysis
The full-length 5′ ends of mEH cDNA were analyzed using a GeneRacer kit (Invitrogen), according to protocols supplied by the manufacturer. This system specifically targets only 5′-capped mRNAs. Input RNAs were isolated from two human liver samples, two human kidney samples, and one small intestine sample. Tissues were from unrelated donors and were kindly provided by Dr. K. Thummel (University of Washington, Seattle, WA). The 5′-RACE assay was conducted using EPHX1-specific primers: 5′-CGACTGGAGCACGAGGACACTGA-3′ and 5′-GGACACTGACATG-GACTGAAGGAGTA-3′ (nested primer). The DNA sequences of all 5′-RACE products were determined using Applied Biosystems (Foster City, CA) 310 and 377 genetic analyzers.
Northern Blot Analysis of EPHX1 mRNA in Human Tissues
A multitissue Northern blot (BD Biosciences Clontech, Palo Alto, CA) was probed with specific EPHX1 oligonucleotide probes directed to exon 1 (E1 oligomer; 5′-CAGAGGGTGAGAACGT-3′) or to exon 1-b (E1-b oligomers; 5′-GAGCCTGCGAGCCGAGACC-3′). Adult EPHX1 expression levels were evaluated in 12 unique tissues, each composed of pooled isolates obtained from 1 to 20 white persons of both sexes and a wide range of ages (BD Biosciences Clontech). Fetal EPHX1 expression levels were evaluated in four tissues, each composed of pooled isolates obtained from 7 to 38 specimens ranging from 18 to 30 weeks of gestational age.
RT-PCR Analysis of Alternative Splicing Variants of EPHX1 in Cultured Cells
The mRNA levels of EPHX1 E1 and E1-b variants were analyzed by one-step RT-PCR using the Access RT-PCR system (Promega, Madison WI). Total RNA of different cultured cells was prepared using the TRIzol reagent (Invitrogen). The specific E1 and E1-b fragments, 320 and 208 bp, respectively, were amplified by E1 and E1-b forward primers (E1, 5′-CAGAGGGT-GAGAACGTGGAG-3′; E1-b, 5′-GAGCCTGCGAGCCGAGAC), together with a common reverse primer (5′-CGTGGATCTCCTCATCT-GACGTTT-3′) at the end of exon 2.
Analysis of Interindividual Hepatic EPHX1 Expression
Ten micrograms of total RNA isolated from each of 25 human livers were applied to a slot blot and probed with oligomers specific to EPHX1 E1, E1-b, or 18S rRNA. After stringent hybridization and washing, relative mRNA expression levels were quantified from scanned autoradiograms by densitometry and image analysis software produced by Scion Corporation (Frederick, MD).
Plasmids Construction and Mutagenesis
A 2.5-kb DNA fragment immediately upstream of each alternative exon 1 sequence was cloned by high-fidelity PCR from the human liver genomic DNA. This fragment was used as the template for the amplification of 5′ truncation fragments by high-fidelity PCR and subcloned into the luciferase-reporter vector pGL3-basic. Internal deletion and point mutation were conducted by the gene splicing by overlap extension technique as described previously (Horton et al., 1990).
Cell Transfection and Luciferase Activity Assay
HepG2 and 293A cells were grown on 24-well plates to ~60% confluence and transfected with LipofectAMINE 2000 (Invitrogen) and FuGENE 6 (Roche Applied Science, Indianapolis IN), respectively. In transactivation assays, the promoter/luciferase-reporter plasmid was cotransfected with GATA-4 or GATA-6 (kindly provided by Dr. K. Walsh, Boston University, Boston, MA) and/or hepatocyte nuclear factor 3 (HNF3) -α, -β, or -γ (kindly provided by Dr. K. H. Kaestner, University of Pennsylvania, Philadelphia, PA) expression vectors. For all the transfections, the Renilla reniformis luciferase expression plasmid pRL-CMV was added to the mixture to normalize for transfection efficiency. Twenty-four hours after transfection, cells were lysed, and the luciferase activities were analyzed using the Dual-Luciferase reporter assay system (Promega) on a Veritas 96-well plate-reading luminometer (Turner Designs, Sunnyvale, CA).
Electrophoretic Mobility Shift Assays
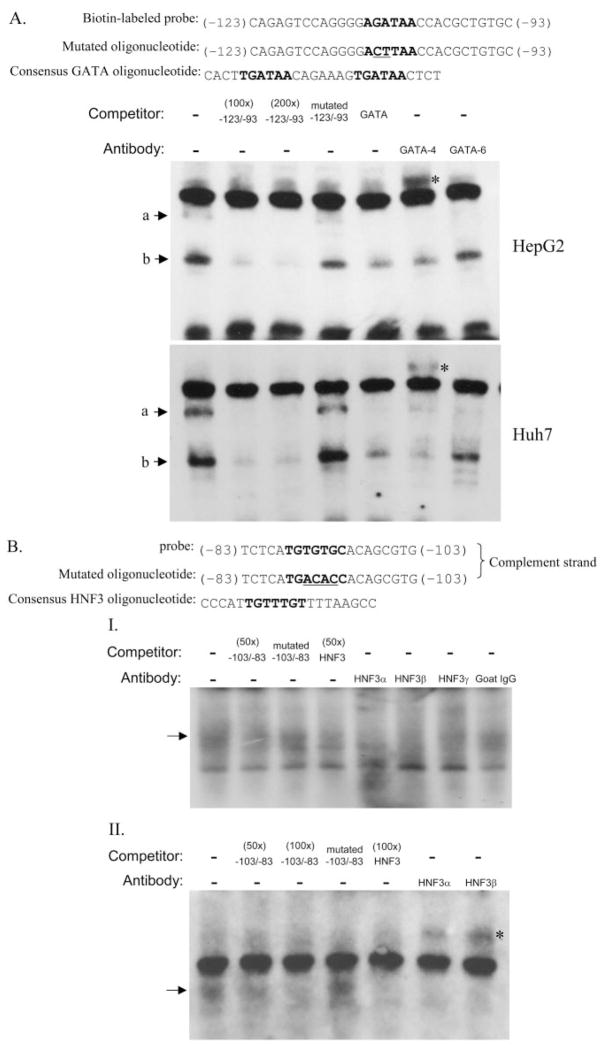
Nuclear extracts were prepared from HepG2, Huh7, 293A, and HeLa cells using NE-PER nuclear extraction reagent (Pierce Chemical, Rockford IL) according to the manufacturer’s instructions. Electrophoretic mobility shift assay (EMSA) for GATA proteins was performed using the nonradioactive LightShift chemiluminescent EMSA kit (Pierce). For the HNF3 protein assays, both standard radioactive and nonradioactive LightShift EMSAs were performed. Binding reactions were performed in a 20-μl mixture containing 1× binding buffer, 1 μg of poly(dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, 5 mM MgCl2, 6 to 8 μg of nuclear extracts, and 20 fmol of biotinylated (for GATA) or ATP[γ-32P] (for HNF3) end-labeled double-stranded probe, with or without unlabeled competitors, for 20 min at room temperature. For the supershift assay, 2 μg of antibodies for human GATA-4, GATA-6, HNF3α or HNF3β, or HNF3γ (Santa Cruz Biotechnology, Inc., Santa Cruz CA) was added to the binding mixture. The total amounts of binding mixtures were separated by electrophoresis using a 5% polyacrylamide gel and transferred to the nylon membrane in 0.5× Tris/borate/EDTA buffer. The biotin-labeled probe was detected using the LightShift reagents and visualized by exposure to X-ray film. The probes and unlabeled competitors used in EMSA are shown in Fig. 8.
Fig. 8.
Gel mobility shift assay for GATA and HNF3 binding sites. A, competition and super-shift assays for the E1-GATA binding site using biotin-labeled GATA probe and nuclear extracts from either HepG2 or Huh7 cells. B, competitive and supershift EMSAs assays were conducted with using HepG2 nuclear extracts and E1-HNF3 DNA targets labeled either with radioactive (I) or biotin (II) methods. Labeled probes were mixed with different unlabeled competitors including GATA or HNF3 site-mutated oligonucleotides and consensus binding oligonucleotides whose sequences are shown on the top of each gel. The core binding sequences are shown in boldface letters, and the mutated bases are underlined. Two micrograms of the respective antibodies was added to the reaction mixture for supershift analysis. ☆, supershift complexes.
Immunoblot Analysis
HepG2, Huh7, and 293A cells were lysed by radioimmunoprecipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1× phosphate-buffered saline, pH 8.0) containing 1× protease inhibitor cocktail (EMD Biosciences, San Diego, CA). Total cell lysates were obtained after centrifugation at 14,000 rpm for 20 min at 4°C, and the protein concentration was determined by the BCA protein assay reagent (Bio-Rad, Hercules, CA). Equal amounts of the protein lysates were separated by SDS electrophoresis on 10% polyacrylamide gels and transferred to a polyvinylidene difluoride membrane. Primary antibodies used for immunoblotting included goat anti-GATA-4, HNF3α, HNF3β, HNF3γ (Santa Cruz Biotechnology), and mouse anti-β-actin (Sigma-Aldrich, St. Louis, MO) antibodies. The immunoreactive signals were detected by the Lumi-Light reagent (Roche Applied Science).
Results
Alternative Exon Usage for Human EPHX1
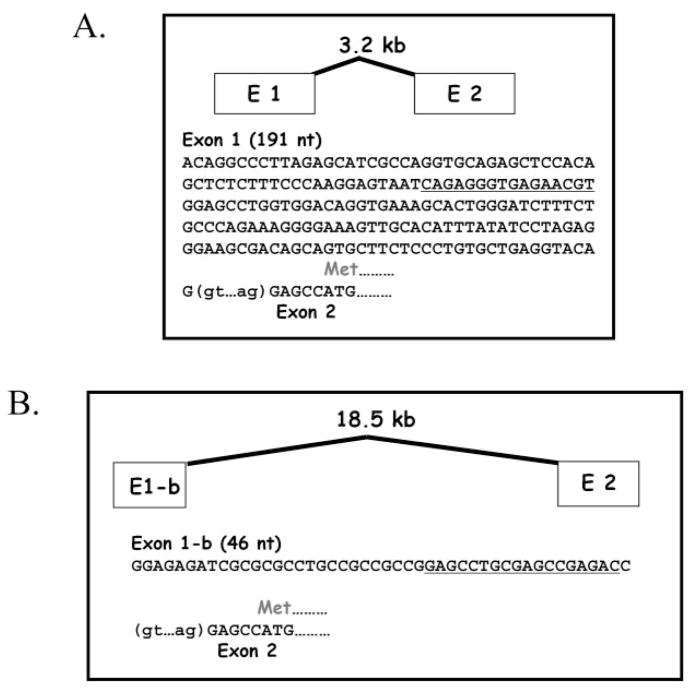
It was reported previously that human EPHX1 undergoes complex splicing processes at the exon 1/2 boundary to create transcript variants with alternative exon 1 sequence (Gaedigk et al., 1997). Using in silico analyses, we discovered that several of these putative exon 1 sequences were identical with elements of the signal recognition particle gene (SRP9; DNA contig: AC099066.3.1.149549) that resides relatively far upstream of the EPHX1 structural region (Fig. 1). Therefore, to reexamine this issue, we used a more stringent 5′-RACE protocol, one that is highly specific for 5′-capped molecules and therefore selective for only fully processed mRNAs, to more definitively characterize expressed EPHX1 transcripts. Two unique first exons were identified from RNA derived from human liver samples. These include 1) a 191-bp sequence with identity to the E1 sequence previously reported in several investigations (Skoda et al., 1988; Hassett et al., 1994b); and 2) a shorter 46-bp sequence that shared identity with a previously identified E1-b (Fig. 2). In contrast, 5′-RACE analysis performed using RNA derived from human kidney or small intestine samples revealed the presence of only E1-b exon 1 transcripts (data not shown). As determined from a compiled genomic contig containing these sequences, E1 is derived from genomic sequence 3.2-kb 5′ of exon 2, whereas E1-b resides much further upstream, ~18.5 kb from the start of exon 2. The base-numbering scheme we use in this investigation is consistent with that reported earlier (Hassett et al., 1994a), as viewed in the National Center for Biotechnology Information Locus Link accession L25879. The start position of the respective exon E1 and E1-b transcripts, as identified in this investigation, is indicated as +1.
Fig. 1.
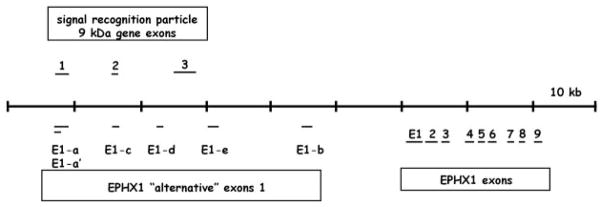
Genomic structural map encompassing the human EPHX1. Genomic contig entries (AC099066.3.1.149549, AL591895.9.134507) were compiled that together contain the previously described EPHX1 structural gene, as well as each of the reported alternative EPHX1 exon 1 sequences. BLAST searches against the nonredundant database demonstrated identities of E1-a/E1-a′ and E1-c to exons 1 and 2 of the signal recognition particle 9-kDa gene, respectively. Dashes representing exons are not drawn to scale, but relative positions are accurate.
Fig. 2.
Exon 1 sequences of the human EPHX1. Two unique first exons, E1 and E1-b, were identified by 5′-RACE analysis. Translation of the mEH protein begins in exon 2, six bases from the intron/exon boundary. The oligomers used as hybridization probes to assess expression of transcripts containing either E1 or E1-b are underlined; nt, nucleotide.
Selective Expression of EPHX1 Exon 1 Variants in Adult and Fetal Tissues
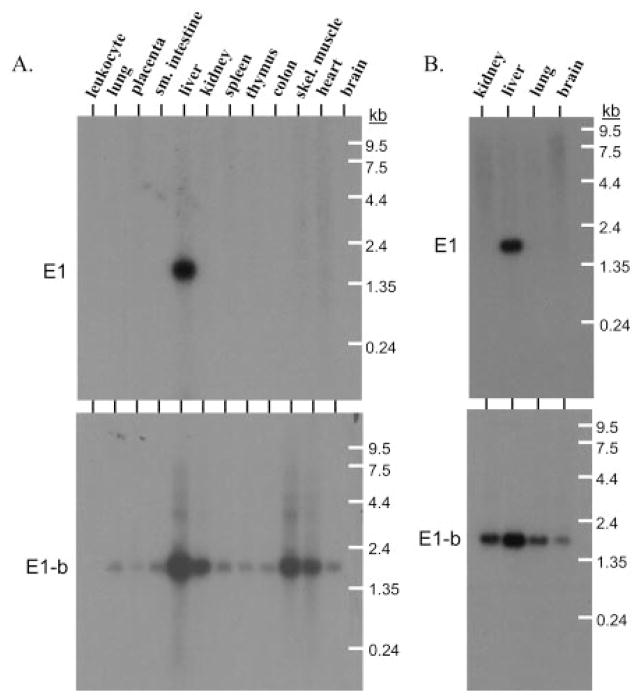
The 5′-RACE experiments verified the use of only two alternative EPHX1 exon 1 5′ sequences, in contrast to the data reported previously (Gaedigk et al., 1997), in which four additional alternative exon 1 sequences were identified that shared sequence identities with the SRP9 gene. To better characterize the respective expression patterns of the two bona fide expressed EPHX1 exon 1 transcripts, multitissue Northern blots composed of mRNAs from both adult and fetal tissue sources were used in hybridization studies. Specific oligonucleotide probes were used that recognized either E1- or E1-b–containing transcripts, respectively. In 12 adult tissues, mRNA containing E1 was expressed only in the liver, whereas E1-b–containing mRNA was clearly present in all of the tissues assessed except for peripheral blood leukocytes (Fig. 3A). The corresponding expression patterns in fetal tissues were identical, with the EPHX1 E1 transcript only detected in liver tissues, whereas the E1-b transcript was expressed in the liver, kidney, lung, and brain (Fig. 3B). These results demonstrate that the expression of EPHX1 transcript variants is subjected to differential tissue-specific regulation.
Fig. 3.
Expression of E1 and E1-b EPHX1 transcripts in adult and fetal tissues. Northern blots containing mRNAs of varied adult (A) and fetal (B) tissues were probed with transcript-specific oligomers (as indicated in Fig. 2) targeted to either mEH E1 or E1-b sequences.
Interindividual Expression of EPHX1 Exon 1 Variants in the Human Liver
Rates of xenobiotic metabolism in humans are subject to large interindividual differences. Because mEH is an important enzyme in the biotransformation process, we assessed the relative expression levels of the alternatively processed EPHX1 transcripts across a bank of 25 human livers. Purified total RNA was analyzed on slot blots and hybridized with either E1- or E1-b–specific oligonucleotide probes (Fig. 4A). The levels of E1 transcript varied up to 5-fold between these 25 human livers (Fig. 4B). The levels of E1-b transcript varied over a wider range, approximately 14-fold, with only low-level expression occurring in several samples. These data demonstrate that variable expression of EPHX1 transcripts is relatively common among individuals.
Fig. 4.
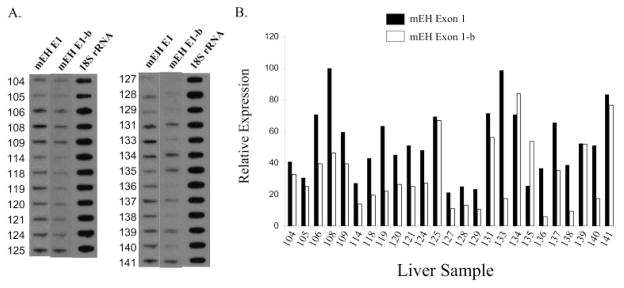
Interindividual differences in the expression of hepatic EPHX1 alternative transcripts. A, total RNAs of 25 human livers were analyzed for the amount of E1 or E1-b EPHX1 transcripts by slot blots probed with exon 1-specific oligomers. B, quantitative data of slot blots after normalization to the 18S rRNA level.
Characterization of Promoter Regions Conferring Differential Expression of EPHX1 Exon 1 Variants
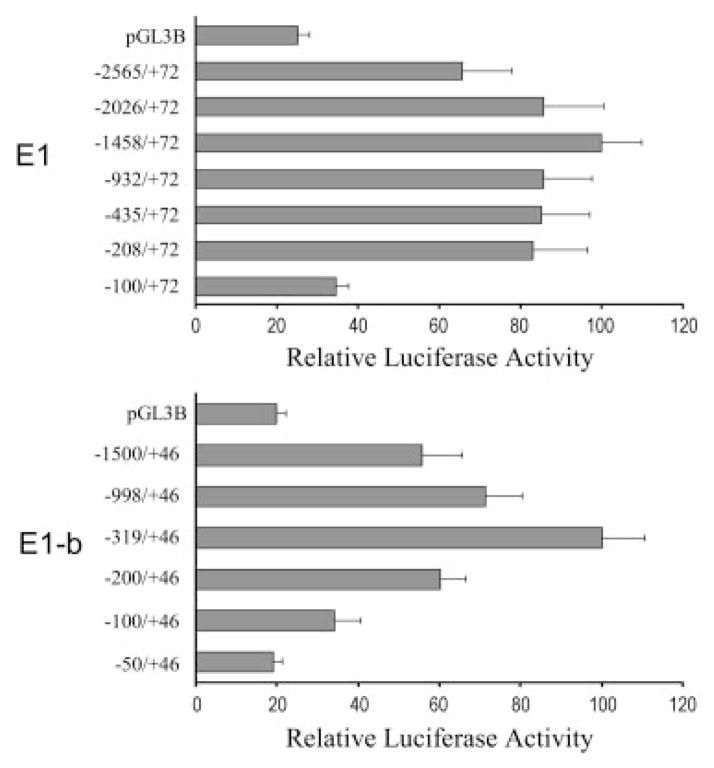
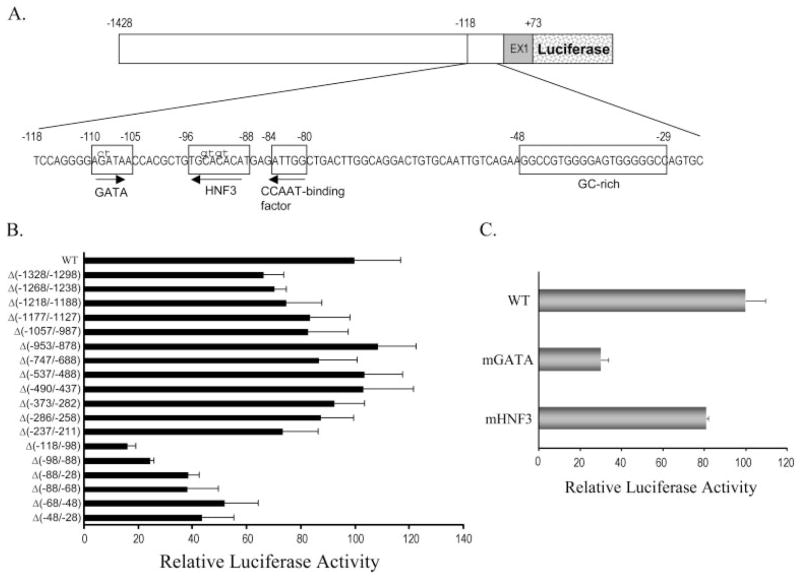
The finding that E1 and E1-b EPHX1 transcripts exhibit unique expression patterns led us to hypothesize that the EPHX1 transcription is controlled by the unique 5′-proximal promoter region of each exon 1. To explore the hypothesis further, we first isolated the respective 2.5-kb upstream regions of E1 and E1-b from genomic DNA derived from human liver. A series of 5′-deleted DNA fragments were then subcloned into a luciferase reporter vector, and the promoter activity of each construct was analyzed using transient transfection assays in HepG2 cells. The results indicated that the −1428/+72 region of E1 and the −319/+46 region of E1-b contributed the highest transcriptional activities for the respective promoters (Fig. 5). In both the E1 and E1-b promoter assays, inclusion of longer DNA fragments resulted in a reduction of transcriptional activities (Fig. 5), suggesting the presence of repressor elements in these upstream regions. Minor decreases in E1 promoter activity were observed by deletions of the −1428- to −208-bp region, whereas a substantive reduction in transcriptional activation resulted from the further deletion of the −208- to −100-bp region (Fig. 5). Likewise, deleting the −319- to −50-bp region of E1-b produced a marked decrease of the corresponding promoter activity (Fig. 5). These results suggest that positive regulatory elements are localized within the proximal 200- and 300-bp upstream promoter regions of E1 and E1-b, respectively (Fig. 5).
Fig. 5.
Identification of the potential promoter sequences in the 5′-flanking regions of E1 and E1-b. Various 5′ truncated DNA fragments were linked to the firefly luciferase reporter in pGL3-basic vector and cotransfected with the R. reniformis luciferase reporter pRL-CMV into the HepG2 cells. The promoter activity of each construct was represented by the firefly luciferase activity after normalization to the R. reniformis luciferase activity. The data were graphed to show the promoter activities relative to the highest one in their group.
Characterization of cis-Acting Elements Directing Liver-Specific Transcription of the EPHX1 E1 Variant
The EPHX1 E1 transcript is specifically expressed in the human adult and fetal liver, suggesting that appropriate control of E1 transcript expression is important in the maintenance of liver function throughout the development. To examine the potential regulatory mechanisms directing liver-specific usage of the E1 promoter, we conducted more detailed functional analysis of this promoter region. Using the −1428/+72 E1 DNA fragment as the full-length reference, we made a series of internal deletion of the regions containing putative transcription factor binding sites, identified with computer sequence analysis. The relative promoter activities were then analyzed after transient transfection of HepG2 cells. Similar to the results obtained from our 5′-deletion assays (Fig. 5A), several internal deletions within the −1428 bp to −211 bp E1 promoter region resulted in a modest reduction of transcriptional activity (Fig. 6B). However, each of the internal deletions from −118 bp and the beginning of E1 demonstrated a substantial reduction of transcriptional activity (Fig. 6B), indicating that essential cis-regulatory elements were contained in this region. Further sequence analysis revealed the potential presence of multiple transcription factor binding motifs, including those for GATA, HNF3, CCAAT binding factors, and a GC box (Fig. 6A). Internal deletion of these sites all exhibited a marked reduction (at least 50%) of promoter activity, with the deletion of putative GATA site resulting in the largest reduction of E1 promoter function (Fig. 6B). Considering that GATA and HNF3 factors have been reported previously to control liver-specific transcription, we further examined the functional contribution of these elements with site-specific mutagenesis of the respective core motifs. Mutation of the GATA site produced an approximately 70% decrease of E1 promoter activity, whereas the HNF3 site mutation yielded only a modest impact on promoter function (Fig. 6C). These data indicate that GATA protein(s) are probably important positive regulators of the E1 promoter.
Fig. 6.
Identification of cis-elements involved in the regulation of E1 promoter activities. A, a diagram showing the sequences of the region containing potential transcriptional factor binding sites. The mutated bases (shown in lowercase letters) were constructed to analyze the effect of GATA and HNF3 protein binding sites on the E1 promoter activities. B, various internal deletions were made in the −1428/+72 E1 fragment as shown in A to scan for the elements involved in controlling E1 promoter activities. C, the effect of GATA and HNF3 sites mutation on E1 promoter activities. The quantitative data from triplicate transfections in HepG2 cells was representative of three independent experiments. The promoter activities of different mutated constructs were normalized and relative to those of the wild type (WT).
EMSA Analysis of the GATA and HNF3 E1 Promoter Elements
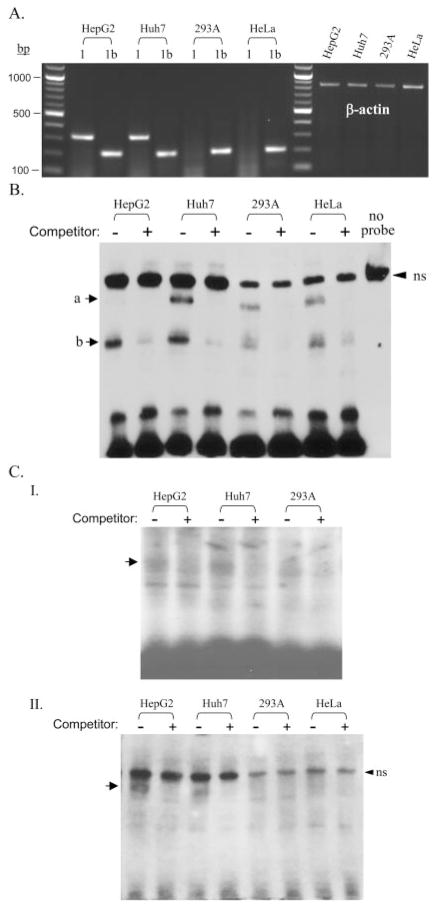
To characterize the function of the GATA and HNF3 binding sites directly in cells, we applied EMSA to examine the interaction of nuclear proteins with the respective elements. Nuclear extracts were prepared from two human hepatoma cell lines, HepG2 and Huh7, and two human nonhepatoma cell lines, 293A and HeLa, and analyzed for interaction with double-strand oligonucleotides containing either the GATA or HNF3 binding elements. Our RT-PCR assays using specific E1 or E1-b primers demonstrated that the E1-b transcript was expressed in all cell lines tested, whereas the E1 transcript was expressed specifically in HepG2 and Huh7 hepatoma cells (Fig. 7A). Therefore, these cell lines were used as models for more detailed analysis of the liver-specific activity of the E1 promoter. We found that two complexes were apparent using the GATA probe (−113 to −93 bp) and nuclear extracts of all the cell lines examined (Fig. 7B). Although the band intensity noted for the larger complex was weak in the HepG2 cell extracts, an excess of unlabeled GATA probe competed out the formation of both complexes (Fig. 7B), indicating that they were specific. Except for the larger complex present with HepG2 cell extracts, the GATA-specific complexes were enriched in the hepatoma cells versus nonhepatoma cells (Fig. 7B). Likewise, HNF3-specific binding complexes were enriched in the hepatoma cell lines when using the HNF3 probe (−103 to −83 bp), as shown by the use of two alternative EMSA protocols (Fig. 7C).
Fig. 7.
Analysis of nuclear protein bindings to the GATA and HNF3 sites of E1 promoter in hepatoma and nonhepatoma cells. A, RT-PCR analysis of the expression of E1 and E1-b transcripts in HepG2, Huh7, 293A, and HeLa cells. The E1- (320 bp) and E1-b–(208 bp) specific PCR products together with referenced β-actin fragments were visualized by ethidium bromide staining. B, analysis of binding complexes for the E1-GATA site by nonradioactive chemiluminescent EMSA using a biotin-labeled −123/−93 GATA probe. C, analysis of binding complexes for the E1-HNF3 site by either radioactive (I) or nonradioactive (II) EMSA, using 32P- and biotin-labeled −103/−83 HNF3 probes, respectively. A 100× molar excess amount of unlabeled probes were used as competitors. The arrow indicates the specific binding complexes for each probe. A nonspecific (ns) band was present in all of the nonradioactive chemiluminescent EMSA assays. Its nonspecificity was demonstrated in HepG2 nuclear extracts run without the addition of any DNA probe in the binding reaction.
To confirm the involvement of GATA and HNF3 proteins in the formation of these complexes, excess amounts of consensus sequence GATA and HNF3-binding oligonucleotides were added to the mixtures. In both cases, the formation of specific binding complexes was competed by consensus oligonucleotides (Fig. 8). Furthermore, the addition of the oligonucleotides containing mutations within the core binding motifs for the respective GATA or HNF3 probes caused them to be unable to compete with the wild-type probes (Fig. 8). Together, these data demonstrate that the putative GATA and HNF3 sites within the E1 promoter are capable of interaction with cellular GATA and HNF3 nuclear factors.
To identify which GATA and HNF3 isoforms interact with these elements, supershift assays were performed using anti-GATA-4 and GATA-6 or HNF3α, HNF3β, and HNF3γ antibodies. We focused on GATA-4 and GATA-6 because these proteins have been shown by others to regulate the liver-specific gene expression (Davis and Burch, 1996; Bossard and Zaret, 1998; Denson et al., 2000). Addition of the GATA-4 antibody substantially reduced the formation of both GATA probe-specific binding complexes in hepatoma cells, accompanied by the formation of a supershifted complex (Fig. 8A). However, although the addition of the GATA-6 antibody seemed to decrease the intensity of the larger GATA complex, no supershifted bands were observed with this antibody under our experimental conditions (Fig. 8A). Whereas GATA-4 seems involved in the formation of two binding complexes using the E1-GATA probe, GATA-6 only participates in the formation of the larger complex. The results suggest that GATA-4 is the major protein interacting with the GATA-region within the E1 promoter (−113 to −93 bp). On the other hand, the complex formed between the E1 HNF3 probe and the HepG2 nuclear proteins was substantially depleted by the addition of either HNF3α or HNF3β antibodies but only partially depleted by HNF3γ antibodies, as shown by two complementary EMSA protocols (Fig. 8B). The super-shifted complexes were clearly identified upon the addition of HNF3α and HNF3β antibodies using the nonradioactive chemiluminescent EMSA protocol (Fig. 8B, II). These data suggest that HNF3α and HNF3β are the major isoforms interacting with the −103/−83 E1 oligonucleotide. The binding complex thus formed probably represents a mixture of HNF3α and HNF3β binding to the −103/−83 E1 oligonucleotide.
Western immunoblot analyses demonstrated that the respective protein levels of GATA-4, HNF3α, and HNF3β were all enriched in the hepatoma cells used in this study (Fig. 9). These results are consistent with the enhanced formation of binding complexes observed with both the GATA and HNF3 probes when incubated with nuclear extracts derived from these liver-derived cells (Fig. 7, B and C).
Fig. 9.
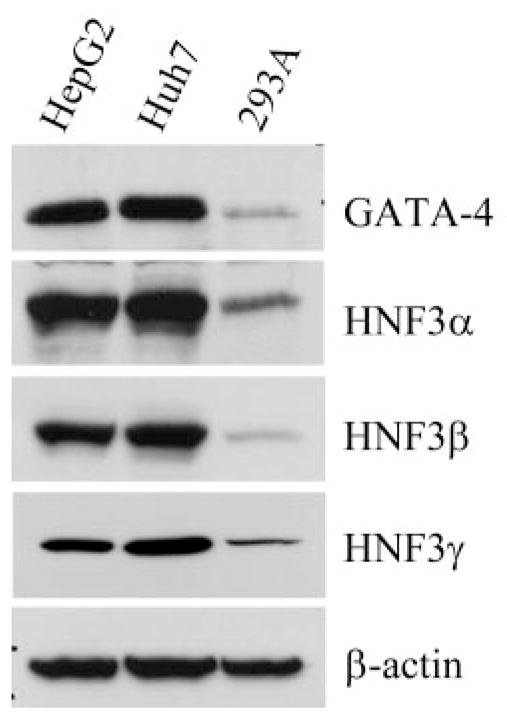
Comparison of GATA-4 and HNF3 protein amounts in hepatoma and nonhepatoma cells. GATA-4, HNF3α, HNF3β, and HNF3γ proteins in HepG2, Huh7, and 293A cells were analyzed by immunoblotting. The β-actin blot was shown as a reference for the equal loading of total proteins (40 μg) of the tested samples.
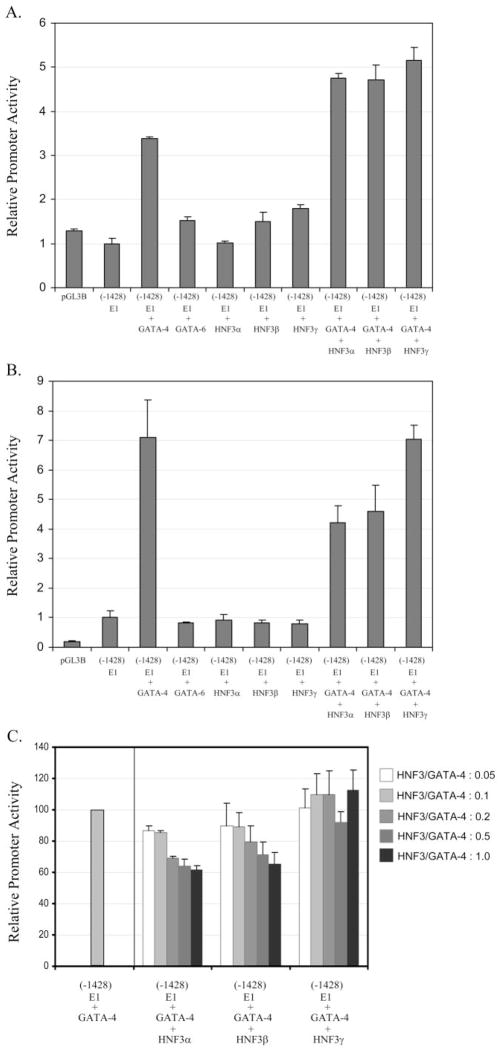
HNF3 Negatively Regulates the Transactivation of E1 Promoter by GATA-4 in Hepatoma Cells
To further assess the functional roles of GATA and HNF3 in regulating expression of the EPHX1 E1 transcript, we cotransfected 293A or HepG2 cells with E1 promoter constructs and GATA and/or HNF3 expression plasmids. No transcriptional activity was detected when the E1 promoter reporter was transfected into the 293A cells (Fig. 10A), results that are in accordance with our RT-PCR analysis demonstrating no expression of the E1 transcript in this cell line. However, the promoter activities were induced more than 3-fold in GATA-4–cotransfected cells (Fig. 10A). In contrast, when the E1 promoter was cotransfected with either GATA-6, HNF3α, HNF3β, or HNF3γ expression plasmids, no transcriptional activation was observed. However, coexpression of HNF3 and GATA-4 further amplified the induction of E1 promoter over that achieved by GATA-4 expression alone (Fig. 10A). In HepG2 cells, overexpression of GATA-4 induced the E1 promoter activities even more markedly (7-fold) than in 293A cells (Fig. 10B). In addition, in contrast to the results obtained with the 293 cells, HNF3α or HNF3β cotransfection combined to decrease the reporter activities induced by GATA-4, whereas individually, neither of the HNF3 isoforms affected E1 promoter activity (Fig. 10B). The inhibitory effects on GATA-4 E1 promoter activation produced by HNF3α and HNF3β cotransfection were further confirmed in concentration-effect studies with the respective expression constructs (Fig. 10C). Together, these data indicate that GATA-4 is a positive regulator of the promoter activity of liver-specific E1 transcript and that HNF3 proteins act to modify the function of GATA-4 differentially on the basis of cell lineage. In HepG2 cells, HNF3α and HNF3β seem to act in concert with hepatocyte-specific factors to negatively regulate the GATA-4 function within the E1 promoter.
Fig. 10.
Transactivation of E1 promoter activities by GATA and HNF3 proteins. The 293A (A) and HepG2 (B) cells were transiently transfected with the E1 promoter (−1428/+72)/luciferase construct and different combination of plasmids expressing GATA-4, GATA-6, and HNF3 proteins. The promoter activities measured in cotransfected samples were shown relative to those transfected with E1 promoter alone. Transfection with promoter-lacking plasmid, pGL3B, served as the negative control for background activities. C, effects of HNF3 on GATA-4–induced E1 promoter activities in hepatoma cells. A fixed amount of GATA-4 plasmids were cotransfected with different concentrations of HNF3 plasmids together with the E1 promoter construct into the HepG2 cells.
Discussion
Microsomal epoxide hydrolase is an important contributor to xenobiotic metabolism, is expressed widely across different cell types, and exhibits high protein sequence conservation in vertebrate species (Hassett et al., 1997). Investigations using EPHX1 knockout mice have demonstrated convincingly that this enzyme is a key risk factor contributing to polyaromatic hydrocarbon-induced carcinogenesis (Miyata et al., 1999; Bauer et al., 2003).
It is well established that RNA diversity in mammals is expanded markedly through the use of alternative promoters and differential RNA splicing mechanisms (Landry et al., 2003; Zhu et al., 2003). A previous report suggested a total of eight potential exon 1 structures within the composite of EPHX1 mRNAs expressed in human tissues, generated through alternative promoter/splicing mechanisms (Gaedigk et al., 1997). Upon closer examination of the human genomic contig, AC099066.3.1.149549, comprising this region of chromosome 1 (the human genome draft sequence was not available at the time of the initial investigations), we determined that many of the previously characterized alternative exon 1 sequences shared identity with sequences derived from the 9-kDa SRP9 structural gene, a gene that exists upstream of the EPHX1 structural sequence (Fig. 1). We questioned whether these SRP9 sequences may have arisen artifactually in the initial PCR/RACE studies. In the current investigation, we used a 5′-RACE protocol modified for highly specific detection of only 5′-capped and therefore mature mRNAs, together with DNA sequence analyses, to reinvestigate potential alternative splicing for human EPHX1. In our 5′-RACE studies, we could identify only two alternative EPHX1 exon 1 structures, termed E1 and E1-b, in mRNA samples derived from human livers, and only one EPHX1 exon 1 structure, that of E1-b, in mRNAs derived from human kidney or small intestine. In hybridization experiments with specific oligomer probes, we further characterized unique tissue-selective expression patterns of EPHX1 mRNAs containing the respective alternative exon 1 sequences.
The existence of two alternatively spliced EPHX1 transcripts is intriguing in several respects. In this investigation, we localized the respective E1 and E1-b promoters on chromosome 1 and found that the distal promoter is separated from its more proximal counterpart by approximately 18.5 kb. From our studies, it is clear that the distal E1-b promoter serves as the primary promoter for the gene, driving EPHX1 expression in a variety of tissues, including hepatic tissues. In contrast, the proximal E1 promoter drives gene expression exclusively in the liver.
Using transient transfection assays of serially deleted E1 promoter constructs, we identified a 100-bp region immediately upstream of the E1 exon as containing important functional elements necessary for regulating the basal transcription of E1 mRNA. Upon further analysis of this region, GATA and HNF3 transcription factor motifs were prominently identified. These factors have been shown previously to be important in liver-selective gene regulation. The GATA family transcription factors consist of six members, with GATA-4, -5, and -6 implicated as regulators of gene expression in the heart, liver, gonad, gut, and lung (Arceci et al., 1993). GATA-4 and GATA-6, in particular, were reported to regulate the expression of liver-specific genes, including albumin, vitellogenin II, and the liver-enriched homeobox gene, Hex (Davis and Burch, 1996; Bossard and Zaret, 1998; Denson et al., 2000). The HNF3 proteins (α, β, and γ) regulate the transcription of numerous genes in adult liver and pancreas, and they also play a pivotal role in the development of metabolic tissues (Kaestner, 2000). Although functioning predominantly as positive regulators, HNF3 factors may also exhibit repressive effects, down-regulating expression of genes such as glucagon, α-fetoprotein, and aldolase B (Gregori et al., 1994; Philippe et al., 1994; Huang et al., 2002). In the E1 promoter region of EPHX1, GATA and HNF3 motifs are adjacent within a confined stretch of 25 nucleotides, and we examined whether these factors acted in concert to regulate the expression of the liver-specific E1 transcript.
After a series of in vitro analyses, we concluded that GATA-4 is the major nuclear protein in hepatoma cells to interact with the EPHX1 E1 GATA element. Overexpression of GATA-4 increased E1 promoter activities in both hepatoma and nonhepatoma cells, demonstrating that GATA-4 is a positive regulator for E1 mRNA transcription. While this article was in preparation, another laboratory also reported that GATA-4 was an important factor regulating the transcriptional activity of the EPHX1 E1 promoter (Zhu et al., 2004). These investigators did not identify the alternate E1-b promoter usage. However, in the comparative aspects of GATA-4 regulation of the E1 promoter, certain differences are apparent in the respective studies. Zhu et al. (2004) reported two functional GATA-4 binding sites, with the −110/−105-bp GATA-4 (our numbering) consistent with that described in this report. The second GATA-4 site was localized further upstream at −158/−153 bp (our numbering). The −110/−105 GATA site was described as having no effect on basal EPHX1 transcription upon mutation, whereas mutational analysis of the −158/−153 GATA site resulted in a 65% loss of promoter activity (Zhu et al., 2004). We observed that a truncated construct (−138/+72), deleted for the putative −158/−153 GATA site while maintaining the −110/−105 site, conferred no detectable alteration of promoter activity compared with the full-length promoter fragment (data not shown). We have regularly used a more extended 5′-flanking fragment, −1428/+72, as the reference promoter region for either deletion or site-mutation analyses, compared with a −593/−54 fragment (or −159/−54 fragment in the GATA site-mutation experiments) used by Zhu et al. (2004). We hypothesize that inclusion of the extended 5′- and 3′-flanking DNA regions may confer additional regulatory control affecting the activity of the −158/−153 and −110/−105 GATA elements.
GATA-4 has been described previously as a ubiquitously expressed regulator able to control tissue-specific gene expression through interaction with other tissue-restricted transcription factors (Molkentin, 2000). This paradigm may apply to the regulation of liver-specific expression of E1 mRNA by GATA-4, because the E1 promoter was more readily transactivated in HepG2 cells by GATA-4 (~7-fold) than in the 293A kidney cell line (~3.5-fold) (Fig. 10). We hypothesize that interactions with hepatocyte-specific coactivators facilitate the GATA-4 regulation of EPHX1 E1 expression. It is interesting that our EMSA results indicated that GATA-4 and GATA-6 together formed a larger molecular weight shift complex with the single GATA-containing oligonucleotide. A previous study in cardiac myocytes demonstrated that GATA-4 and GATA-6 may physically interact (Charron et al., 1999). However, we were unsuccessful in detecting a GATA-6–mediated transactivation of EPHX1 E1 promoter activity (Fig. 10) or enhancement of GATA-4 activation of this promoter (data not shown). Therefore, we were unable to detect a functional involvement of GATA-6 in the regulation of E1 EPHX1 transcription.
Despite the indication that both HNF3α and HNF3β interacted in vitro with an HNF3 element adjacent to the GATA site (Fig. 8B), disruption of these interactions via site-directed mutagenesis had no substantial impact on resultant E1 promoter activity (Fig. 6C). Furthermore, all three isoforms of HNF3 were unable to transactivate the E1 promoter in either hepatoma and nonhepatoma cells (Fig. 10). However, we did observe that an internal deletion from −98/−88, containing an HNF3 site, resulted in the dramatic reduction of promoter activity (Fig. 6B). This deletion may affect the function of other essential regulatory elements in the −98/−88 region. Because HNF3 and GATA-4 have been reported to act synergistically to induce the expression of the liver-enriched homeobox gene (Denson et al., 2000), we conducted cotransfection assays to examine possible functional interactions between these factors in E1 promoter regulation. Surprisingly, we found that HNF3 proteins, particularly HNF3α and HNF3β, could either amplify or reduce GATA-4–mediated induction of E1 promoter activity in 293A and HepG2 cells, respectively. Thus, although HNF3 proteins in isolation did not seem to transactivate the liver-specific E1 promoter, they may function to cooperatively interact with GATA factors, and probably other regulatory partners present in HepG2 cells, to down-regulate E1 mRNA expression in this cell type.
The results presented here define two alternative promoters and two distinct noncoding exon 1 structures for human EPHX1 and therefore raise the question as to the biological importance for the dual promoter system, particularly for the liver, which expresses both mRNA variants simultaneously. The ubiquitous pattern of E1-b expression suggests that the use of the E1-b promoter may program cells with an essential basal level of mEH across a variety of human tissues. Although the tissue profile of E1-b expression is broad, E1-b may be subjected to tissue-specific regulation. Evidence supporting this concept is derived from Northern blot analysis indicating that liver expresses higher levels of E1-b mRNA relative to other tissues (Fig. 3). Furthermore, in preliminary studies using cotransfection approaches, we have observed that GATA-4 and HNF3 proteins also seem to modulate E1-b promoter activities (data not shown), suggesting the possibility of liver-specific regulation of both the E1 and E1-b promoters. Because the liver is exposed to a high concentration of dietary chemicals and is extremely active in generating epoxide intermediates via cytochrome P450-dependent metabolism pathways, one potential explanation for the use of a second promoter that drives additional expression of hepatic mEH is for simple supplementation of epoxide-metabolizing activity in this organ. The liver is an essential organ catalyzing detoxification of potentially reactive substances, and therefore high levels of liver-selective mEH expression probably provide an enhanced threshold of protection for this organ (Oesch et al., 2000). On the other hand, examples of other genes have been described that use alternative RNA splicing to generate structurally distinct mRNAs with distinct 5′-untranslated regions that confer differences in RNA stability (Chen et al., 1998), cap-dependent and -independent translation (Nanbru et al., 1997; Stein et al., 1998), and RNA polymerase II processivity (Wolf et al., 1995). Although additional studies are required to assess these functions with respect to EPHX1, it is clear that in humans, control of EPHX1 RNA expression has evolved that entails the use of widely separated promoters to drive transcription of the gene according to tissue-selective programs. Detailed analysis of the regulatory mechanisms underlying the control of EPHX1 expression in different cell types should ultimately enhance our abilities to predict organ-specific toxicities and to identify individuals with altered sensitivities to xenobiotic exposure.
Acknowledgments
We gratefully acknowledge Dr. Andrea Gaedigk for providing helpful suggestions and insights. We thank Dr. Kenneth Walsh for the gift of GATA-4 and GATA-6 expression vectors; Dr. Klaus Kaestner for the HNF3α, β, and γ expression vectors; and Dr. Kenneth Thummel for generously providing several human tissue samples.
This research was supported by a United States Public Health Service grant from the National Institute of Environmental Health Sciences (ES-04978).
ABBREVIATIONS
- mEH
microsomal epoxide hydrolase
- HNF3
hepatocyte nuclear factor 3
- EMSA
electrophoretic mobility shift assay
- EPHX1
human microsomal epoxide hydrolase gene
- RACE
rapid amplification of cDNA ends
- SRP9
9-kDa signal recognition particle
- E1
exon 1
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- bp
base pair
- kb
kilobase
References
- Alves C, von Dippe P, Amoui M, Levy D. Bile acid transport into hepatocyte smooth endoplasmic reticulum vesicles is mediated by microsomal epoxide hydrolase, a membrane protein exhibiting two distinct topological orientations. J Biol Chem. 1993;268:20148–20155. [PubMed] [Google Scholar]
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AK, Faiola B, Abernethy DJ, Marchan R, Pluta LJ, Wong VA, Gonzalez FJ, Butterworth BE, Borghoff SJ, Everitt JI, et al. Male mice deficient in microsomal epoxide hydrolase are not susceptible to benzene-induced toxicity. Toxicol Sci. 2003;72:201–209. doi: 10.1093/toxsci/kfg024. [DOI] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 MRNA by the C-Jun NH2-terminal kinase pathway. Science (Wash DC) 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- Cho MK, Kim SG. Differential induction of rat hepatic microsomal epoxide hydrolase and RGSTA2 by diazines: the role of cytochrome P450 2E1-mediated metabolic activation. Chem Biol Interact. 1998;116:229–245. doi: 10.1016/s0009-2797(98)00093-3. [DOI] [PubMed] [Google Scholar]
- Davis DL, Burch JB. The chicken vitellogenin II gene is flanked by a GATA factor-dependent estrogen response unit. Mol Endocrinol. 1996;10:937–944. doi: 10.1210/mend.10.8.8843410. [DOI] [PubMed] [Google Scholar]
- Denson LA, McClure MH, Bogue CW, Karpen SJ, Jacobs HC. HNF3beta and GATA-4 transactivate the liver-enriched homeobox gene, Hex. Gene. 2000;246:311–320. doi: 10.1016/s0378-1119(00)00082-2. [DOI] [PubMed] [Google Scholar]
- Fandrich F, Degiuli B, Vogel Bindel U, Arand M, Oesch F. Induction of rat liver microsomal epoxide hydrolase by its endogenous substrate 16 alpha, 17 alpha-epoxyestra-1,3,5-trien-3-ol. Xenobiotica. 1995;25:239–244. doi: 10.3109/00498259509061848. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Leeder JS, Grant DM. Tissue-specific expression and alternative splicing of human microsomal epoxide hydrolase. DNA Cell Biol. 1997;16:1257–1266. doi: 10.1089/dna.1997.16.1257. [DOI] [PubMed] [Google Scholar]
- Gregori C, Kahn A, Pichard AL. Activity of the rat liver-specific aldolase b promoter is restrained by HNF3. Nucleic Acids Res. 1994;22:1242–1246. doi: 10.1093/nar/22.7.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner TM, Cai D, Wallin R. Co-purification of microsomal epoxide hydrolase with the warfarin-sensitive vitamin K1 oxide reductase of the vitamin K cycle. Biochem Pharmacol. 1998;55:169–175. doi: 10.1016/s0006-2952(97)00431-0. [DOI] [PubMed] [Google Scholar]
- Hardwick JP, Gonzalez FJ, Kasper CB. Transcriptional regulation of rat liver epoxide hydratase, NADPH-cytochrome P-450 oxidoreductase and cyto-chrome P-450b genes by phenobarbital. J Biol Chem. 1983;258:8081–8085. [PubMed] [Google Scholar]
- Hartsfield JK, Jr, Sutcliffe MJ, Everett ET, Hassett C, Omiecinski CJ, Saari JA. Assignment 1 of microsomal epoxide hydrolase (EPHX1) to human chromosome 1q42.1 by in situ hybridization. Cytogenet Cell Genet. 1998;83:44–45. doi: 10.1159/000015164. [DOI] [PubMed] [Google Scholar]
- Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994a;3:421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett C, Lin J, Carty CL, Laurenzana EM, Omiecinski CJ. Human hepatic microsomal epoxide hydrolase: comparative analysis of polymorphic expression. Arch Biochem Biophys. 1997;337:275–283. doi: 10.1006/abbi.1996.9794. [DOI] [PubMed] [Google Scholar]
- Hassett C, Robinson KB, Beck NB, Omiecinski CJ. The human microsomal epoxide hydrolase gene (EPHX1): complete nucleotide sequence and structural characterization. Genomics. 1994b;23:433–442. doi: 10.1006/geno.1994.1520. [DOI] [PubMed] [Google Scholar]
- Hassett C, Turnblom SM, DeAngeles A, Omiecinski CJ. Rabbit microsomal epoxide hydrolase: isolation and characterization of the xenobiotic metabolizing enzyme cDNA. Arch Biochem Biophys. 1989;271:380–389. doi: 10.1016/0003-9861(89)90287-7. [DOI] [PubMed] [Google Scholar]
- Honscha W, Oesch F, Friedberg T. Tissue-specific expression and differential inducibility of several microsomal epoxide hydrolase mRNAs which are formed by alternative splicing. Arch Biochem Biophys. 1991;287:380–385. doi: 10.1016/0003-9861(91)90493-3. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Hsu K, Chang DY, Maraia RJ. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J Biol Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- Huang MC, Li KK, Spear BT. The mouse alpha-fetoprotein promoter is repressed in HepG2 hepatoma cells by hepatocyte nuclear factor-3 (FOXA) DNA Cell Biol. 2002;21:561–569. doi: 10.1089/104454902320308933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab. 2000;11:281–285. doi: 10.1016/s1043-2760(00)00271-x. [DOI] [PubMed] [Google Scholar]
- Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Brennan P, Boffetta P, London SJ, Benhamou S, Rannug A, To-Figueras J, Ingelman-Sundberg M, Shields P, Gaspari L, et al. Microsomal epoxide hydrolase polymorphisms and lung cancer risk: a quantitative review. Biomarkers. 2002;7:230–241. doi: 10.1080/13547500210121882. [DOI] [PubMed] [Google Scholar]
- McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoebonnie A, Oforiadjei D, et al. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci USA. 1995;92:2384–2387. doi: 10.1073/pnas.92.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertes I, Fleischmann R, Glatt HR, Oesch F. Interindividual variations in the activities of cytosolic and microsomal epoxide hydrolase in human liver. Carcinogenesis. 1985;6:219–223. doi: 10.1093/carcin/6.2.219. [DOI] [PubMed] [Google Scholar]
- Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5 and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, Prats AC. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- Oesch F, Herrero ME, Hengstler JG, Lohmann M, Arand M. Metabolic detoxification: implications for thresholds. Toxicol Pathol. 2000;28:382–387. doi: 10.1177/019262330002800305. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Aicher L, Holubkov R, Checkoway H. Human Peripheral lymphocytes as indicators of microsomal epoxide hydrolase activity in liver and lung. Pharmacogenetics. 1993;3:150–158. doi: 10.1097/00008571-199306000-00005. [DOI] [PubMed] [Google Scholar]
- Philippe J, Morel C, Prezioso VR. Glucagon gene expression is negatively regulated by hepatocyte nuclear factor 3 beta. Mol Cell Biol. 1994;14:3514–3523. doi: 10.1128/mcb.14.5.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer JM, Yagi H, van Bladeren PJ, Levin W, Jerina DM. Stereoselectivity of microsomal epoxide hydrolase toward diol epoxides and tetrahydroepoxides derived from benz[a]anthracene. J Biol Chem. 1985;260:1630–1640. [PubMed] [Google Scholar]
- Skoda RC, Demierre A, McBride OW, Gonzalez FJ, Meyer UA. Human microsomal xenobiotic epoxide hydrolase. Complementary DNA sequence, complementary DNA-directed expression in COS-1 cells and chromosomal localization. J Biol Chem. 1988;263:1549–1554. [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich CM, Bigler J, Whitton JA, Bostick R, Fosdick L, Potter JD. Epoxide hydrolase Tyr113His polymorphism is associated with elevated risk of colorectal polyps in the presence of smoking and high meat intake. Cancer Epidemiol Biomarkers Prev. 2001;10:875–882. [PubMed] [Google Scholar]
- Wolf DA, Strobl LJ, Pullner A, Eick D. Variable pause positions of RNA polymerase II lie proximal to the c-myc promoter irrespective of transcriptional activity. Nucleic Acids Res. 1995;23:3373–3379. doi: 10.1093/nar/23.17.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shendure J, Mitra RD, Church GM. Single molecule profiling of alternative pre-MRNA splicing. Science (Wash DC) 2003;301:836–838. doi: 10.1126/science.1085792. [DOI] [PubMed] [Google Scholar]
- Zhu QS, Qian B, Levy D. Regulation of human microsomal epoxide hydrolase gene (EPHX1) expression by the transcription factor GATA-4. Biochim Biophys Acta. 2004;1676:251–260. doi: 10.1016/j.bbaexp.2004.01.002. [DOI] [PubMed] [Google Scholar]