Abstract
Below a certain level, table salt (NaCl) is beneficial for animals whereas excessive salt is harmful. However, it remains unclear how low and high salt perceptions are differentially encoded. Here, we identified a salt taste coding mechanism in Drosophila melanogaster. Flies use distinct types of gustatory receptor neurons (GRNs) to respond to different concentrations of salt. We demonstrated that a member of the newly discovered ionotropic glutamate receptor (IR) family, IR76b, functioned in detection of low salt, and was a Na+ channel. Loss of IR76b selectively impaired the attractive pathway, leaving salt-aversive GRNs unaffected. Consequently, low salt became aversive. Our work demonstrated that the opposing behavioral responses to low and high salt were determined largely by an elegant bi-modal switch system operating in GRNs.
To address the fundamental question as to how low- and high-salt taste perceptions are differentially encoded in GRNs in insects we chose the fruit fly as a model. We first tested the animal’s behavioral responses to different salt concentrations ranging from 1 mM to 1000 mM using a robust, food-color based preference assay (fig. S1A, B). Akin to mammals, flies preferred low-salt food (1 mM to 100 mM) with a maximal preference at 50 mM NaCl, whereas they rejected high-salt food (≥ 200 mM) (Fig. 1A) (1–3). This pattern differs from both sugar and bitter taste in that flies prefer sweet and dislike bitter compounds regardless of the concentration.
Fig. 1. Wild-type responses to different concentrations of salt.
(A) Behavioral responses to 1 – 1000 mM salt. The dashed line indicates no preference. n = 10. (B) Cartoon showing the distribution of L-, I- and S-type sensilla in the labellum. (C and D) Tip recordings using L4 and S6 sensilla in response to low salt. n = 10. The arrows indicate application of the recording electrode to the sensilla. SEMs. *p < 0.01. (E) Schematic depicting how the L- and S-type sensilla differentially mediated attractive and aversive salt taste.
In Drosophila, the primary taste sensory organ, the labellum contains 31 sensilla, which are further classified by size as small (S), intermediate (I), and large (L) sensilla (Fig. 1B) (4). Sensilla contain multiple GRNs, which respond to distinct stimuli, including bitter, sweet and salty tastants (4–6). We surveyed the physiological responses of sensilla to low salt (50 mM) and high salt (500 mM) by performing tip recordings (7) (fig. S2). The GRNs housed by two L-type sensilla (L4 and L6) produced the most robust firings in response to low salt, while the GRNs in three S-type sensilla (S4, S6 and S8) displayed the strongest responses to high salt (fig. S2, A and C). These three S-type sensilla respond differentially to bitter tastants (8), suggesting that each gustatory sensillum has a unique taste tuning profile. GRNs in I-type sensilla responded to salt (9), but none as robustly as the most sensitive L- or S-type sensilla (fig. S2). With the exception of S4 and S8, the S-type sensilla show robust responses to the broadest array of aversive tastants (5, 8), while L-type sensilla produce the strongest physiological responses to attractive tastants, such as sugars (6, 8, 10, 11). Thus, we deduced that the responses to low and high salt (5, 6, 12) were likely to be controlled by a balance between the GRNs housed in L- and S-type sensilla.
We focused on L4 and S6 sensilla, using NaCl ranging from 1 mM to 1000 mM (Fig. 1, C and D). The firing of salt GRNs in the L4 sensilla increased progressively at low concentrations and peaked at 100 mM. In contrast, the salt GRNs in S6 sensilla were much less active than the salt GRNs in L4 sensilla, suggesting that these latter sensilla played a predominant role in low salt response. As the salt concentration increased above 100 mM, the firing of salt GRNs in L4 sensilla gradually declined. In contrast, the action potentials produced by S6 salt GRNs exhibited a remarkable increase (≥100 mM) with a maximal response at 500 mM. At high salt concentrations, the firing of S6 salt GRNs far exceeded L4 salt GRNs, indicating that the high salt response was controlled predominantly by salt GRNs in S-type sensilla.
We therefore propose a model in which competition between GRNs in the S- and L-type sensilla accounts for the bidirectional behavioral responses to salt. At low concentrations, the low-salt GRNs dominate over the high-salt GRNs, thereby causing the animals to prefer low salt (Fig. 1E). At high salt levels, the high-salt GRNs overwhelm the low-salt GRNs, resulting in salt rejection.
We next tested several candidate salt receptors and channels, none of which affected salt taste (supplementary online text). Ionotropic receptors (IRs) comprise a class of olfactory receptors in Drosophila, which are distantly related to mammalian ionotropic glutamate receptors (iGluRs) (13). Several Ir genes, such as Ir25a and Ir76b, also appear to be expressed in gustatory sensilla (13–15). The Ir25a2 mutant had no obvious deficits in sensing either low salt or high salt (fig S3, A and C). We then carried out a genetic analysis of Ir76b, and generated two null alleles, Ir76b1 and Ir76b2 by P-element-mediated imprecise excision (Fig. 2A). We also retained a revertant line that underwent a precise P-element excision (Ir76bR1). Loss of Ir76b did not impair the responses to KCl, sucrose, water or bitter tastants (fig. S4, A–C).
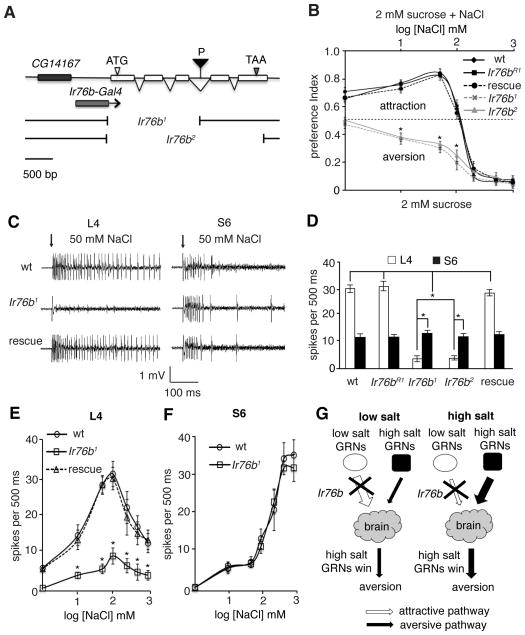
Fig. 2. Ir76b required for low salt preference.
(A) Genomic organization of Ir76b. Shown are a P-element (P) inserted in Ir76b, the deletions in Ir76b1 and Ir76b2, and the genomic region included in the Ir76b-Gal4. (B) Two-way choice tests to sucrose versus sucrose plus salt. The “rescue” were Ir76b1 flies harboring the UAS-Ir76b and the Ir76b-Gal4 transgenes. n = 10. (C and D) 50 mM NaCl-induced action potentials in L4 and S6 sensilla. n = 15. (E and F) Action potential frequencies produced by L4 (n = 5) and S6 (n = 3) sensilla across different NaCl concentrations. ANOVA tests. (G) Cartoon depicting that loss of Ir76b selectively disrupted the attractive salt taste pathway. SEMs. *p < 0.01.
The Ir76b deletions resulted in severe defects in the attraction to low-salt concentrations (1 – 100 mM; Fig. 2B). In contrast, the Ir76b mutants showed the same aversion to high salt as the Ir76b+ control (w1118). Ir76bR1 behavior was indistinguishable from the “wild-type” Ir76b+ control (w1118) at all salt concentrations (Fig. 2B).
We next performed tip recordings to monitor physiological abnormalities in the GRNs. The Ir76b mutations caused a decease in the number of action potentials by salt GRNs in L4 sensilla in response to 50 mM salt (Fig. 2, C and D), demonstrating a functional defect in the GRNs. There were no significant changes in the firing frequencies of S6 GRNs in the Ir76b mutants, compared with wild-type (Fig. 2, C and D). Using the Ir76b-Gal4 and UAS-Ir76b transgenes, we restored normal attractive responses to low salt in the Ir76b1 mutant (Fig. 2, C–E).
We also examined the physiological responses to different salt concentrations (1 – 1000 mM NaCl). Loss of Ir76b caused significant reductions in the firing frequencies of the salt GRNs in L4 sensilla at all salt concentrations (Fig. 2E). Firing of salt GRNs in L6 sensilla were also impaired (fig. S4D). However, there were no effects on the physiological responses of high-salt GRNs in S4 or S6 sensilla (Fig. 2F and fig. S4E).
Taken together, our studies indicated that removal of Ir76b selectively disrupted the attractive salt pathway, while leaving the aversive salt pathway intact (Fig. 2G). Consequently, Ir76b mutant animals avoided rather than preferred low salt, while they retained aversion to high salt.
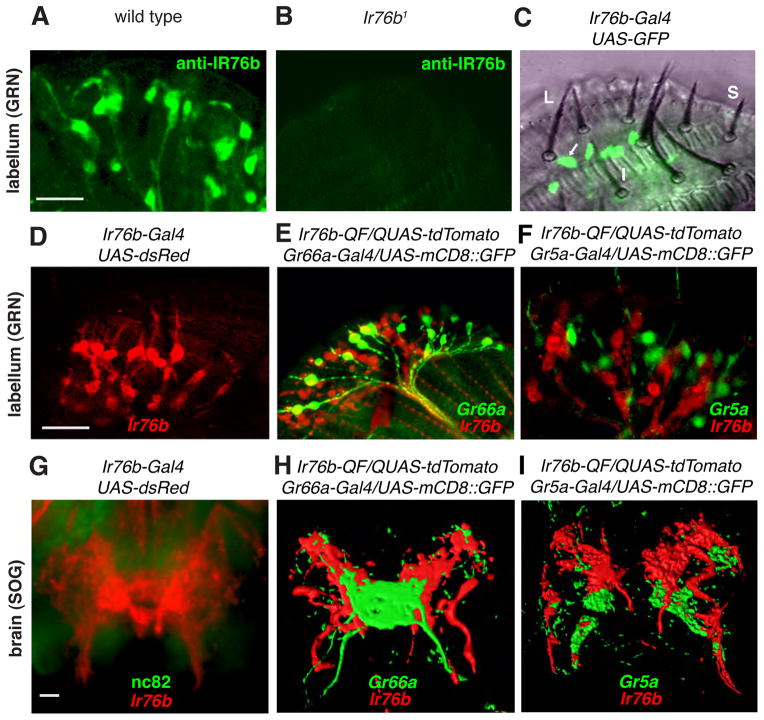
To examine the cellular distribution pattern of IR76b, we raised antibodies against IR76b. The antibodies marked GRNs in the labellum, and the staining was virtually eliminated in Ir76b mutants (Fig. 3, A and B, and fig. S5A). We also generated flies expressing an Ir76b reporter (Ir76b-Gal4). In combination with UAS-mCD8::GFP or UAS-dsRed, we detected prominent GRN staining in the proboscis (Fig. 3, C and D). Ir76b-expressing GRNs were in all L-type sensilla, including L4 and L6 (Fig. 3C). We also detected Ir76b reporter expression in GRNs in the leg tarsi and wing margins, which sent projections to the ventral nerve cord (fig. S5, E–G). Ir76b reporter expression largely overlapped with the anti-IR76b staining, suggesting that the reporter reflected the bona fide cellular distribution of Ir76b (fig. S5, B–D)
Fig. 3. Expression of Ir76b in GRNs.
(A and B) Anti-IR76b staining of wild-type and Ir76b1 labella. (C) GFP fluorescence superimposed on a bright field view of a Ir76b-Gal4/UAS-mCD8::GFP labellum. The arrow indicates a GFP-positive GRN within an L4 sensillum. (D) Ir76b reporter staining a labellum. (E) Labellum expressing Ir76b (red) and Gr66a (green) reporters. (F) Labellum expressing Ir76b (red) and Gr5a (green) reporters. (G) Projections of GRNs in the SOG expressing an Ir76b reporter. (H and I) Three-dimensional reconstructions of the GRN projections in the SOG using flies expressing Ir76b (red), Gr66a (green), and Gr5a (green) reporters as indicated. The scale bars represent 10 μm.
To determine if Ir76b-positive GRNs overlapped with Gr66a expressing bitter- or Gr5a expressing sugar-responsive GRNs, we generated an Ir76b reporter using the Q system (Ir76b-QF) (16). Double-labeling showed that Ir76b-positive GRNs were distinct from either Gr66a or Gr5a GRNs, (Fig. 3, E and F). Gr66a- and Gr5a-positive GRNs project their axons from the labellum to non-overlapping regions in the brain, the subesophageal ganglion (SOG) (17, 18). The projections of Ir76b GRNs in the SOG (Fig. 3G and Fig. S5H) showed minimal overlap with regions innervated by the axons of Gr5a and Gr66a GRNs (Fig. 3, H–I). Thus, Ir76b GRNs represented a class of GRNs distinct from sugar or bitter-responsive GRNs.
In olfactory receptor neurons (ORNs), IRs function either alone or in conjugation with other IRs (13, 19). We tested whether misexpression of IR76b alone conferred salt taste when introduced in non-salt responsive GRNs. Because Ir76b and Gr5a are expressed in different GRN populations, we introduced Ir76b in Gr5a-sugar neurons in an Ir76b1 background. We recorded from L2 sensilla, which showed few responses to low salt even in wild type (Fig. 4A). In response to NaCl, there was a robust train of action potentials produced by these Gr5a GRNs in L2 sensilla (Fig. 4, A and B, and fig. S6A). In contrast, these same GRNs did not induce a response to NMDGCl or potentiate the response to sucrose (Fig. 4, A and B). Thus, the action potentials were due to Na+ and not Cl, and was not a consequence of nonspecific elevation of Gr5a GRN activity. The behavioral deficit in low salt preference in Ir76b mutants was rescued by misexpressing Ir76b in Gr5a GRNs (Fig. 4C).
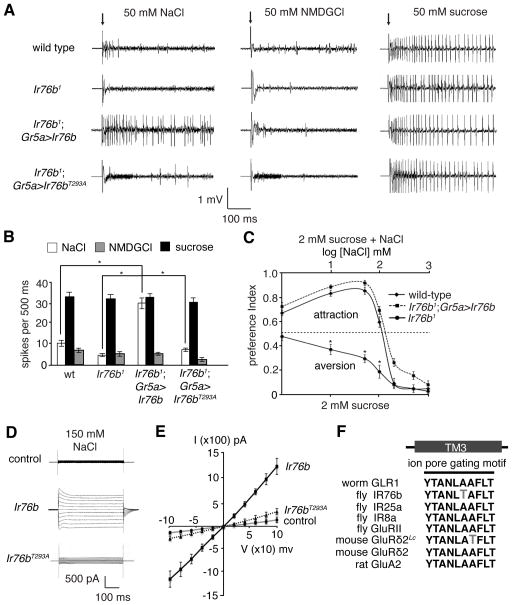
Fig. 4. Ir76b was sufficient to function as a salt sensor.
(A and B) Tip recordings and quantification showing action potentials triggered by 50 mM NaCl after misexpression of Ir76b in Gr5a GRNs. NMDGCl and sucrose were negative and positive controls, respectively. n = 10. (C) Two-way choice tests (2 mM sucrose versus 2 mM sucrose plus different concentrations of NaCl) after misexpression of UAS-Ir76b using the Gr5a-Gal4. n = 5. (D) Whole-cell voltage clamp recordings of HEK293T cells expressing wild-type IR76b or IR76bT293A (150 mM NaCl in the bath). The cells were stimulated with voltage steps of 500 ms duration (100 mV to +100 mV in 10 mV increments). (E) Current and voltage relationships of cells expressing IR76b or IR76bT293A (150 mM NaCl in the bath). SEMs. *p < 0.01. (F) Sequences of the ion pore gating motif in TM3 of the indicated iGluRs and IRs.
To test if IR76b was capable of functioning as a Na+ permeable channel, we performed whole-cell recordings after expressing IR76b in HEK293T cells (fig. S7, A and B). The IR76b expressing cells showed increased current (IIR76b) relative to control cells (Fig. 4D). The nearly linear current-voltage (I–V) relationship indicated that IIR76b was not strongly voltage-dependent (Fig. 4E). Replacement of the external Cl− with gluconate anions had little effect on IIR76b (fig. S7D). Using bionic conditions, the relative ion selectivity of IR76b was PNa (1.0) = PCs (1.0) > PK (0.4) (Fig. 4E and fig. S7C and E). The Na+ conductance properties of IR76b were similar to NALCN—a mouse Na+ leak channel (20), suggested that IR76b was in a constitutively open state.
The ion conductance of iGluRs is controlled by residues in the third transmembrane (TM3) region that includes YTANLAAFLT (21). In the absence of ligand, the channels are closed. A spontaneous A288T mutation in TM3 of mouse GluRδ2 (Lurcher mutation; GluRδ2Lc) disrupts the closed conformation, resulting in a constitutive Na+ conductance (22). Notably, IR76b harbored a threonine (T293), in nearly the same position as the Lurcher substitution (A288T; Fig. 4F, fig. S7, F – G). A threonine is absent in corresponding positions of other fly IRs and mammalian iGluRs (Fig, 4F). Therefore, we postulated that T293 enabled IR76b to be fixed in an open Na+-permeable state. To test this idea, we replaced IR76b with IR76bT293A and determined the effects of this substitution in vivo and in vitro. When expressing UAS-Ir76bT293A using the Gr5a-Gal4, we did not detect a salt response in L2 sensilla (Fig. 4A). Moreover, the T293A mutation greatly attenuated the constitutive current in HEK293T cells (Fig. 4, D and E).
To explain how the fly uses IR76b to detect salt, we propose that IR76b is a Na+ leak channel, and is effective due to the unusual extracellular cation composition bathing the GRNs. Different from the body hemolymph that contains high Na+, the endolymph that bathes insect chemosensory neurons appears to have a lower Na+ concentration that is similar to the intracellular levels (23). Under resting conditions, there may be little Na+ conductance. After consuming Na+-containing foods, the Na+ levels in the endolymph rises, driving Na+ influx through IR76b. Excitation of salt-attractive GRNs induces the animals to consume salt. Loss of IR76b selectively impaired the attractive pathway, making the otherwise attractive low salt become to become aversive.
Our work establishes that the salt attractive pathway relies on a type of Na+ permeable channel not previously known to function in taste, and this channel, IR76b, bears no relationship to ENaC channels that are required for sensing low salt in mice (24). Some ENaC channels may be constitutively active (25), and lead to depolarization of taste receptor cells following a rise in cation levels at the cell surface (26). Thus, despite the divergence between fly IRs and mammalian ENaC channels, they may mediate salt taste through similar mechanisms. Our competition model for low and high salt taste detection may represent a widely used mechanism for salt taste coding in other animals, including mammals.
Supplementary Material
Acknowledgments
This project was supported by a grant to C.M from the NIDCD (DC007864).
Footnotes
References and notes
- 1.Nakamura M, Baldwin D, Hannaford S, Palka J, Montell C. J Neurosci. 2002;22:3463. doi: 10.1523/JNEUROSCI.22-09-03463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan R, Rodrigues V. J Exp Biol. 1991;157:161. doi: 10.1242/jeb.157.1.161. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, et al. Neuron. 2003;39:133. doi: 10.1016/s0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 4.Shanbhag SR, Park SK, Pikielny CW, Steinbrecht RA. Cell Tissue Res. 2001;304:423. doi: 10.1007/s004410100388. [DOI] [PubMed] [Google Scholar]
- 5.Hiroi M, Meunier N, Marion-Poll F, Tanimura T. J Neurobiol. 2004;61:333. doi: 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- 6.Vosshall LB, Stocker RF. Annu Rev Neurosci. 2007;30:505. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 7.Hodgson ES, Lettvin JY, Roeder KD. Science. 1955;122:417. doi: 10.1126/science.122.3166.417-a. [DOI] [PubMed] [Google Scholar]
- 8.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. Neuron. 2011;69:258. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meunier N, Marion-Poll F, Rospars JP, Tanimura T. J Neurobiol. 2003;56:139. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- 10.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Neuron. 2007;56:503. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montell C. Curr Opin Neurobiol. 2009;19:345. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujishiro N, Kijima H, Morita H. J Insect Physiol. 1984;30:317. [Google Scholar]
- 13.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Cell. 2009;136:149. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croset V, et al. PLoS genetics. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron P, Hiroi M, Ngai J, Scott K. Nature. 2010;465:91. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. Cell. 2010;141:536. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Singhvi A, Kong P, Scott K. Cell. 2004;117:981. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Thorne N, Chromey C, Bray S, Amrein H. Curr Biol. 2004;14:1065. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Abuin L, et al. Neuron. 2011;69:44. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu B, et al. Cell. 2007;129:371. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Sobolevsky AI, Rosconi MP, Gouaux E. Nature. 2009;462:745. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo J, et al. Nature. 1997;388:769. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 23.Kaissling K-E, Thorson J. In: Receptors for Neurotransmitters, Hormones, and Pheromones in Insects. Sattelle DB, Hall LM, Hildebrand JG, editors. Elsevier; Amsterdam: 1980. pp. 261–282. [Google Scholar]
- 24.Chandrashekar J, et al. Nature. 2010;464:297. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald FJ, Price MP, Snyder PM, Welsh MJ. Am J Physiol. 1995;268:C1157. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- 26.DeSimone JA, Heck GL, DeSimone SK. Science. 1981;214:1039. doi: 10.1126/science.7302576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.