Abstract
Perlecan/HSPG2, a heparan sulfate proteoglycan typically found at tissue borders including those separating epithelia and connective tissue, increases near sites of invasion of primary prostatic tumors as previously shown for other proteins involved in desmoplastic tissue reaction. Studies of prostate cancer cells and stromal cells from both prostate and bone, the major site for prostate cancer metastasis, showed that cancer cells and a subset of stromal cells increased production of perlecan in response to cytokines present in the tumor microenvironment. In silico analysis of the HSPG2 promoter revealed two conserved NFκB binding sites, in addition to the previously reported SMAD3 binding sites. By systematically transfecting cells with a variety of reporter constructs including sequences up to 2.6 kb from the start site of transcription, we identified an active cis element in the distal region of the HSPG2 promoter, and showed that it functions in regulating transcription of HSPG2. Treatment with TNF-α and/or TGFβ1 identified TNF-α as a major cytokine regulator of perlecan production. TNF-α treatment also triggered p65 nuclear translocation and binding to the HSPG2 regulatory region in stromal cells and cancer cells. In addition to stromal induction of perlecan production in the prostate, we identified a matrix-secreting bone marrow stromal cell type that may represent the source for increases in perlecan in the metastatic bone marrow environment. These studies implicate perlecan in cytokine-mediated, innate tissue responses to cancer cell invasion, a process we suggest reflects a modified wound healing tissue response co-opted by prostate cancer cells.
Keywords: Perlecan/HSPG2, Prostate Cancer, TNF-α, TGFβ, Cytokines, Gene Expression
Introduction
Prostate cancer progression is marked by increasing interaction of stroma with the tumor, and an accompanying phenotypic shift of associated stromal cells first in the prostate then at sites of metastasis [Dakhova et al., 2009]. Activated stromal cells involved in such desmoplastic reactions are variously named cancer-associated fibroblasts, tumor-associated fibroblasts, or reactive stromal cells [Olumi et al., 1999]. A hallmark of desmoplastic stroma in cancer [Coulson-Thomas et al.] is deposition of a fibrotic matrix that closely mimics the wound healing response including the heparan sulfate proteoglycan, perlecan, encoded by the gene HSPG2 [Yamazaki et al., 2004]. The parallels between the actions of the fibroblasts in wound healing with the persistence of reactive stromal cells during disease has led some to describe cancer as “the wound that never heals” [Schafer and Werner, 2008]. Inflammatory cell recruitment is crucial to the process of wound healing [DiPietro, 1995], and is inextricably woven into tumor progression in prostate cancer [Comito et al., 2013]. Cytokines and growth factors produced at the wound [Kohyama et al., 2004; Siegbahn et al., 1990] or the tumor [Shaw et al., 2009] recruit and activate stromal cells whose role is to produce extracellular matrix (ECM) to encase the site of the perceived wound. The ECM deposition profile of proteins present in reactive stroma includes fibronectin, collagens, and various proteoglycans [Brown et al., 1999; Lagace et al., 1985]. Heparan sulfate proteoglycans, such as perlecan, facilitate growth factor delivery during tissue remodeling or repair [Jung et al., 2013; Zcharia et al., 2005] in addition to filling various extracellular scaffolding [Farach-Carson and Carson, 2007], adhesive [Chen et al., 2005], and boundary setting [Farach-Carson et al., 2013] roles that establish tissue function.
Perlecan is a large (~200 nm, 400–800 kDa) [Farach-Carson and Carson, 2007] heparan sulfate proteoglycan found in all basement membranes [Yurchenco et al., 2002]. It is particularly abundant in the bone marrow, where it is the predominant heparan sulfate proteoglycan [Schofield et al., 1999], and in cartilage, where it resists vascular invasion [Brown et al., 2008]. The perlecan core protein has a modular structure that shares homology with other ECM proteins [Murdoch et al., 1992] while the attached heparan sulfate chains function as a reservoir for growth factors useful in wound healing [D'Souza et al., 2008; Savore et al., 2005; Yang et al., 2005]. The scaffolding function of the core protein [Behrens et al., 2012; Farach-Carson and Carson, 2007] reinforces the barrier function of the basement membrane – important to denying invasion and metastasis of carcinoma in situ [Terranova et al., 1986]. Perlecan helps promote prostate cancer cell viability [Savoré et al., 2005] and is part of the reactive stroma gene expression profile seen in some cancers [Sabit et al., 2001]. HSPG2 gene expression also increases in bone after fracture [Wang et al., 2006]. Given that bone marrow is both a perlecan-rich environment [Schofield et al., 1999] and the predominant site of prostate cancer metastasis [Bubendorf et al., 2000], it is of interest to study regulation of HSPG2 gene expression in the context of bone marrow stromal cells, as well as in prostate stromal cells near sites of tumor. Because prostate cancer cells encountering tumor stroma undergo epithelial mesenchymal transformation (EMT) [Zhau et al., 2008], and EMT is associated with expression of ECM [Freire-de-Lima et al., 2011], perlecan production by the prostate cancer cells themselves also is of interest.
A previous study examined the role of local cytokines such as transforming growth factor beta (TGFβ) or tumor necrosis factor alpha (TNF-α) on HSGP2 expression in the tumor microenvironment [Iozzo et al., 1997], a finding not yet examined in prostate cancer although the role of these cytokines in this disease is clear [Dayyani et al., 2011; Yang et al., 2010]. The proximal promoter region of the perlecan gene contains a functional binding site for SMAD3 [Iozzo et al., 1997]. In the context of bone metastasis, we previously found that TGFβ signaling is dynamically regulated in the bone marrow tumor stroma through down regulation of the TGFβ co-receptor endoglin, which also reduces signaling through SMAD2/3, suggesting that it is unlikely that SMAD3 elements in the perlecan promoter are directly responsible for increases in perlecan expression in the desmoplastic tumor microenvironment [O'Connor et al., 2007]. Both TGFβ and TNF-α can recruit and activate immune and stromal cells at primary and secondary tumor sites [Andrades et al., 1999; Postlethwaite and Seyer, 1995], triggering further increases in inflammatory cytokines and increasing ECM production in an attempt to heal the perceived wound. TNF-α, produced by many immune cells, induces apoptosis in the human bone marrow hematopoietic cell line, HS-5, and in some cancer cells, but not in the HS-27a bone marrow stromal cell line [Byun et al., 2005; Sumitomo et al., 1999]. TNF-α signaling, acting through NFκB, activates transcription of many target genes including that encoding the ECM protein, tenascin [Nakoshi et al., 2008], but its ability to activate HSPG2 gene expression has never been examined. In this study, we systematically examined HSPG2 expression in the tumor microenvironment and by prostate and bone marrow stromal cells in the presence of cytokines, TNF-α and/or TGFβ. We present evidence for inflammatory cytokine-mediated stimulation of HSPG2 expression and perlecan secretion, with implications for wound healing and cancer progression.
Materials & Methods
Immunohistochemistry
Formalin fixed, paraffin embedded prostate cancer and normal adjacent tissue sections were processed as previously described [Tuxhorn et al., 2002] with the following modifications. Antigen retrieval was achieved by incubating the tissue in 0.1 mg/mL bovine testicular hyaluronidase (Sigma-Aldrich - St. Louis, MO) and tissue was blocked with Background Punisher (BioCare Medical, Concord, CA). Anti-perlecan HSPG2 Prestige Rabbit polyclonal antibody (Sigma) was diluted 1:100 in antibody diluent (Ventana Medical Systems - Tucson, AZ) and incubated overnight at 4°C. Tissue was incubated with the Diagnostic Biosystems (Pleasanton, CA) Polymer Penetration Enhancer and incubated with Anti-mouse/Rabbit PolyVue HRP (Diagnostic Biosystems). Tissues were mounted in Cytoseal 60 (Fisher Scientific, Hampton, NH).
Cell Culture
The bone marrow stromal cell lines HS-5 and HS-27a, the prostate cancer cell lines LNCaP and C4-2B and the prostate stromal cell line WPMY-1 were cultured as previously described [O'Connor et al., 2007; Webber et al., 1999]. Although isolated from the same bone marrow aspirate, the hematopoietic stem cell-supporting HS-5 cell line secretes many growth factors and cytokines, and the HS-27a cell displays a structural secretory profile, including rich production of ECM components [Roecklein and Torok-Storb, 1995]. The prostate stromal cell line, WPMY-1, (ATCC), which expresses α- smooth muscle actin characteristic of reactive stromal cells [Webber et al., 1999], was used to represent stromal cells at the site of the primary tumor. LNCaP cells originate from a lymph node metastasis of prostate cancer and C4-2B cells are adapted to growing in the bone marrow [Thalmann et al., 2000].
For all treatments, cells were plated into 6-well plates and allowed to grow for 48 hr (until 90% confluent). Media was then changed to 1% (v/v) serum and antibiotic-free medium overnight, and then treatments initiated the next day. Recombinant TNF-α produced by E. coli (Roche Applied Science – Indianapolis, IN - #11371843001) was stored at −20°C at 10 ng/µL stock solution, and cell treatments used a final concentration of 10 ng/mL. TGFβ1 (R&D Systems Inc. – Minneapolis, MN. #100-B) purified from human platelets was reconstituted in 4 mM HCl and 0.1% (w/v) bovine serum albumin (BSA) at 10 ng/mL. TGFβ1 treatments were carried out at 10 ng/mL. BSA was dissolved in phosphate buffered saline (pH 7.4) at 1 mg/mL to serve as the vehicle control.
Plasmid Construction
Genomic DNA was purified from HS27a cells using the DNeasy Blood & Tissue kit (Qiagen - Valencia, CA). The 2627 and 2,393 base pair upstream region (hereafter referred to as the 2.6 and 2.4 kb HSPG2 promoter constructs) of the HSPG2 proximal promoter was amplified by genomic polymerase chain reaction (PCR) (for primer pairs see supplementary table I). This PCR product then was cloned into pCR®2.1 TOPO (Life Technologies). This insert ligated into the promoterless pGL4.10 firefly luciferase reporter vector (Promega - Madison, WI). Site directed mutagenesis was performed using the Quickchange II Site-Directed Mutagenesis Kit (Agilent – Santa Clara, CA) according to the manufacturer’s instructions (for primers see supplementary table I).
Transient Transfection
Cells were transiently transfected with luciferase constructs using Lipofectamine™ 2000 reagent (Invitrogen) according to the manufacturer’s instructions. A ratio of 1 µg of total DNA to 2 µL (for HS-27a and HS-5 cells) or 3 µL (for WPMY-1, C4-2B, and LNCaP cells) of Lipofectamine was used for each well in six-well culture plates. For experiments using the perlecan promoter constructs, the control Renilla luciferase was driven by the thymidine kinase promoter in a pHRG-TK vector (Promega). For experiments using insertions of HSPG2 promoter elements into pGL4.74 with the HSV-TK promoter driving expression of the Renilla luciferase, firefly luciferase expression driven by the CMV promoter was used as the control luciferase.
Luciferase reporter assay
The Dual-Luciferase® Reporter Assay system (Promega) was used to measure luciferase activity in transiently transfected cells, according to the manufacturer’s instructions.
Quantitative PCR
Quantitative PCR was performed as previously described [Savore et al., 2005]. Reverse transcription reactions were performed using the QScript cDNA Supermix kit (Quanta Biosciences, Gaithersburg, MD). The final concentration of primers (sequences found in supplementary table I) was 200 nM (HSPG2) and 400 nM (ACTB). IQ SYBR Mix (Quanta Biosciences) was used according to the manufacturer’s instructions, to equal half of the final reaction volume. The thermal cycling program was as follows: 95°C for 3 min, then 40 cycles of the following two steps: 95°C for 30 seconds, 63°C for 45 seconds. Relative transcript levels were determined by the Pfaffl method, using ACTB as a constitutively expressed gene control.
Immunoblotting for secreted perlecan protein
Dot blotting was performed to analyze accumulation of extracellular perlecan as previously described [Savore et al., 2005]. For dot blotting, equal volumes of medium were loaded into each well. Medium was diluted 1:10 in phosphate buffered saline (PBS), and 100 µL of the diluted solution were loaded into each well. Perlecan antibody A7L6 (Santa Cruz Biotechnology, Santa Cruz, CA) was diluted 1:5,000 in blocking buffer and incubated with the membrane overnight at 4°C while shaking.
WST-1 assay for metabolically active cell content
Assays were performed in parallel with conditioned medium collection. First, conditioned medium was removed for use in dot blotting for perlecan secretion. Serum- and phenol red-free medium was returned to the well, with 10% (v/v) WST-1 reagent (Roche) added. Cells were incubated for 1 hr at 37°C and then medium was transferred to a 96-well plate, where dual wavelength measurements were taken at 440 nm (measurement wavelength) and 610 nm (reference wavelength). Per the manufacturer’s instructions, only metabolically active cells will metabolize this salt at the cell surface, resulting in a spectrophotometric shift in the medium. This allows determination of a ratio of metabolically viable cells during treatment compared to cells treated with the vehicle control.
TNF-α enzyme-linked immunosorbent assay (ELISA)
Cells were grown to 80% confluence in normal growth medium, then medium was removed and cells were prepared for medium collection during which time they typically reached confluency. Cells in 6-well culture plates continued to grow for 24 hrs in 1 mL serum-free medium, and this conditioned medium was collected and assayed for TNF-α production. At the same time, the cells were trypsinized from the plates, and counted using a hemocytometer. TNF-α levels were assayed and quantified using the human TNF-α ELISA kit (Thermo Scientific) according to the manufacturer’s instructions.
Immunostaining for NFkB
Cells were grown in 8-well chamber slides (NUNC – Thermo Scientific) in normal growth medium and subsequently treated in serum-free medium supplemented with TNF-α (10 ng/mL) for 30 min at 37°. Cells were rinsed with PBS and fixed with paraformaldehyde. After rinsing, they were permeabilized with Triton X-100 (0.2% v/v) at room temperature for 10 min, blocked with 3% (w/v) BSA for 40 min at room temperature then incubated with anti-p65 (Santa Cruz Biotechnology – sc109) in a 1:200 dilution at 4°C overnight. Slides then were washed and incubated with a goat anti-rabbit Alexa-Fluor 488 secondary antibody (Invitrogen) in a 1:200 dilution, DAPI (Invitrogen) in a 1:500 dilution, and Phalloidin 633 (Invitrogen) at a 1:500 dilution for 40 min at room temperature. Slides were mounted in SlowFade Gold (Invitrogen) and imaged using a LSM 710 confocal microscope (Zeiss, Oberkochen, Germany).
Chromatin Immunoprecipitation (ChIP)
The Chromatrap® kit (Porvair Sciences – Leatherhead, Surrey, England) was used for ChIP assays. LNCaP cells were grown to ~70% confluency and serum-withdrawn overnight, then treated for 24 h with TNF-α (10 ng/mL) or vehicle control prior to collection of chromatin. Fixation, DNA shearing and ChIP were performed following the manufacturer’s instructions. Antibodies included the anti-p65 SC-109X antibody (Santa-Cruz), as well as the control antibodies anti-polII SC-9001 (Santa Cruz) and anti-H3K4me3 39159 (Active Motif – Carlsbad, CA). Primer sequences used to amplify various gene regulatory regions are included in supplementary table 1. The NFKBIA gene promoter served as a positive control for p65 immunoprecipitation, and the GAPDH promoter was used a positive control for RNA polymerase II immunoprecipitation. The HBB gene was the negative control for nonspecific pulldown by either the antibody or adsorption by the column beads.
Statistics
All reporter assays, quantitative PCR and dot blotting were analyzed using a one-way ANOVA followed by Tukey’s post-test. Tissue microarray data were analyzed by the Cedars Sinai statistics group funded through the NIH/NCI P01CA098912. Stain concentration was normalized using square root transformation. These values were analyzed by ANOVA and two-sample test.
Results
Perlecan accumulates in desmoplastic stroma of PCa
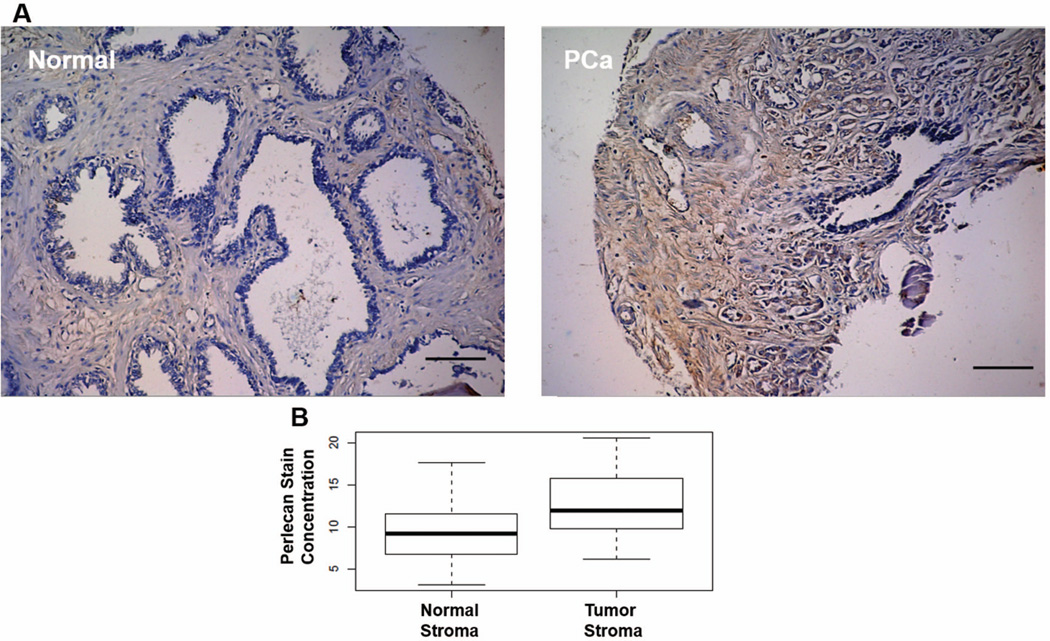
Figure 1A shows immunostaining for perlecan in paraffin-embedded sections of PCa (right panel) and normal adjacent tissue (left panel) from the same patient. In sections of normal tissue, perlecan is expressed diffusely in the stroma. In this section containing Gleason grade 4 cancer, stromal cells surround and infiltrate the tumor, a hallmark of reactive stroma. Perlecan immunostaining surrounding these stromal cells is intense, noted by the rich brown color, in contrast to what is seen in normal stroma. The increase in perlecan deposition in tumor stroma versus normal stroma was statistically significant in this representative microarray (p=.0018, Figure 1B) comprising tissue from fifty-seven patients. This finding adds perlecan to the growing list of ECM proteins deposited into the desmoplastic stroma formed at primary sites of prostate cancer.
Figure 1. Perlecan accumulates in the reactive stroma of prostate cancer.
Perlecan staining (brown) with nuclear counterstain (blue) in Gleason grade 4 prostate cancer (PCa, right panel) and normal adjacent tissue (Normal, left panel) from the same patient. Increased deposition of perlecan is observed in the regions surrounding PCa when compared to regions of normal morphology. Quantification of perlecan staining in the tissue microarray is shown in figure 1B. This microarray included 34 sections of normal prostate and 23 sections of prostate cancer. Perlecan staining in normal stroma was compared to perlecan staining in tumor stroma in each of these sections. Perlecan staining was significantly higher in tumor stroma than in normal stroma with a p value of 0.0018. Scale bar = 100 µm.
Perlecan secretion and HSPG2 mRNA expression increase in response to cytokine treatment
Cells representing tumor and stromal compartments of the tumor microenvironment were treated with TNF-α and TGFβ, alone and in combination, and perlecan production and HSGP2 gene expression were assessed. Because of the size and heterogeneity of the perlecan proteoglycan (~900 kDa) western blots are very difficult to use for quantification of perlecan protein and dot blotting is the preferred method.
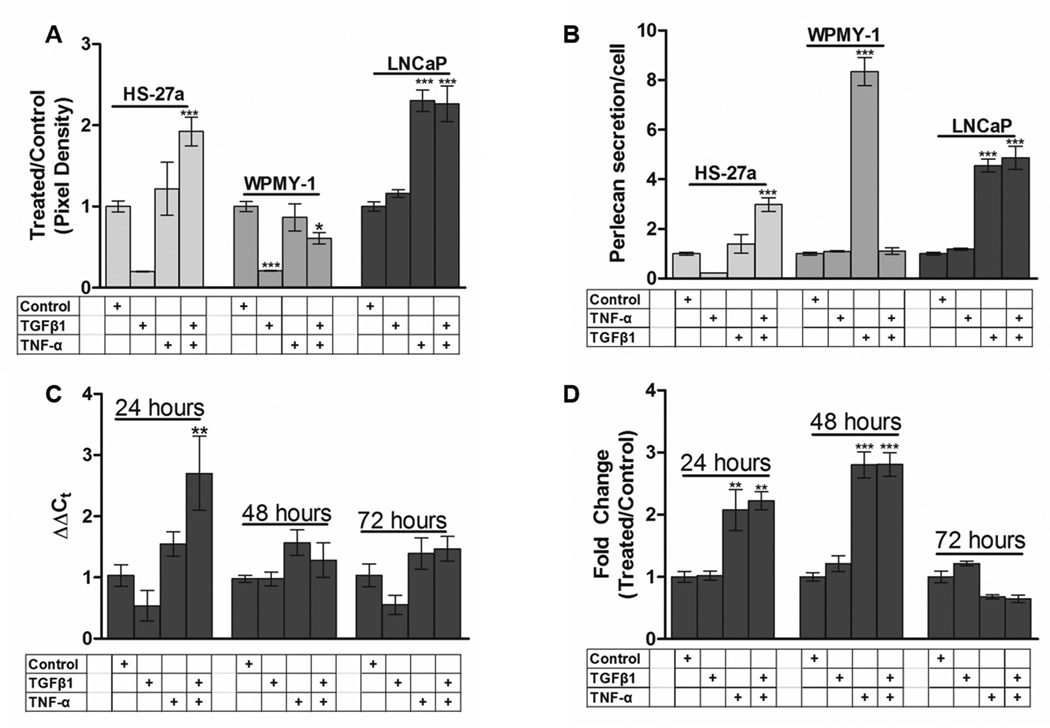
When cell populations of the prostate stromal cell line, WPMY-1, were treated with TNF-α and TGFβ (Figure 2A), we observed a dramatic loss of viable cells compared to controls (supplementary figure S1). For those surviving WPMY-1 cells, there was a clear increase in perlecan production on a per cell basis in response to TNF-α, but not TGFβ, treatment. In HS-27a bone marrow stromal cells, perlecan secretion was increased in response to TNF-α treatment alone, and even more so when cells received both TNF-α and TGFβ (Figures 2A,B). Thus, for both cells resembling the stromal cells present in the prostate (WPMY-1) or bone marrow (HS-27a), TNF-α increased perlecan secretion within 48 hr of treatment.
Figure 2. Expression of HSPG2 mRNA and perlecan increase in HS-27a and LNCaP cells during TGFβ1 and TNF-α treatment.
Medium was collected and perlecan content assessed by quantitative dot blot as described in Materials and Methods (A). The fold change in perlecan secretion shown in (A) was divided by the live cell number to determine perlecan secretion/cell (B). A time-course treatment was performed using LNCaP cells and cells were collected every 24 hr for 72 hr. Steady state HSPG2 mRNA levels were assessed (C) by qRT-PCR as described in Materials and Methods. Conditioned medium was collected from LNCaP cells over the same time periods, and protein assessed by dot blot as above (D). Data is presented as the ratio of densitometric value of treated medium over the value for vehicle controls set to 1.0 (A,B,D) or Delta delta Ct (C). Bars represent the mean +/− SEM, *, p<.05 vs control, **, p<.01 vs control, ***, p<.001 vs control. At least three biological replicates were performed for each condition.
During the course of these experiments, we observed that the stromal cell response to TNF-α in terms of perlecan protein production, mRNA levels and reporter activity diminished over time with increasing passage number, e.g. see response level in figure 4 vs response level in figure 6. These non-transformed stromal cells appear to change phenotypically in cell culture during prolonged passage, making this response difficult to measure once outside the responsive window. For this reason, experiments were performed routinely with lower passage (<20) cells.
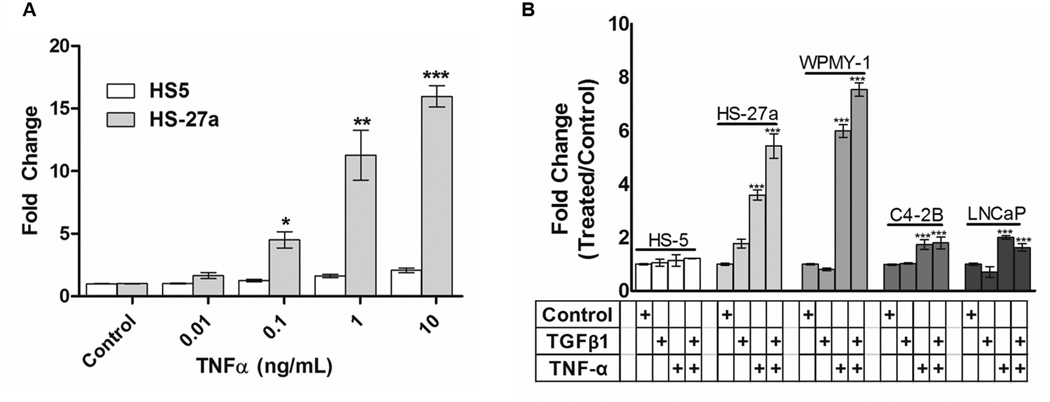
Figure 4. HSPG2 promoter activity increases in response to TNF-α in a dose-dependent manner.
Cells were transfected with the 2.6 kb reporter plasmid shown in 3A, and treated for 24 hr with TNF-α at the indicated final concentrations (A), before measuring luciferase activity in lysed cells as described in the text. Concentrations above10 ng/ml did not further increase activity (not shown). For experiments shown in (B), cells were treated with TGFβ1 (10 ng/ml), TNF-α (10 ng/ml) or a combination of the two, then luciferase activity was measured as described in Materials and Methods. Data are reported as the fold change observed between cytokine treated and vehicle treated cells. Bars represent the mean +/− SEM, *, p<.05 vs .01, control, **, p<.001 vs .1, .01, control, ***, p<.001 vs 1, 0.1, 0.01, control. At least three biological replicates were performed for each condition.
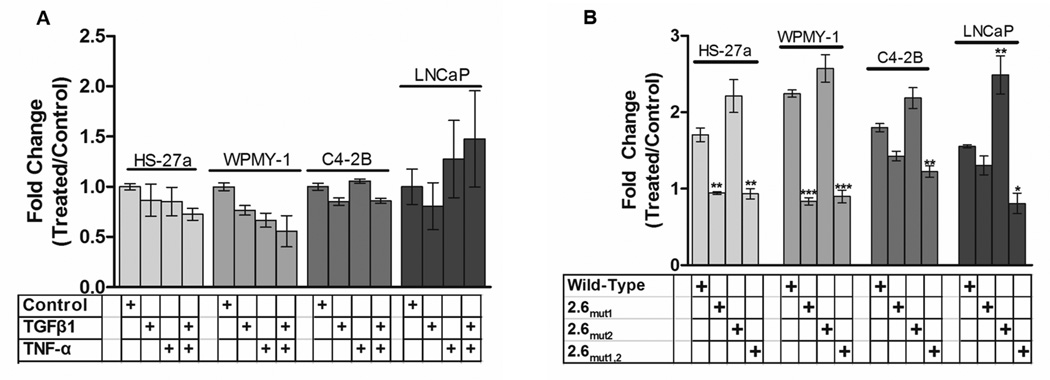
Figure 6. Deletion of the distal 200 base pairs of the HSPG2 promoter or mutation of NFκB consensus elements eliminates responsiveness to TNF-α and TGFβ1.
Cells in culture were transfected with the 2.4 kb promoter seen in Figure 3B, or constructs in which the two NFκB sites were mutated as described above and responses to each cytokines alone (10 ng/ml) or in combination (each at 10 ng/ml) were measured at 24 hr of treatment. Panel A shows that removal of the entire region containing both sites eliminates responses to cytokines in all four cell lines shown. Panel B shows that 2.6mut1 or 2.6mut1,2, but not 2.6mut2, loses activity, hence response to TNF-α is associated with the more distal of the two putative NFκB sites. Bars represent the mean +/− SEM. **, p<.01 vs wild-type, ***, p<.001 vs wild type. At least three biological replicates were performed for each condition.
LNCaP prostate cancer cells increased perlecan protein secretion during 48 hr treatment with TNF-α (10 ng/mL - 588 pM) but not TGFβ (10 ng/mL - 781 pM) (figure 2A,B), concentrations consistent with those used in other studies[Iozzo et al., 1997; Schmidt et al., 2006; Yagi et al.]. Per cell secretion of perlecan by LNCaP cells was not increased further by TGFβ either alone or when TNF-α was present. LNCaP cells treated with TNF-α or TNF-α combined with TGFβ increased HSPG2 mRNA levels after 24 hr, which returned to basal levls by 72 hr (figure 2C). The addition of TGFβ magnified the mRNA response at 24 hr, but this effect was not seen at the level of absolute protein secretion at either 24 or 48 hr (figure 2D). As with the mRNA levels, protein secretion returned to basal levels by 72 hr. Thus for LNCaP cells, TNF-α is able to acutely and transiently increase perlecan production with increases in mRNA preceding changes at the protein level that are best seen within 48 hr of treatment.
The HSPG2 promoter has conserved NFκB binding elements more than 2.4 kb upstream of the start site of transcription
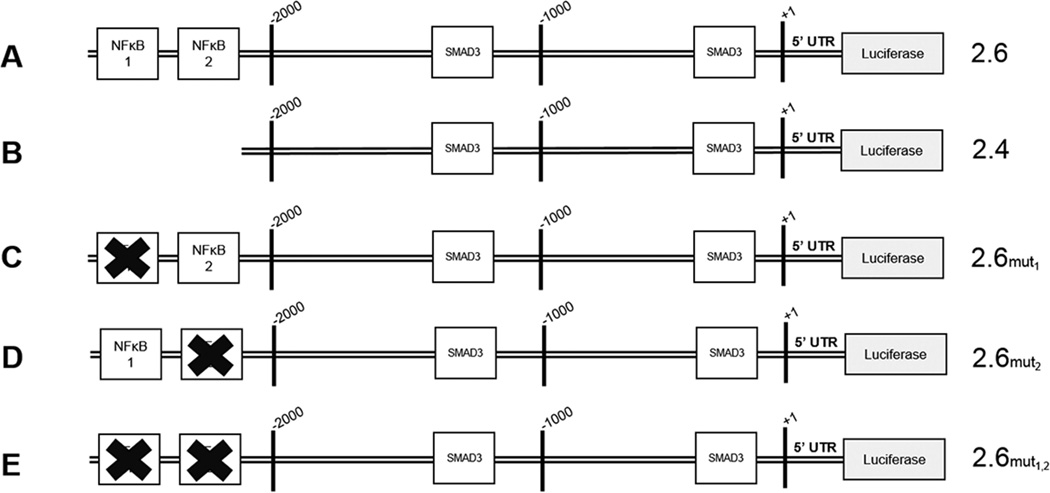
In silico analyses of the HSPG2 promoter revealed multiple potential transcription factor binding sequences within 2.6 kb upstream of the start site of transcription including those for NFκB and Smad3 (Figure 3). This 2.6 kb of upstream promoter sequence plus 79 bp of the 5' untranslated region of the HSPG2 transcript were cloned into the reporter vector used for the majority of these experiments (Figure 3A). Given the responses to TNF-α seen in a variety of cells, it was interesting to find two conserved putative binding sites for NFκB that were located more than 2.4 kb upstream of the transcriptional start site. These two consensus sequences are separated by 28 bp which were both deleted to produce the 2.4 construct (Figure 3B). To determine if these sites are functional NFκB sites, each was mutated singly (Figure 3C,D) then in tandem (Figure 3E) to create three new promoter constructs, labeled 2.6mut1, 2.6mut2, and 2.6mut1,2. As expected, two conserved putative SMAD3 binding sites also were present, one site more than 1 kb upstream of the transcriptional start site and one site approximately 200 bp upstream of the transcriptional start site.
Figure 3. Conserved NFκB consensus elements are present in the HSPG2 promoter.
The schematic depicts the HSPG2 promoter that was used in the luciferase promoter-reporter assays. The full-length reporter used in these studies (A) extends more than 2.6 kb upstream of the transcriptional start site, and includes two putative NFκB binding sites in the most distal 200 base pairs. The 2.4 kb deletion construct (B) was created by removing the distal 200 base pairs of the 2.6 kb promoter. Directed mutants of the putative NFκB sites include mutation of NFκB 1 (C), NFκB 2 (D) or both NFκB sites (E).
Bone marrow stromal, prostate stromal and prostate cancer cell lines secrete very low levels of TNF-α
Although it is expected that the bulk of the TNF-α in the tumor microenvironment is produced by infiltrating immune cells, we sought to measure the baseline levels of cytokine produced by the various cancer and stromal cells used in this study (supplementary figure S2). Over 24 hr, HS-27a, WPMY-1 and C4-2B cells each secreted TNF-α at levels near 40 pg/mL (2.35 pM), a concentration near the lower limit of detection for the assay. LNCaP cells secreted slightly more at ~60 pg/mL (3.5 pM), and HS-5 cytokine-secretory cells secreted ~70 pg/mL TNF-α (4.1 pM). Cells were counted in order to calculate per cell secretion of TNF-α, and each of these cell lines secreted less than 10 fg/mL TNF-α per cell per day. These very low levels of TNF-α secretion were well below those required to increase expression of perlecan. We previously published 24 hr baseline levels of TGFβ produced by these lines: prostate cancer cells (0.4–4.8 pM, 0.01–0.12 pg/mL); bone marrow stromal cells (2.5–7 pM, 0.06–0.175 pg/mL), or prostate fibroblasts (5 pM – 0.125 pg/mL) [Chung et al., 2009]. The endogenous production of TNF-α and TGFβ was more than 100-fold lower than the concentrations of these cytokines used in our routine treatments.
The HSPG2 promoter is activated by TNF-α in a dose-dependent manner
In untreated cells, the 2.6 kb promoter construct shown in Figure 3A demonstrated the highest level of activity in the prostate stromal cell line, WPMY-1, and the bone marrow stromal cell line, HS-27a, and the lowest activity in the prostate cancer cell line, LNCaP (supplementary figure S3). Responsiveness of the HSGP2 2.6 kb promoter to TNF-α treatment was measured by luciferase assay using HS-27a and HS-5 cells (Figure 4A). These data revealed that promoter activity steadily increased during treatment of HS-27a cells with up to 10 ng/mL (588 pM) TNF-α in HS-27a cells. At concentrations above 10 ng/mL, no further increase in promoter-reporter response was observed (data not shown). In contrast, HS-5 cells did not respond to TNF-α at any concentration, consistent with the lack of effect on perlecan mRNA and protein levels (data not shown). We used these cells as a negative control for NFκB activation in immunofluorescence assays and reporter assays. We also studied the response of the HSPG2 promoter to a variety of other growth factors and cytokines (interleukin-1β, insulin-like growth factor 1, β-2 microglobulin, receptor activator of NFκB ligand, interferon γ, and a synthetic Hedgehog pathway activator Hh-Ag1.5) as well as co-culture with other prostate cancer and stromal cell lines. None of these treatments or co-cultures provoked a significant perlecan response in any of the cells tested here (data not shown).
The HSPG2 promoter is active in stromal and prostate cancer cells
Next, TNF-α or TGFβ (each at 10 ng/mL) were added singly or in combination to relevant prostate and stromal cell lines transfected with the 2.6 kb HSPG2 promoter (figure 4B). TNF-α treatment increased HSPG2 promoter activity in HS-27a cells and in the WPMY-1 prostatic stromal cells, but not in the growth factor and cytokine producing bone marrow stromal cells, HS-5, used as a negative control. TNF-α addition to prostatic cancer cell lines, LNCaP and C4-2B, also increased reporter activity, but to a lesser extent than seen in the responsive stromal cell lines. TGFβ alone did not increase promoter activity in any of these cell lines to a statistically significant extent. When added in combination with TNF-α, TGFβ increased the HSPG2 promoter activity in the two responsive stromal cell lines, but not in the cancer cells. Interestingly, the stromal cell response to TNF-α increased in the presence of TGFβ whereas prostate cancer cells showed no additional response to TGFβ.
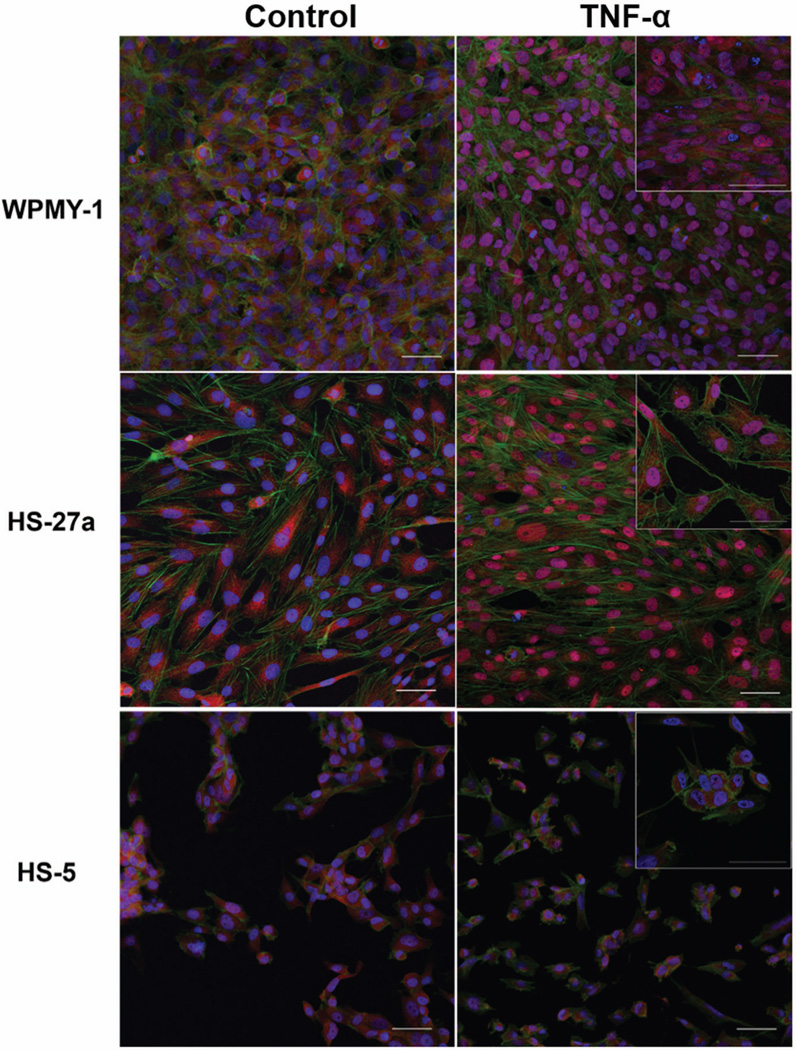
TNF-α treatment of HS-27a and WPMY-1, but not HS-5, stromal cell lines increases NFκB nuclear localization
Given the differential responses of the 2.6 kb promoter in HS-27a, WPMY-1 and HS-5 stromal cells, we sought to determine if this was attributable to inherent cell-specific differences in transcriptional context for the promoter or to differences in the ability to respond to the cytokine itself. As seen in Figure 5, cells in which the HSPG2 promoter responded to TNF-α (HS-27a, WPMY-1) also showed nuclear translocation of NFκB in response to the cytokine. In contrast, HS-5 cells which did not show HSPG2 promoter responsiveness to TNF-α also failed to show a nuclear translocation of NFκB. The antibody blocking peptide and secondary antibody-only background controls were negative for fluorescence, indicating that this signal is specific to p65 (data not shown).
Figure 5. TNF-α treatment triggers nuclear translocation of NFκB in HS-27a and WPMY-1, but not HS-5, cells.
Cells were treated with TNF-α (10 ng/mL) in serum-free medium for 1 hr, permeabilized, and then stained with anti-NFκB as described in Materials and Methods. Cells were imaged by confocal microscopy. Red, NFκB. Green, phalloidin. Blue, DAPI. Scale bars, 50 µm.
HSPG2 promoter responsiveness to TNF-α resides more than 2.4 kb upstream of the start site of transcription in the more distal NFκB binding site
The constructs shown in Figure 3B were used to dissect the nature of the cis element(s) in the HSPG2 promoter that are responsible for the TNF-α response. As shown in Figure 6, neither treatment with TNF-α nor TGFβ nor a combination of the two activated the 2.4 kb promoter-reporter in any of the four cell lines that demonstrated responsiveness of the 2.6 kb promoter (Figure 6A). In these experiments, the full-length promoter construct responded to TNF-α similarly to that seen in figure 4 (data not shown).
To identify the functional cis element(s) in the distal promoter region, the three mutant constructs shown in Figure 3 (2.6mut1, 2.6mut2, 2.6mut1,2) were transfected into the same four cell lines and responses to cytokines measured. In both stromal cell lines, mutation of NFκB site 1 (2.6mut1) or mutation of both NFκB sites completely abrogated the response to TNF-α (figure 6B). In contrast, mutating NFκB site 2 alone (2.6mut2) did not decrease the promoter response, and actually increased the promoter response over that of the wild-type promoter in each of the cell lines (figure 6B). The mutation of both NFκB sites (2.6mut1,2) decreased responsiveness to TNF-α in the prostate cancer cell lines LNCaP and C4-2B but mutation of NFκB site 1 alone (NFκBmut1).
The NFκB HSPG2 regulatory region is bound by p65
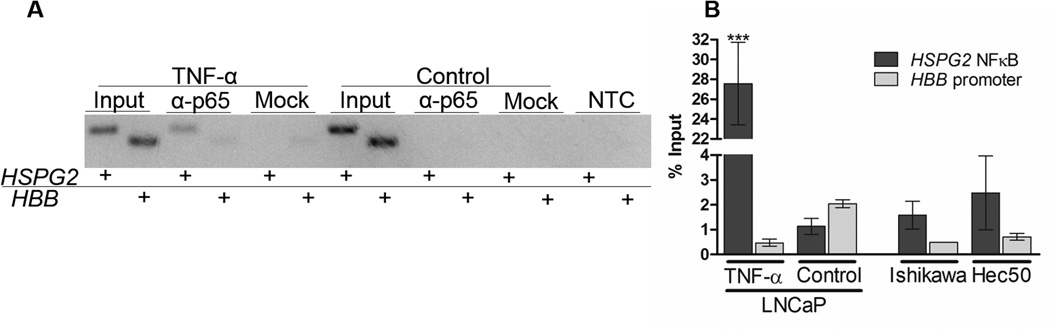
Chromatin immunoprecipitation (ChIP) was performed to validate binding of NFκB to the elements identified above. LNCaP cells were treated with TNF-α for 24 h. After immunoprecipitating with anti-p65 antibody, the HSPG2 promoter was significantly enriched compared to either the internal background control (HBB gene promoter) or relative to cells treated with vehicle alone. Representative endpoint PCR is shown in figure 7A and quantitative PCR is shown in figure 7B. Quantitative PCR data is presented as a percentage of the input chromatin control. The column binding control mock immunoprecipitation and no template controls are shown in the replicate of endpoint PCR, and demonstrate very little amplification. Chromatin from two endometrial cancer cell lines were used as positive controls for NFκB activation, and the HSPG2 regulatory region was immunoprecipitated in these samples as well. Collectively these data demonstrate that NFκB binds to the same region identified in the reporter assays described above.
Figure 7. p65 binds the HSPG2 5' regulatory region in LNCaP cells during TNF-α treatment.
Chromatin immunoprecipitation was used to detect p65 binding the perlecan promoter. The HSPG2 regulatory region is significantly enriched compared to the background control (the HBB promoter) in LNCaP cells treated with TNF-α. Endpoint PCR (panel A) indicates the HSPG2 regulatory region is amplified when p65 is pulled down in the chromatin of TNF-α treated cells, but not in that of cells treated with the vehicle control. This amplification was quantified using quantitative PCR. The HSPG2 regulatory region was also pulled down in Ishikawa and Hec-50 endometrial cancer cells, used as positive controls for NFκB activation. Three biological replicates were performed using LNCaP cells, and one replicate was used for each of the endometrial cancer cell lines.
Discussion
Metastatic tumors may be perceived by the host defense system in similar ways as certain pathogens or foreign bodies that result in in encapsulation of the tumor with dense matrix that resists tissue penetration. The large proteoglycan, perlecan, is a key component of this host defense, and has evolved a unique multi-domain structure that allows it to perform a variety of functions in the desmoplastic stroma [Farach-Carson et al., 2013; Sabit et al., 2001; Terranova et al., 1986]. Here we examined the actions of cytokines present in the tumor microenvironment that are most likely to contribute to perlecan production. We found that prostate cancer cells, prostate stromal cells, and a structural subpopulation of bone marrow stromal cells are programmed to increase HSPG2 mRNA expression and perlecan production in response to s pro-inflammatory cytokine stimulus. While TGFβ, a key factor in connective tissue growth, plays a major role in the stromal response to cancer and induction of fibrosis [Ronnov-Jessen and Petersen, 1993], our work suggests that TNF-α is the primary effector of increased HSPG2 expression in the invasive prostate cancer microenvironment both in the prostate and at sites of metastasis.
While resting stroma has low levels of perlecan, tumor stroma shows high levels of perlecan deposition. In the context of the bone microenvironment, it is interesting that the stromal cell line with a phenotype supporting the structure of the bone marrow microenvironment, represented by HS-27a cells, rather than the cell line with a phenotype supporting hematopoietic progenitor cells, represented by HS-5 cells, were the ones most responsive to TNF-α induction of HSPG2. Bone marrow stromal cells with the structural phenotype (HS-27a) also are resistant to TNF-α induced cell death (Figure S1), in contrast to the secretory bone marrow stromal cell line HS-5 [Byun et al., 2005]. This finding is consistent with these cells being the primary producers of perlecan-rich matrix at sites of bone metastases. Additional perlecan deposition may be provided by the cancer cells themselves, especially if they have undergone epithelial mesenchymal transformation (EMT). This latter notion is supported by our findings that the 2.6 kb perlecan promoter is five times more active (at baseline levels compared to internal controls) in C4-2B cells derived from bone-metastasis than in the parental LNCaP line, despite the fact that basal HSPG2 mRNA levels are similar in these cell lines [Savore et al., 2005].
The highest basal activity of the 2.6 kb promoter was found in the two stromal lines, WPMY-1 and HS-27a. Deletion of the distal 200 base pairs of the promoter decreased baseline reporter activity in all cell lines (ten-fold reduction in stromal cells; five-fold reduction in prostate cancer cells), highlighting the importance of this previously unexplored region in regulation of HSPG2 transcription. The upstream 2.5 kb of the HSPG2 promoter was examined previously and determined to contain all of the elements of the core promoter region [Iozzo et al., 1997]. Since this early report, the human genome has been better annotated and it now made sense to reexamine the promoter using currently available information and tools. Using the ENCODE data track available on the UCSC genome browser (genome.UCSC.edu) we were able to make new inferences concerning the location of the HSPG2 promoter (supplementary figure S4). Using the H3K27Ac mark track (part of the ENCODE data set), it is evident that there are activated histones (associated with activated enhancer elements) located throughout the 40 kilobase intron 1 of the human HSPG2 gene, as well as up to 7 kilobases upstream of the start site of transcription, well beyond the core promoter region denoted by the H3K4me3 histone mark. This suggests that complete regulation of the HSPG2 gene cannot be fully understood until transcriptional regulatory activities in this entire region are appreciated in cell-specific context. It also may explain why addition of the distal 200 base pairs to the promoter construct used in this study increased baseline reporter activity so greatly: important regulatory sequences exist in this heretofore unstudied upstream region.
Along with other inflammatory cytokines, TGFβ and TNF-α both have been implicated in modulation of stromal cell activity at tumor sites and in other pathologies [Rodriguez Perez et al., 2011; Ronnov-Jessen and Petersen, 1993]. While TGFβ is produced locally by stromal cells and other connective tissue cells, TNF-α is secreted by nearby tumor-associated macrophages that appear in the tumor microenvironment in response to tissue stress from the perceived wound. The HSPG2 gene promoter regulating perlecan expression responds to TGFβ [Iozzo et al., 1997], and we expect that local secretion of TGFβ maintains a basal level of perlecan production in stromal and connective tissue cells through an autocrine signaling loop.
NFκB activity plays a role in prostate cancer progression and formation of reactive stroma, but direct effects of this transcription factor on perlecan production at the level of the promoter have never been reported. Here we showed that a consensus NFκB binding sequence in the HSPG2 promoter is a key driver of the TNF-α response in bone marrow stromal and prostate cancer cells. We also showed that in stromal cell lines that respond to TNF-α treatment by increasing perlecan expression, NFκB localizes to the nucleus, whereas in the unresponsive stromal cell line, HS-5, there was no nuclear translocation of NFκB in response to TNF-α treatment. While the reason for the lack of TNF-α responsiveness in the HS-5 cell line is unclear, it nonetheless accounts for the lack of effect on perlecan expression in these cells.
It is of note that the per-cell increase in perlecan secretion in the WPMY-1 cells that survived treatment with TNF-α was much greater than the increase in perlecan secretion in the whole population which was dying off throughout the experiment (Figure 2 panels A & B). This distinction may have important implications in the formation of reactive stroma during cancer invasion. It is appreciated that a subpopulation of stromal fibroblasts reacting to wounding can differentiate into myofibroblasts, which increases their desmoplastic ECM deposition profile [Tuxhorn et al., 2002]. Future work might establish whether the subset of stromal cells in the prostate producing large amounts of perlecan are indeed myofibroblasts.
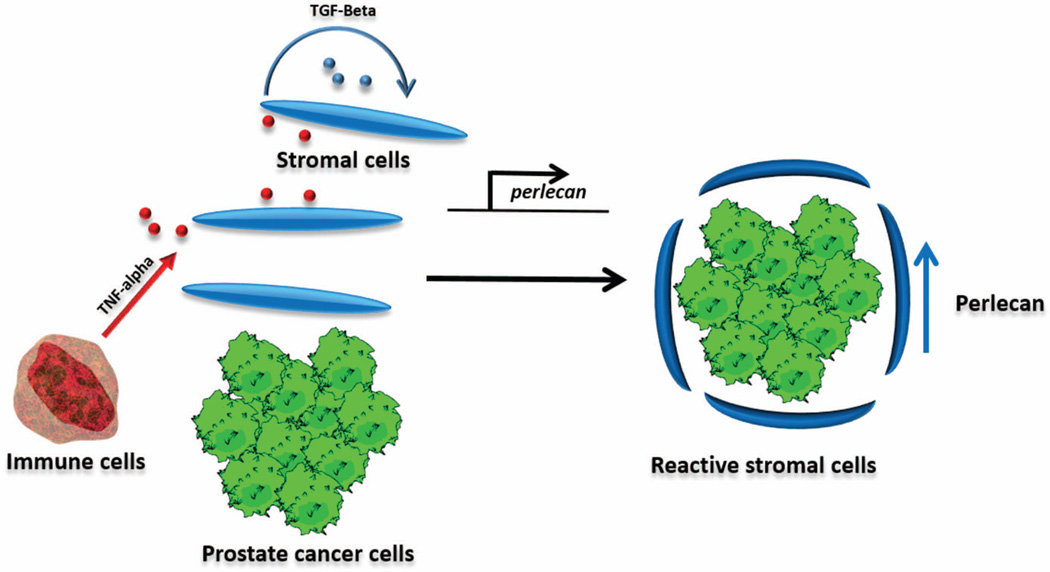
Figure 8 shows a model that explains the up-regulation of HSPG2 in the stromal environment. Our data indicate that none of these stromal or cancer cell types can be the source of TNF-α in the microenvironment. Tumor associated macrophages are known to secrete inflammatory cytokines such as TNF-alpha, and are integral components of tumor progression. This activity in concert with stromal cell autocrine TGFβ signaling could activate stromal cells to secrete perlecan and perform the other functions of cancer-associated fibroblasts. This model may further inform strategies for targeting reactive stroma in prostate cancer.
Figure 8. Model for HSPG2 induction by tumor and immune cell interaction.
In our proposed model, tumor recruitment of immune cells including macrophages results in inflammatory cytokine release. This amongst other cancer cell-derived factors induces stromal activation. Part of this phenotypic program is perlecan secretion.
Supplementary Material
Acknowledgments
We are grateful to Dr. Majd Zayzafoon and the core pathology facility at the University of Alabama at Birmingham for the tissue specimens. We appreciate the support of the core statistics team at Cedars-Sinai Hospital in experimental planning and analysis especially Dr. André Rogatko, Ph.D. (Director, Biostatistics and Bioinformatics, Cedars Sinai, Los Angeles, CA). The authors benefitted greatly from the input and lively biweekly discussions of the entire PO1 team, led by Dr. Leland Chung. Porvair Sciences graciously provided sample Chromatrap® kits for performing the chromatin immunoprecipitation assay. The authors thank Ms. Sharron Kingston for her help and assistance with the preparation and submission of the manuscript.
Grant Support Information: This work was funded by the National Institutes of Health/National Cancer Institute P01CA098912 (MCFC Project 2), and by funds and equipment from Rice University.
References
- Andrades JA, Han B, Becerra J, Sorgente N, Hall FL, Nimni ME. A recombinant human TGF-beta1 fusion protein with collagen-binding domain promotes migration, growth, and differentiation of bone marrow mesenchymal cells. Exp Cell Res. 1999;250:485–498. doi: 10.1006/excr.1999.4528. [DOI] [PubMed] [Google Scholar]
- Behrens DT, Villone D, Koch M, Brunner G, Sorokin L, Robenek H, Bruckner-Tuderman L, Bruckner P, Hansen U. The epidermal basement membrane is a composite of separate laminin- or collagen IV-containing networks connected by aggregated perlecan, but not by nidogens. J Biol Chem. 2012;287:18700–18709. doi: 10.1074/jbc.M111.336073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Alicknavitch M, D'Souza SS, Daikoku T, Kirn-Safran CB, Marchetti D, Carson DD, Farach-Carson MC. Heparanase expression and activity influences chondrogenic and osteogenic processes during endochondral bone formation. Bone. 2008;43:689–699. doi: 10.1016/j.bone.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LF, Guidi AJ, Schnitt SJ, Van De Water L, Iruela-Arispe ML, Yeo TK, Tognazzi K, Dvorak HF. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin Cancer Res. 1999;5:1041–1056. [PubMed] [Google Scholar]
- Bubendorf L, Schopfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31:578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- Byun CH, Koh JM, Kim DK, Park SI, Lee KU, Kim GS. Alpha-lipoic acid inhibits TNF-alpha-induced apoptosis in human bone marrow stromal cells. J Bone Miner Res. 2005;20:1125–1135. doi: 10.1359/JBMR.050302. [DOI] [PubMed] [Google Scholar]
- Chen Y, Shi-Wen X, van Beek J, Kennedy L, McLeod M, Renzoni EA, Bou-Gharios G, Wilcox-Adelman S, Goetinck PF, Eastwood M, Black CM, Abraham DJ, Leask A. Matrix contraction by dermal fibroblasts requires transforming growth factor-beta/activin-linked kinase 5, heparan sulfate-containing proteoglycans, and MEK/ERK: insights into pathological scarring in chronic fibrotic disease. Am J Pathol. 2005;167:1699–1711. doi: 10.1016/s0002-9440(10)61252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SW, Miles FL, Sikes RA, Cooper CR, Farach-Carson MC, Ogunnaike BA. Quantitative modeling and analysis of the transforming growth factor beta signaling pathway. Biophys J. 2009;96:1733–1750. doi: 10.1016/j.bpj.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comito G, Giannoni E, Segura CP, Barcellos-de-Souza P, Raspollini MR, Baroni G, Lanciotti M, Serni S, Chiarugi P. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene. 2013 doi: 10.1038/onc.2013.191. [DOI] [PubMed] [Google Scholar]
- Coulson-Thomas VJ, Coulson-Thomas YM, Gesteira TF, de Paula CA, Mader AM, Waisberg J, Pinhal MA, Friedl A, Toma L, Nader HB. Colorectal cancer desmoplastic reaction up-regulates collagen synthesis and restricts cancer cell invasion. Cell Tissue Res. 346:223–236. doi: 10.1007/s00441-011-1254-y. [DOI] [PubMed] [Google Scholar]
- D'Souza S, Yang W, Marchetti D, Muir C, Farach-Carson MC, Carson DD. HIP/RPL29 antagonizes VEGF and FGF2 stimulated angiogenesis by interfering with HS-dependent responses. J Cell Biochem. 2008;105:1183–1193. doi: 10.1002/jcb.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhova O, Ozen M, Creighton CJ, Li R, Ayala G, Rowley D, Ittmann M. Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res. 2009;15:3979–3989. doi: 10.1158/1078-0432.CCR-08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayyani F, Gallick GE, Logothetis CJ, Corn PG. Novel therapies for metastatic castrate-resistant prostate cancer. J Natl Cancer Inst. 2011;103:1665–1675. doi: 10.1093/jnci/djr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPietro LA. Wound healing: the role of the macrophage and other immune cells. Shock. 1995;4:233–240. [PubMed] [Google Scholar]
- Farach-Carson MC, Carson DD. Perlecan--a multifunctional extracellular proteoglycan scaffold. Glycobiology. 2007;17:897–905. doi: 10.1093/glycob/cwm043. [DOI] [PubMed] [Google Scholar]
- Farach-Carson MC, Warren CR, Harrington DA, Carson DD. Border patrol: Insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. 2013 doi: 10.1016/j.matbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-de-Lima L, Gelfenbeyn K, Ding Y, Mandel U, Clausen H, Handa K, Hakomori SI. Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process. Proc Natl Acad Sci U S A. 2011;108:17690–17695. doi: 10.1073/pnas.1115191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Pillarisetti J, Sharma B, Murdoch AD, Danielson KG, Uitto J, Mauviel A. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming growth factor-beta via a nuclear factor 1-binding element. J Biol Chem. 1997;272:5219–5228. doi: 10.1074/jbc.272.8.5219. [DOI] [PubMed] [Google Scholar]
- Jung M, Lord MS, Cheng B, Lyons JG, Alkhouri H, Hughes JM, McCarthy SJ, Iozzo RV, Whitelock JM. Mast cells produce novel shorter forms of perlecan that contain functional endorepellin: a role in angiogenesis and wound healing. J Biol Chem. 2013;288:3289–3304. doi: 10.1074/jbc.M112.387811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama T, Liu X, Wen FQ, Kobayashi T, Abe S, Rennard SI. IL-4 and IL-13 induce chemotaxis of human foreskin fibroblasts, but not human fetal lung fibroblasts. Inflammation. 2004;28:33–37. doi: 10.1023/b:ifla.0000014709.47056.a9. [DOI] [PubMed] [Google Scholar]
- Lagace R, Grimaud JA, Schurch W, Seemayer TA. Myofibroblastic stromal reaction in carcinoma of the breast: variations of collagenous matrix and structural glycoproteins. Virchows Arch A Pathol Anat Histopathol. 1985;408:49–59. doi: 10.1007/BF00739962. [DOI] [PubMed] [Google Scholar]
- Murdoch AD, Dodge GR, Cohen I, Tuan RS, Iozzo RV. Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). A chimeric molecule with multiple domains homologous to the low density lipoprotein receptor, laminin, neural cell adhesion molecules, and epidermal growth factor. J Biol Chem. 1992;267:8544–8557. [PubMed] [Google Scholar]
- Nakoshi Y, Hasegawa M, Sudo A, Yoshida T, Uchida A. Regulation of tenascin-C expression by tumor necrosis factor-alpha in cultured human osteoarthritis chondrocytes. J Rheumatol. 2008;35:147–152. [PubMed] [Google Scholar]
- O'Connor JC, Farach-Carson MC, Schneider CJ, Carson DD. Coculture with prostate cancer cells alters endoglin expression and attenuates transforming growth factor-beta signaling in reactive bone marrow stromal cells. Mol Cancer Res. 2007;5:585–603. doi: 10.1158/1541-7786.MCR-06-0408. [DOI] [PubMed] [Google Scholar]
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwaite AE, Seyer JM. Identification of a chemotactic epitope in human transforming growth factor-beta 1 spanning amino acid residues 368–374. J Cell Physiol. 1995;164:587–592. doi: 10.1002/jcp.1041640317. [DOI] [PubMed] [Google Scholar]
- Rodriguez Perez CE, Nie W, Sinnett-Smith J, Rozengurt E, Yoo J. TNF-alpha potentiates lysophosphatidic acid-induced COX-2 expression via PKD in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2011;300:G637–G646. doi: 10.1152/ajpgi.00381.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- Ronnov-Jessen L, Petersen OW. Induction of alpha-smooth muscle actin by transforming growth factor-beta 1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab Invest. 1993;68:696–707. [PubMed] [Google Scholar]
- Sabit H, Tsuneyama K, Shimonishi T, Harada K, Cheng J, Ida H, Saku T, Saito K, Nakanuma Y. Enhanced expression of basement-membrane-type heparan sulfate proteoglycan in tumor fibro-myxoid stroma of intrahepatic cholangiocarcinoma. Pathol Int. 2001;51:248–256. doi: 10.1046/j.1440-1827.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Savore C, Zhang C, Muir C, Liu R, Wyrwa J, Shu J, Zhau HE, Chung LW, Carson DD, Farach-Carson MC. Perlecan knockdown in metastatic prostate cancer cells reduces heparin-binding growth factor responses in vitro and tumor growth in vivo. Clin Exp Metastasis. 2005;22:377–390. doi: 10.1007/s10585-005-2339-3. [DOI] [PubMed] [Google Scholar]
- Savoré C, Zhang C, Muir C, Liu R, Wyrwa J, Shu J, Zhau HE, Chung LW, Carson DD, Farach-Carson MC. Perlecan knockdown in metastatic prostate cancer cells reduces heparin-binding growth factor responses in vitro and tumor growth in vivo. Clin Exp Metastasis. 2005;22:377–390. doi: 10.1007/s10585-005-2339-3. [DOI] [PubMed] [Google Scholar]
- Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9:628–638. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Lorkowski S, Seidler D, Breithardt G, Buddecke E. TGF-beta1 generates a specific multicomponent extracellular matrix in human coronary SMC. Eur J Clin Invest. 2006;36:473–482. doi: 10.1111/j.1365-2362.2006.01658.x. [DOI] [PubMed] [Google Scholar]
- Schofield KP, Gallagher JT, David G. Expression of proteoglycan core proteins in human bone marrow stroma. Biochem J. 1999;343(Pt 3):663–668. [PMC free article] [PubMed] [Google Scholar]
- Shaw A, Gipp J, Bushman W. The Sonic Hedgehog pathway stimulates prostate tumor growth by paracrine signaling and recapitulates embryonic gene expression in tumor myofibroblasts. Oncogene. 2009;28:4480–4490. doi: 10.1038/onc.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegbahn A, Hammacher A, Westermark B, Heldin CH. Differential effects of the various isoforms of platelet-derived growth factor on chemotaxis of fibroblasts, monocytes, and granulocytes. J Clin Invest. 1990;85:916–920. doi: 10.1172/JCI114519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo M, Tachibana M, Nakashima J, Murai M, Miyajima A, Kimura F, Hayakawa M, Nakamura H. An essential role for nuclear factor kappa B in preventing TNF-alpha-induced cell death in prostate cancer cells. J Urol. 1999;161:674–679. [PubMed] [Google Scholar]
- Terranova VP, Hujanen ES, Loeb DM, Martin GR, Thornburg L, Glushko V. Use of a reconstituted basement membrane to measure cell invasiveness and select for highly invasive tumor cells. Proc Natl Acad Sci U S A. 1986;83:465–469. doi: 10.1073/pnas.83.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann GN, Sikes RA, Wu TT, Degeorges A, Chang SM, Ozen M, Pathak S, Chung LW. LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate. 2000 Jul 1;44:91–103. doi: 10.1002/1097-0045(20000701)44:2<91::aid-pros1>3.0.co;2-l. 44(2). [DOI] [PubMed] [Google Scholar]
- Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8:2912–2923. [PubMed] [Google Scholar]
- Wang K, Vishwanath P, Eichler GS, Al-Sebaei MO, Edgar CM, Einhorn TA, Smith TF, Gerstenfeld LC. Analysis of fracture healing by large-scale transcriptional profile identified temporal relationships between metalloproteinase and ADAMTS mRNA expression. Matrix Biol. 2006;25:271–281. doi: 10.1016/j.matbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Webber MM, Trakul N, Thraves PS, Bello-DeOcampo D, Chu WW, Storto PD, Huard TK, Rhim JS, Williams DE. A human prostatic stromal myofibroblast cell line WPMY-1: a model for stromal-epithelial interactions in prostatic neoplasia. Carcinogenesis. 1999;20:1185–1192. doi: 10.1093/carcin/20.7.1185. [DOI] [PubMed] [Google Scholar]
- Yagi H, Soto-Gutierrez A, Navarro-Alvarez N, Nahmias Y, Goldwasser Y, Kitagawa Y, Tilles AW, Tompkins RG, Parekkadan B, Yarmush ML. Reactive bone marrow stromal cells attenuate systemic inflammation via sTNFR1. Mol Ther. 18:1857–1864. doi: 10.1038/mt.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Cheng J, Hao N, Takagi R, Jimi S, Itabe H, Saku T. Basement membrane-type heparan sulfate proteoglycan (perlecan) and low-density lipoprotein (LDL) are co-localized in granulation tissues: a possible pathogenesis of cholesterol granulomas in jaw cysts. J Oral Pathol Med. 2004;33:177–184. doi: 10.1111/j.0904-2512.2004.00087.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WD, Gomes RR, Jr, Alicknavitch M, Farach-Carson MC, Carson DD. Perlecan domain I promotes fibroblast growth factor 2 delivery in collagen I fibril scaffolds. Tissue Eng. 2005;11:76–89. doi: 10.1089/ten.2005.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD, Smirnov S, Mathus T. Analysis of basement membrane self-assembly and cellular interactions with native and recombinant glycoproteins. Methods Cell Biol. 2002;69:111–144. doi: 10.1016/s0091-679x(02)69010-7. [DOI] [PubMed] [Google Scholar]
- Zcharia E, Zilka R, Yaar A, Yacoby-Zeevi O, Zetser A, Metzger S, Sarid R, Naggi A, Casu B, Ilan N, Vlodavsky I, Abramovitch R. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. FASEB J. 2005;19:211–221. doi: 10.1096/fj.04-1970com. [DOI] [PubMed] [Google Scholar]
- Zhau HE, Odero-Marah V, Lue HW, Nomura T, Wang R, Chu G, Liu ZR, Zhou BP, Huang WC, Chung LW. Epithelial to mesenchymal transition (EMT) in human prostate cancer: lessons learned from ARCaP model. Clin Exp Metastasis. 2008;25:601–610. doi: 10.1007/s10585-008-9183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.