Summary
Candida albicans is a diploid fungal pathogen lacking a defined complete sexual cycle, and thus has been refractory to standard forward genetic analysis. Instead, transcription profiling and reverse genetic strategies based on Saccharomyces cerevisiae have typically been used to link genes to functions. To overcome restrictions inherent in such indirect approaches, we have investigated a forward genetic mutagenesis strategy based on the UAU1 technology. We screened 4700 random insertion mutants for defects in hyphal development and linked two new genes (ARP2 and VPS52) to hyphal growth. Deleting ARP2 abolished hyphal formation, generated round and swollen yeast phase cells, disrupted cortical actin patches and blocked virulence in mice. The mutants also showed a global lack of induction of hyphae-specific genes upon the yeast-to-hyphae switch. Surprisingly, both arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants were still able to endocytose FM4-64 and Lucifer Yellow, although as shown by time-lapse movies internalization of FM4-64 was somewhat delayed in mutant cells. Thus the non-essential role of the Arp2/3 complex discovered by forward genetic screening in C. albicans showed that uptake of membrane components from the plasma membrane to vacuolar structures is not dependent on this actin nucleating machinery.
Introduction
Candida albicans is among the leading causes of hospital-acquired mycosis with an estimated mortality rate of 38–49% (Pfaller and Diekema, 2007; Koh et al., 2008; Leroy et al., 2009). Although found as a normal commensal in the gastrointestinal tracts and mouths of 70% of the healthy human population, C. albicans can become life-threatening to the increasing population of immunocompromized individuals that result from conditions such as HIV infections and organ transplatation and from patients undergoing broad-spectrum-antibiotic or chemotherapy treatments (Ruhnke and Maschmeyer, 2002; Pfaller and Diekema, 2007). Identifying new genes involved in the virulence of this fungus remains a challenge, especially because genetic manipulation and functional characterization studies in C. albicans have been limited because of its diploidy, the absence of a true sexual cycle and the pathogen’s use of a non-canonical genetic code that translates CUG into a serine instead of a leucine (Kurtz et al., 1988).
The well-studied yeast Saccharomyces cerevisiae has frequently been used as a model to identify functions in C. albicans. For instance, our understanding of morphogenesis, signal transduction, mating and drug resistance in S. cerevisiae has led to successful gene discovery and subsequently to functional analysis in the fungal pathogen (Berman and Sudbery, 2002; Casamayor and Snyder, 2002; Schwartz and Madhani, 2004; Bennett and Johnson, 2005; Berman, 2006; Whiteway and Bachewich, 2007; Cowen, 2008). There are, however, evident limitations to using yeast to define C. albicans processes. For example, clear homologues of many S. cerevisiae proteins have not been identified in C. albicans (Weig et al., 2004; Bennett and Johnson, 2005; Braun et al., 2005). In other cases a homologue of a S. cerevisiae gene was identified in C. albicans, but no functional conservation could be observed (Nicholls et al., 2004; Santos et al., 2004). Moreover, determining function based on homology can become particularly challenging in cases where the yeast model lacks the process under investigation.
One such example is hyphal morphogenesis in C. albicans. Although for many years researchers have relied on principles of bud emergence in S. cerevisiae in an attempt to understand hyphal formation of filamentous fungi such as C. albicans, several features of hyphal growth cannot be explained by extrapolating findings from the model yeast (Harris and Momany, 2004). Overall, this reversible yeast-to-hyphal switch has been intensively studied in the polymorphic fungus (Liu, 2001; Whiteway and Bachewich, 2007). Well-known environmental signals that trigger the morphological transition involve high temperature (37°C), serum, neutral pH, starvation, CO2, adherence and N-acetylglucosamine (GlcNAc) (Gow, 1997; Sudbery et al., 2004). Multiple pathways, for instance the cAMP protein kinase, MAP kinase or the pH-responsive pathways then transmit these signals to activate expression of hyphal-specific genes (Biswas et al., 2007). The importance of the yeast-to-hyphae transition in C. albicans is highlighted by its implication in virulence; C. albicans mutants unable to switch between a yeast and hyphal growth mode are greatly reduced in virulence in mouse infection models (Lo et al., 1997; Laprade et al., 2002; Kumamoto and Vinces, 2005). It has been proposed that while the yeast form contributes to the dissemination of an infection in the host, the hyphal form facilitates penetrating tissue surfaces and escaping host cell internalization (Gow et al., 2002; Whiteway and Oberholzer, 2004).
With the goal of identifying new genes involved in the yeast-to-hyphae transition in C. albicans, we have used an unbiased approach for randomly generating homozygous null mutants directly in the pathogen. Screening 4700 random transposon insertion mutants identified two new genes, ARP2 and VPS52, which are both required for hyphal formation. Surprisingly, in contrast to many other organisms, the highly conserved Arp2/3 complex was not essential for viability or endocytosis in C. albicans, while structural differences in actin organization support the importance of a functional cytoskeletal architecture in permitting morphological switching. These findings underline the potential for forward genetics in the pathogen itself to link functions to genes.
Results
Strategy for an in vivo random forward mutagenesis screen in C. albicans
The UAU1 marker cassette (Enloe et al., 2000) allows the selection of homozygosed mutants in the diploid organism C. albicans. This insertional mutagenesis strategy employs a single transformation in an arg4/arg4, ura3/ura3 double auxotrophic background selecting initially for Arg+/Ura− colonies (genotype orf::UAU1/ORF), then allowing mitotic recombination to homozygose the insertion (genotype orf::UAU1/orf::UAU1), and following this with a second round of selection yielding Arg+/Ura+ segregants (genotype orf::UAU1/orf::URA3) that potentially carry a homozygous disruption at the site of the initial insertion. Mitchell et al. (Davis et al., 2002) have used this UAU1 marker cassette to randomly mutagenize in vitro the genome of C. albicans. This in vitro random mutagenesis resulted in a pool of plasmids each carrying the UAU1 marker cassette flanked by a specific C. albicans genomic DNA sequence. We used the entire pool, rather than a chosen set of characterized UAU1 plasmids, to randomly mutagenize the genome of C. albicans in vivo, and then screened the potentially homozygous orf::UAU1/orf::URA3 inserts for a phenotype of interest.
To assess whether it is possible to extract a desired phenotype by using the entire UAU1 plasmid pool, we created a preliminary set of 300 orf::UAU1/orf::URA3 random insertion mutants and screened for specific auxotrophic mutants. We found one mutant that was unable to grow on SD-Trp media (Fig. 1A). To map this mutation we performed inverse PCR (Ochman et al., 1988) and found the transposon insertion in TRP1 (ORF19.6096), the phosphoribosylanthranilate isomerase gene. TRP1 is an essential enzyme for tryptophan biosynthesis, and mutating this gene results in tryptophan auxotrophy (Ostrander and Gorman, 1994).
Fig. 1. Phenotypic hits resulting from forward genetics screening.

A. Screening for auxotrophs identified TRP1. The picture shows replica-plating of WT (BWP17) and an insertion in TRP1 (Tn::trp1) on SD-complete (SD-c) versus SD-c minus tryptophan (SD-trp) media.
B. Screening for glycerol-sensitive mutants identified ORF19.875. An over night culture was adjusted to OD600 of 0.1 and then 10-fold serially diluted. Two microlitres was then spotted, and plates were incubated for 2 days before being photographed.
C. Examples of phenotypic hits involved in hyphal formation. Hyphae formation was assessed by spotting 10 μl of an over night culture diluted to OD600 0.1 on different hyphae inducting media (10% FBS and Spider) or YPD media. Plates were incubated at 37°C for 2–3 days and photographed. Transposon insertions in CDC39 (Tn::cdc39), VPS52 (Tn::vps52) and ARP2 (Tn::arp2) blocked wrinkle formation on the cell surface on all inducing media. The last column indicates the site of insertion, relative to the start codon, over the entire length of the ORF. See also Fig. S1 for morphology phenotypes associated with the remaining nine potential insertion mutant hits.
In an attempt to link genes to more challenging and less conserved phenotypes between fungal species, such as carbon source utilization (Martchenko et al., 2007a,b; Askew et al., 2009), we expanded the number of orf::UAU1/ORF insertions to about 5000. From these we obtained about 4700 (94%) that generated Arg+Ura+ segregants that represent potential orf::UAU1/orf::URA3 derivatives. Among them we found one insertion mutant (Tn-orf19.875) that cannot grow on glycerol (Fig. 1B). A deletion mutant and revertant strain confirmed the link between growth defects on glycerol and this uncharacterized C. albicans gene that has no obvious homologues in budding and fission yeast (Arnaud et al., 2007). Thus, this preliminary screen for auxotrophs and mutants that can not grow on media containing glycerol as the sole carbon source showed that it is possible to directly link functions to a gene by using the whole pool of UAU1 plasmids.
Screening for random mutants involved in hyphal formation
In order to link genes to a phenotype that is absent S. cerevisiae, we phenotypically analysed the 4700 candidate homozygotes for hyphae formation defects. No convincingly hyper-filamentous colonies were identified. By contrast, we isolated a total of 20 mutants with a strong and consistent hypo-filamentous phenotype and confirmed these phenotypes by retesting a single colony from each mutant. Mapping the 20 hits by inverse PCR showed that 11 insertions were directly in an ORF, while one insertion was mapped in the promoter of ORF19.860. In the remaining eight events we either could not map the insertion because of repeated sequences (six events) or they fell in an intergenic region (two events). Mutants that had a transposon insertion in an intergenic region were excluded from further analysis as it was unclear if the observed phenotype was linked to one of the genes adjacent to the transposon insertion. Figures 1C and S1 summarize hypo-filamentation phenotypes of the 11 ORF and one promoter insertion mutant hits as determined by plate assays. To distinguish between hypo-filamentation and non-filamenting phenotypes, we analysed the 11 ORF and one promoter hypo-filamentation insertion mutants under liquid hyphal inducing conditions [Yeast extract, peptone, dextrose (YPD) media + 10% fetal bovine serum (FBS) at 37°C for 3 h] and found that insertions in CDC39, VPS52 and ARP2 failed to form true hyphae in liquid.
VPS52 and ARP2 deletion mutants do not form hyphae
CDC39 was previously linked to a hypo-filamentation phenotype (Uhl et al., 2003), while ARP2 and VPS52 were newly identified mutants that could not form hyphae under liquid-inducing conditions. We validated these latter two transposon insertion hits by both verifying the absence of the WT band in each transposon mutant and constructing deletion mutants (Fig. S2). To confirm that ARP2 and VPS52 mutants are unable to form true hyphae, we re-analysed polarized morphogenesis under hyphal inducing conditions in liquid culture (Fig. 2). Tn::arp2 (the transposon mutant for ARP2), arp2Δ/Δ (the deletion mutant for ARP2), Tn::vps52 and vps52Δ/Δ did not form true hyphae, while BWP17 (WT strain for all transposon mutants) and SN95 (WT strain for all deletion mutants), both heterozygous mutants (arp2/ARP2, vps52/VPS52) as well as the ARP2 and VPS52 revertants (arp2Δ/Δ + ARP2, vps52Δ/Δ + VPS52) did.
Fig. 2. VSP52 and Arp2/3 complex mutants do not form hyphae.
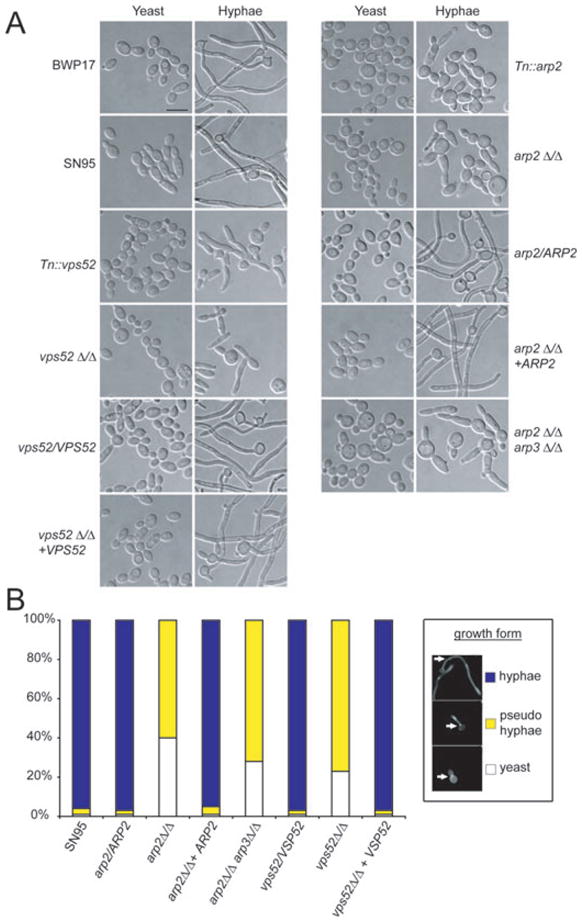
A. Overnight cultures grown in YPD media were diluted 1:200 in YPD (yeast conditions) or YPD + 10% FBS (hyphae conditions) and placed for 3 h at either 30°C (yeast) or 37°C (hyphae). Both transposon and deletion mutants for VSP52 and ARP2 do not form hyphae under these conditions. Heterozgygous mutants and revertants behaved like WT. An arp2Δ/Δarp3Δ/Δ double knockout showed identical behaviour compared with the arp2Δ/Δ single knockout. Bar = 10 μm.
B. Quantifying polarized morphogenesis. Hyphal induction was done as described for (A). While > 96% of WT, heterozygous and revertant strains formed true hyphae, the majority of vps52Δ/Δ, arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants did not form hyphae and grew as pseudohyphae instead. n > 200 for each strain.
To analyse polarized morphogenesis in more detail, we stained strains with calcofluor white (CFW) to assess what percentage of cells were growing as yeast, pseudohyphae or true hyphae (Fig. 2B). Cells that had the first septum/septin ring located in the growing bud tube and showed no constrictions at septal junctions were considered true hyphae (Sudbery et al., 2004). Under the conditions tested, more than 96% of the WT strain and both heterozygotes and revertants were growing as true hyphae. On the other hand, no true hyphal formation was observed in either arp2Δ/Δ or vps52Δ/Δ cells, while 60% of arp2Δ/Δ and 77% of vps52Δ/Δ cells were found to grow as pseudohyphae and 40% of arp2Δ/Δ and 23% of vps52Δ/Δ cells were still in the yeast state.
Taken together, the hyphal-deficient transposon insertion phenotypes for ARP2 and VPS52 were confirmed by deletion mutants and by reverting phenotypes after reintroduction of the WT genes. Comparative genome hybridization analysis further showed that no aneuploidies were found in the arp2Δ/Δ or vps52Δ/Δ strains (Fig. S3). Because other C. albicans vacuolar protein sorting (VPS)-type mutants such as vps11Δ/Δ, vps28Δ/Δ, vps32Δ/Δ and the conditional vps1Δ/VPS1-tetR mutant showed hyphal formation defects similar to vps52Δ/Δ cells (Palmer et al., 2003; Cornet et al., 2005; Bernardo et al., 2008), we decided to focus further investigation on the Arp2/3 complex.
C. albicans Arp2/3 complex mutants show dramatic actin cytoskeleton defects, but are still able to endocytose
ARP2 encodes one of seven evolutionary conserved subunits of the Arp2/3 complex, which nucleates actin filaments (F-actin) into branched networks (Machesky et al., 1994; Welch et al., 1997). Given its conserved association with actin regulation, we asked how deletion of ARP2 in C. albicans affects the actin cytoskeleton by staining logarithmically growing yeast cells with rhodamine/phalloidin (Fig. 3). In contrast to WT stained cells where actin patches are observed as bright dots localizing to sites of polarized growth, the C. albicans arp2Δ/Δ mutant did not show such distinct, bright actin patches. Instead, the most prominent actin structures that formed in arp2Δ/Δ cells were large filamentous aggregates similar to those observed in conditional arp2 and arp3 mutants of S. cerevisiae or in bee1/las17 yeast mutants, which also lack actin patches (Li, 1997; Winter et al., 1997; Martin et al., 2005). On the other hand, other actin-based structures such as actin cables and cytokinetic rings were still observed in arp2Δ/Δ cells (Fig. 3). When only one copy of the ARP2 gene was present (arp2Δ/Δ + ARP2), distinct actin patches were observed and hyphal formation was restored, but these patches appeared less frequent when compared with WT. A double KO mutant, where the two copies of both ARP2 and ARP3 were deleted (arp2Δ/Δarp3Δ/Δ), showed similar morphological phenotypes to the arp2Δ/Δ single mutant. These mutants also exhibited hyphal formation defects, cells clumping together when grown in logarithmic phase, and individual cells appearing phenotypically round and swollen with a wider bud neck as well as similar actin cytoskeleton defects (Figs 2 and 3).
Fig. 3.
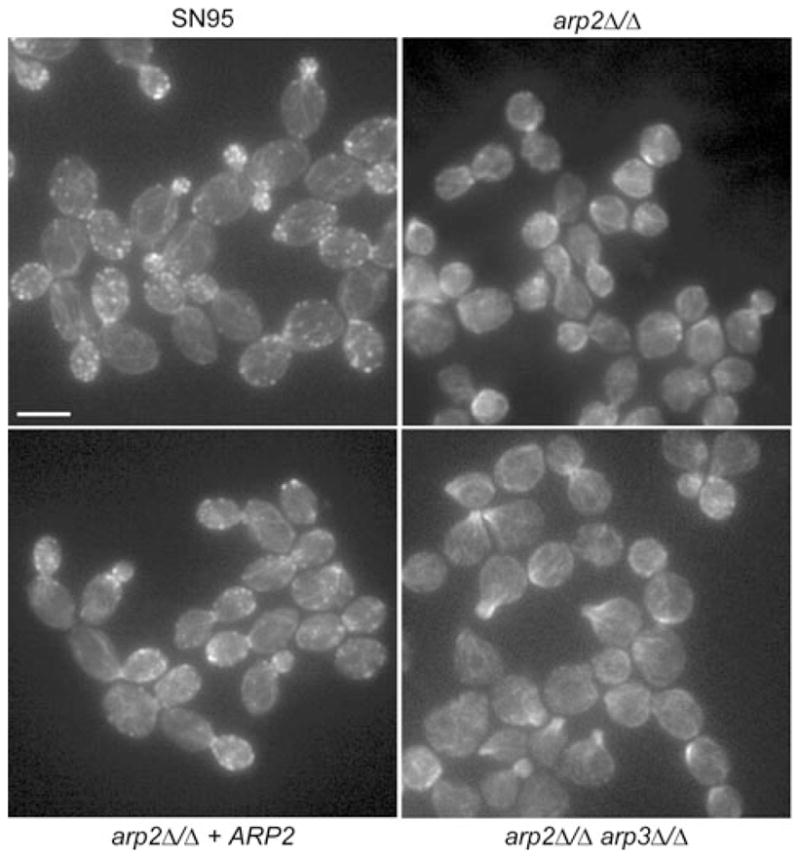
Assaying actin cytoskeleton structures in Arp2/3 complex mutants. Rhodamine/phalloidin staining was used to visualize the actin cytoskeleton in logarithmically growing yeast cells. In WT (SN95) cells, actin patches localize to sites of polarized growth in most phases of the cell cycle. Independent of the cell cycle, distinctive actin patch structures were not observed in arp2Δ/Δ or arp2Δ/Δarp3Δ/Δ cells. Actin cables seemed unaffected in mutant cells. Bar = 5 μm.
Because assembly of actin filaments mediated by the Arp2/3 complex has been shown to be an essential part of endocytosis in a wide range of organisms (for reviews see: Goley and Welch, 2006; Kaksonen et al., 2006; Galletta and Cooper, 2009), we stained C. albicans Arp2/3 complex mutants with the lipophilic dye FM4-64, which is commonly used to visualize membrane internalization and endocytotic delivery to the vacuole (Vida and Emr, 1995). Both arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants were clearly able to deliver FM4-64 to the vacuole as observed by the intracellular appearance of the dye after a 45 min chase period (Fig. 4A). In some Arp2/3 mutant cells, however, the vacuolar morphology appeared to be fragmented, which sometimes resulted in staining throughout the vacuole. While in WT cells typically one to three vacuoles were apparent, four or more smaller vacuoles could be observed in Arp2/3 complex mutant cells. This fragmented vacuolar morphology has also been described for C. albicans wal1Δ/Δ (LAS17 in S. cerevisiae) mutants, an activator of the Arp2/3 complex (Walther and Wendland, 2004).
Fig. 4. Arp2/3 complex mutants can still endocytose FM4-64 and LY.
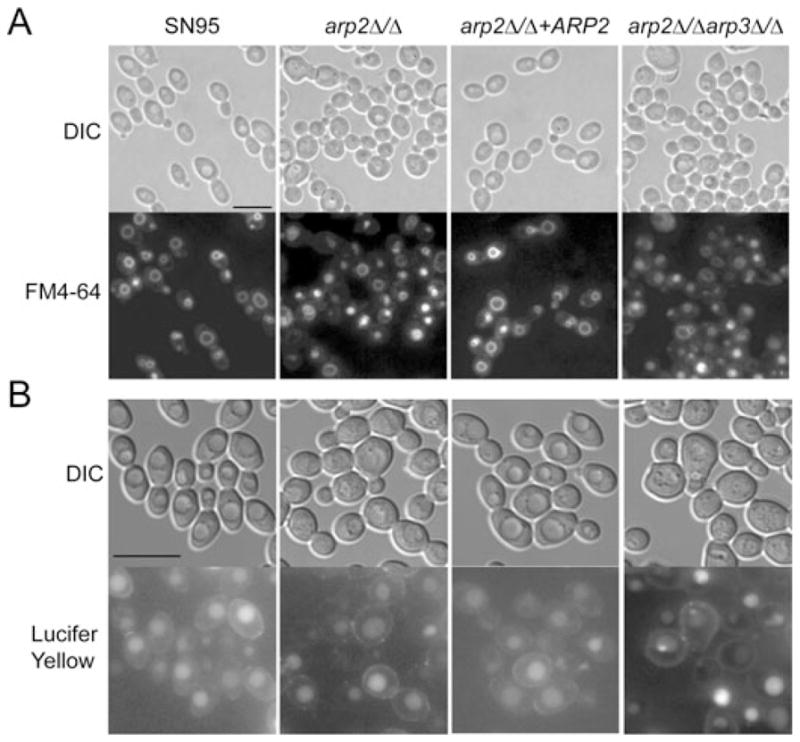
A. Cells in logarithmic phase were incubated for 5 min with 20 μM FM4-64, washed twice, chased for 45 min and visualized by epifluorescence microscopy. Under these conditions, the dye is endocytosed and reliably visualized by vacuolar membrane staining in WT (SN95) and mutant cells.
B. Logarithmically growing cells were incubated with 4 mg ml−1 Lucifer Yellow (LY), incubated for 90 min, washed twice and visualized. LY is endocytosed in both WT and Arp2/3 complex mutants. Bars = 10 μm.
As some S. cerevisiae conditional Arp2/3 complex mutants were partially able to endocytose FM4-64, but not LY (Lucifer Yellow is a dye taken up by fluid phase endocytosis) (Riezman, 1985; Moreau et al., 1996; 1997; Martin et al., 2005; Daugherty and Goode, 2008), we assessed LY uptake in C. albicans Arp2/3 complex mutants. No difference in LY uptake was observed in Arp2/3 mutants when compared with WT after 90 min incubation (Fig. 4B).
To quantitatively assess endocytosis, we performed time-lapse microscopy and included a C. albicans myo5Δ/Δ mutant that has been shown to be endocytosis-defective (Oberholzer et al., 2004). Co-incubation of GFP-labelled WT C. albicans with either arp2Δ/Δ, arp2Δ/Δarp3Δ/Δ or myo5Δ/Δ cells confirmed that endocytosis still occurred in arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants, while myo5Δ/Δ cells did not endocytose FM4-64 within 3 h (see movies 1, 2, 3 and 4 as well as Fig. S4, which can be found in the Supporting information). However, vacuolar staining was somewhat delayed in arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ cells; FM4-64 staining appeared after 20–30 min in WT cells, and it took between 70 and 80 min until the dye reached the vacuole in the arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants. In both arp2Δ/Δ, arp2Δ/Δarp3Δ/Δ and myo5Δ/Δ cells, the dye initially appeared as punctate-like dots apparently stuck in the plasma membrane. As dye-internalization occurred in arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ cells over time, the bright dots at the membrane slowly faded at the same time as vacuolar staining began to emerge. In myo5Δ/Δ cells, on the other hand, bright dot-like structures remained at the cell membrane until the end of our observations with no distinct vacuolar structures appearing. Taken together, these results suggest the Arp2/3 complex plays a crucial role in actin cytoskeleton organization in C. albicans, and while clearly delayed in endocytosis, Arp2/3 complex mutants can still endocytose as assessed by FM4-64 or LY uptake.
Transcriptional analysis of Arp2/3 complex mutants
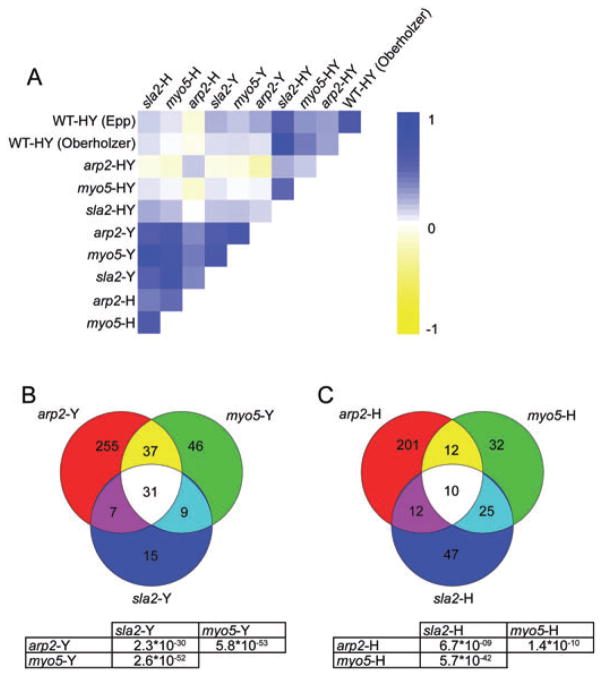
To gain insights into cellular processes affected by disrupting Arp2/3 complex functions, we performed transcriptional profiling under yeast growth conditions (YPD at 30°C for 3 h) or hyphal induction (YPD + 10% FBS at 37°C for 3 h) and compared transcriptional consequences of deleting ARP2 to MYO5 and SLA2 microarray data sets (Oberholzer et al., 2006). MYO5 is an Arp2/3 complex activator, while SLA2 is an actin binding protein that couples actin to the vesicle coat during endocytosis (Robertson et al., 2009a). Both sla2Δ/Δ and myo5Δ/Δ C. albicans mutants suffer similar related complications, such as no hyphal formation, delocalized actin patches and endocytosis defects (Asleson et al., 2001; Oberholzer et al., 2002; 2004). Despite using different chip platforms and different WT strains, there was a good overall correlation of the ARP2 microaray data set and the MYO5 and SLA2 data sets (compare ‘HY Epp’ and ‘HY Oberholzer’ in Fig. 5A, Table S1 shows numerical values for each correlation coefficient). When significantly regulated genes (more than twofold, P < 0.05) were compared, deleting ARP2, MYO5 or SLA2 resulted in similar cellular responses, although the overlap of regulated genes was much more significant under yeast compared with hyphal growth conditions (Fig. 5B and C). To further compare the transcriptional response of arp2Δ/Δ mutants upon hyphal induction, hyphae-specific genes, i.e. genes that are significantly regulated (more than twofold, P < 0.05) in our WT-HY comparison, were clustered in Fig. 6. Many of the genes in that list are known to become highly induced upon activation of the hyphal programme, for example ECE1, ALS3, HYR1, SAP5, SAP6, HWP1, RBT8, IHD1, PST1, CIP1, DCK1 and ORF19.1691 (Nantel et al., 2002; Garcia-Sanchez et al., 2005; Kadosh and Johnson, 2005) (Table S2). Deleting ARP2 resulted in a global lack of hyphal-specific gene induction (arp2-H in Fig. 6). When arp2-Y was compared directly with arp2-H, this observation of improper hyphae-specific gene induction was confirmed as > 92% or 35 out of 38 of the most highly upregulated (more than fourfold) hyphae-specific genes were at least twofold less induced in the absence of ARP2 and > 95% or 75 out of 79 of the remaining upregulated hyphae-specific genes were less induced in arp2Δ/Δ mutants compared WT cells (Fig. 6, Table S2). Deleting myo5Δ/Δ resulted in a comparable response in that some hyphae-specific genes are not properly induced (Oberholzer et al., 2006). However, the lack of proper gene induction was much more pronounced in the absence of ARP2 than in the absence of MYO5 (compare arp2-H vs. myo5-H in Fig. 6, Table S3). Together, these results suggest that while the ARP2 profile showed significant similarities to the MYO5 and SLA2 profiles under yeast growth conditions, there was less correlation under hyphal growth conditions possibly because of the pronounced lack of proper hyphal-specific gene induction in the absence of ARP2.
Fig. 5. Transcript similarities of the ARP2 microaray data set and MYO5/SLA2 data sets.
A. Pearson analysis showing the overall correlation of ARP2 and MYO5/SLA2 microarray data sets. Note that for this analysis no selection of significantly regulated genes was made. The conditions include comparing WT and mutant strains grown under hyphal growth conditions to the same strains grown under yeast growth conditions (HY) and comparing arp2Δ/Δ, myo5Δ/Δ and sla2Δ/Δ mutants against WT cells under yeast (Y) or hyphal (H) growth conditions. ‘WT-HY (Epp)’ and ‘WT-HY (Oberholzer)’ correspond our and the data set obtained by Oberholzer et al. respectively. A value of one (dark blue) corresponds to perfect correlation and −1 (dark yellow) to inverse correlation.
B and C. Venn diagrams showing that deleting ARP2, MYO5 and SLA2 resulted in similar transcriptional consequences when only significantly regulated genes were compared. P-values of overlap are given below the Venn diagram. B shows data from yeast form cells, C shows data from hyphal form cells.
Fig. 6.
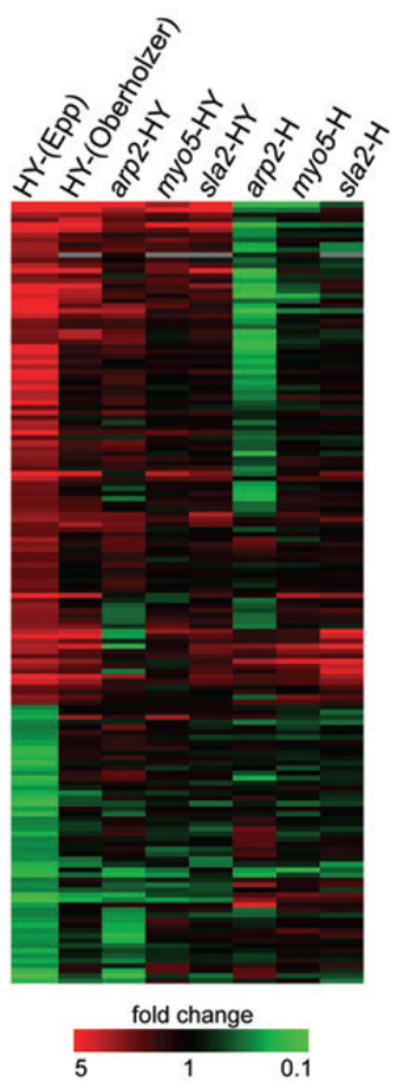
Cluster tree of hyphae-specific genes. While the majority of hyphae-specific genes are not properly induced in the absence of ARP2, MYO5 or SLA2, this lack of hyphal gene induction is most prominent in arp2Δ/Δ cells. Table S2 reports values used to create this cluster tree.
These observations suggest the failure in hyphal growth of Arp2/3 complex mutants could be a result of either impaired endocytosis, problems with the actin cytoskeleton, failure to activate hyphal-specific genes or some combination of these defects. If the hyphal defect was primarily due to failure to activate gene expression, derepressing hyphal-specific gene expression by deleting the NRG1 repressor could potentially suppress the defect, as deletion of NRG1 leads to constitutive filamentous growth even in the absence of any hyphal induction signals (Garcia-Sanchez et al., 2005; Kadosh and Johnson, 2005). We created an nrg1Δ/Δarp2Δ/Δ mutant, which exhibited a doubling time more than twice as long as WT (Fig. S5). When grown under non-inducing conditions, nrg1Δ/Δarp2Δ/Δ cells showed the arp2Δ/Δ mutant morphology of round and swollen cells. When induced for hyphal growth, nrg1Δ/Δarp2Δ/Δ cells also exhibited the arp2Δ/Δ cell morphology and did not form hyphae even after extended overnight incubation times (Fig. 7). We also attempted to create a tup1Δ/Δarp2Δ/Δ mutant, but were not successful.
Fig. 7.
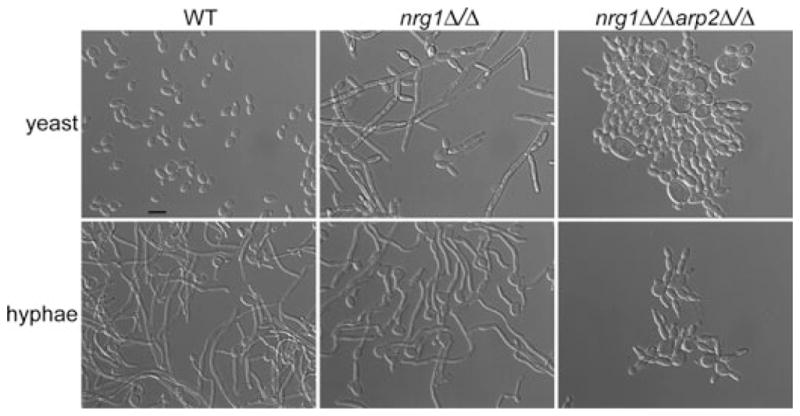
The nrg1Δ/Δarp2Δ/Δ mutant phenocopies arp2Δ/Δ cell morphologies. Hyphal formation was assayed as described in Fig. 2A. Deletion of one transcriptional repressor for hyphal-specific genes, NRG1, leads to constitutive filamentous growth even in the absence of any hyphal signals (yeast). Deleting NRG1 in the arp2Δ/Δ mutant (nrg1Δ/Δarp2Δ/Δ) did not restore filamentous growth even in the presence of hyphal signals (hyphae). Bar = 10 μm.
To determine if the hyphal-specific genes are derepressed in the nrg1Δ/Δarp2Δ/Δ mutant, we performed transcript profiling. We compared the nrg1Δ/Δarp2Δ/Δ mutant grown under hyphal conditions to the arp2Δ/Δ mutant grown under the same conditions (YPD + 10% serum, 37°C, 3 h) and found that a significant number of hyphal-specific genes that are normally induced when WT cells are undergoing the yeast-to-hyphae switch (WT-HY) showed greater expression in the nrg1Δ/Δarp2Δ/Δ mutant compared with arp2Δ/Δ cells (P-value 4.9 × 10−9). When we examined the set of NRG1-dependent hyphal-specific genes previously identified (Kadosh and Johnson, 2005), we found that seven of 28 genes (HYR1, SAP5, SAP4, KIP4, ORF19.6079, ALS3 and UME6) showed significantly increased expression (twofold or more) in nrg1Δ/Δarp2Δ/Δ cells compared with arp2Δ/Δ cells, while a further four genes (IHD1, CBP1, ORF19.6705 and ALS10) showed moderately increased expression between 1.5- and 2-fold (Table S4). Thus, while deleting a transcriptional repressor of the filamentation programme leads to derepression of many hyphal genes, the entire regulated gene set is not derepressed; this presumably reflects the complex interplay that different transcriptional (co-)repressors exert on the yeast-to-hyphae transition (Garcia-Sanchez et al., 2005; Kadosh and Johnson, 2005). We further found that despite the increased induction of some hyphal genes in the nrg1Δ/Δarp2Δ/Δ mutant, a few of those genes are not as highly induced as in WT cells (Table S4). One gene that was induced in both the ‘nrg1Δ/Δarp2Δ/Δ vs arp2Δ/Δ’ and the ‘nrg1Δ/Δarp2Δ/Δ vs WT’ comparisons is UME6, a recently identified key regulator of the hyphal programme (Banerjee et al., 2008; Zeidler et al., 2009). Interestingly, although constitutive overexpression of UME6 in WT cells resulted in constitutive filamentous growth even in the absence of hyphae signals (Carlisle et al., 2009), the increased expression level of UME6 in the nrg1Δ/Δarp2Δ/Δ mutant is not sufficient to restore filamentation in the absence of a functional Arp2/3 complex. Thus despite partial derepression of the hyphal programme, hyphae do not form, making it likely other roles of the Arp2/3 complex, such as its function in actin patch formation and actin branching, are required for hyphal development.
Confirming ‘actin-patch’ phenotypes
Besides defects in filamentous growth, many phenotypes found in myo5Δ/Δ and sla2Δ/Δ mutants have been linked to the Arp2/3 complex and include cell membrane and cell wall defects as well as salt sensitivity (Oberholzer et al., 2006). We tested whether these actin patch-associated phenotypes are also observed in Arp2/3 complex mutants. arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ cells showed typical actin-patch phenotypes such as salt sensitivity as well as cell wall and cell membrane defects, illustrated by increased sensitivity to congo red, CFW, SDS and hygromycin B. Arp2/3 complex mutants also showed abnormal cell wall patterning with aberrant, relatively random chitin deposition (Fig. 8).
Fig. 8. Arp2/3 complex mutants show typical actin patch-associated phenotypes.
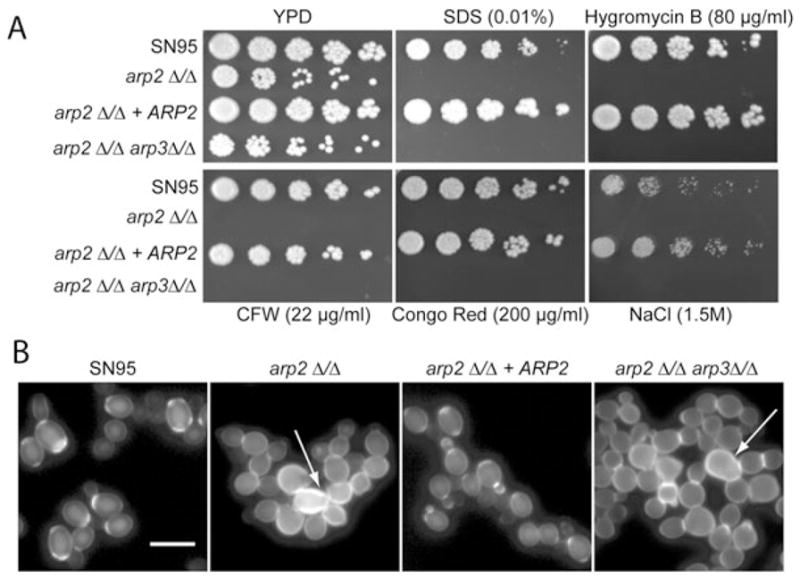
A. Plate spotting assays were done as described in Fig. 1 legend. Arp2/3 complex mutants show similar cell wall and cell membrane defects as well as salt sensitivity as previously described for myo5Δ/Δ mutants.
B. Arp2/3 complex mutants showed aberrant cell wall deposition, indicating defects in cell separation (arrows). See also movie 2 (Supporting information) with arp2Δ/Δ cells that show cell separation defects. Logarithmically growing cells were stained with CFW directly in YPD media for 5 min, washed and visualized. Bar = 10 μm.
Transcriptional analysis also showed that many ergosterol genes (e.g. ERG1, ERG5, ERG6, ERG10, ERG11, ERG27 and ERG252) were downregulated when arp2Δ/Δ cells were compared with WT cells in either yeast or hyphae condition (Table S5). We reasoned that Arp2/3 complex mutants might be more sensitive to drugs targeting this important component of fungal cell membranes. Figure 9 illustrates that both arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants showed increased sensitivity to the ergosterol targeting drug fluconazole, a phenotype that has also been described for mob2Δ/Δ cells, a key component of the RAM pathway that showed additional related actin-patch phenotypes (Song et al., 2008). Taken together, many actin patch-associated phenotypes previously described for myo5Δ/Δ, sla2Δ/Δ, wal1Δ/Δ and mob2Δ/Δ cells could be confirmed with Arp2/3 complex mutants.
Fig. 9.
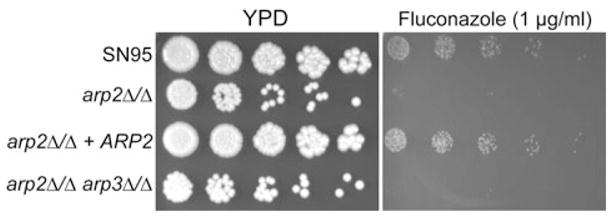
Arp2/3 complex mutants show increased fluconazole sensitivity. arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants are more sensitive to the ergosterol targeting drug fluconazole compared with WT and revertant strains. Serial dilution and spotting was done as describe in Fig. 1 legend.
A functional Arp2/3 complex is required for virulence
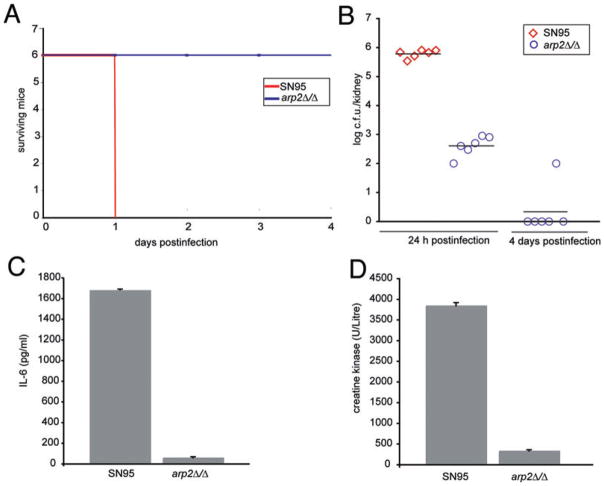
Because the yeast-to-hyphae switch is one important virulence attribute, we tested the arp2Δ/Δ mutant for fungal replication in a complement-5 (C5)-deficient mouse model of disseminated candidiasis (Mullick et al., 2004; 2006; Tuite et al., 2005). Mice infected with the WT strain SN95 were moribund after 24 h post infection, while all mice infected with arp2Δ/Δ cells did not show any clinical signs such as lethargy, ruffled fur or hunched back even on day 4 post infection (Fig. 10A). This observation was confirmed by measuring fungal load from the kidney, the site of highest fungal replication in the A/J mouse model (Mullick et al., 2004). WT-infected mice had a significantly higher fungal burden at 24 h post infection compared with the arp2Δ/Δ-infected mice sacrificed at the same time and at 4 days post infection hardly any fungal cells could be recovered from the kidneys of mice infected with arp2Δ/Δ mutants (Fig. 10B).
Fig. 10.
The Arp2/3 complex is required for virulence in an A/J mouse model. Six mice were infected per group via tail vein with 3 × 105 C. albicans cells.
A. 100% of mice infected with WT C. albicans were moribund at 24 h post infection, while all mice infected with arp2Δ/Δ cells showed no clinical signs at the end of the experiment on day 4. Survival was closely monitored according to approved protocols.
B. Kidney fungal burden was determined at 24 h post infection. Mice infected with arp2Δ/Δ cells showed significantly lower fungal load compared with mice infected with WT cells (student t-test, P < 1 × 10−9). In an independent experiment, fungal load was collected at 4 days post infection from arp2Δ/Δ-infected mice. Bars represent means.
C. and D. IL-6 and CK levels were determined from blood collected at 24 h post infection. The IL-6 and CK levels for mice infected with arp2Δ/Δ cells were significantly lower compared with WT-infected mice (P < 1 × 10−5). Error bars indicate the standard errors of the means.
To gain further insights into the host–pathogen interaction and whether arp2Δ/Δ cells trigger a host response despite their reduced capacity to replicate in the A/J mice background, we biochemically analysed the host blood collected by heart puncture at 24 h post infection. We focused on two metabolic markers that typically show a specific response upon infection with C. albicans (Mullick et al., 2006): levels of interleukin 6 (IL-6), a key inflammatory cytokine, and creatine kinase (CK), a cardiac protein, both of which become highly upregulated upon encounter with C. albicans. The blood from arp2Δ/Δ mutant-infected mice had significantly lower levels of IL-6 compared with WT-infected mice (Fig. 10C). Likewise, significantly lower amounts of CK were found in blood collected from mice infected with arp2Δ/Δ compared with the WT-infected mice (Fig. 10D). These results indicate that arp2Δ/Δ cells do not trigger a normal host response as determined by IL-6 and CK levels. Taken together, based on survival time, fungal burden and the biochemical analysis of the host blood, we conclude that C. albicans arp2Δ/Δ mutants are avirulent in this mouse model of infection.
Discussion
While the complete sequence of the C. albicans genome (assembly 20 became available in 2006), in combination with molecular tools such as epitope-tagging, inducible promoters, reporter genes, auxotrophic and dominant markers, has tremendously facilitated functional analysis in C. albicans (Care et al., 1999; Reuss et al., 2004; Nantel, 2006; Schaub et al., 2006; Lavoie et al., 2008), a comprehensive genome-wide collection of C. albicans mutants is not available, although clearly desirable for functional genomics. The inherent difficulty of genetic manipulation in C. albicans has resulted in only a few large-scale mutagenesis efforts capable of directly linking a gene to a cellular function (Roemer et al., 2003; Uhl et al., 2003; Nobile and Mitchell, 2005; Bruno et al., 2006; Shen et al., 2008). Here, we have investigated a forward genetics approach in C. albicans. Screening of 4700 insertion mutants showed that VPS52 and ARP2 are essential for the yeast-to-hyphae transition, demonstrating that this approach can successfully link genes to a phenotype of interest in C. albicans.
Advantages of forward genetics in C. albicans
Given that mutating any of the seven Arp2/3 complex subunits resulted in severe growth defects or lethality in S. cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Caenorhbditis elegans (Lees-Miller et al., 1992; Schwob and Martin, 1992; Balasubramanian et al., 1996; Mccollum et al., 1996; Winter et al., 1997; 1999; Hudson and Cooley, 2002; Zallen et al., 2002; Sawa et al., 2003), we were surprised that in C. albicans loss of two Arp2/3 complex subunits does not severely compromise viability (Fig. S5). Using reverse genetics by relying on other yeast models might have led to incorrect assumptions that the Arp2/3 complex is essential in C. albicans and therefore it might have escaped functional characterization. Functional analysis based on alternate models would also have proven challenging in linking ORF19.875, an uncharacterized gene in C. albicans with no obvious homologues in S. cerevisiae or S. pombe (Arnaud et al., 2007), to a glycerol growth defect phenotype (Fig. 1B). These two examples demonstrate the utility of unbiased large-scale mutagenesis directly in C. albicans.
Limitations of forward genetics in C. albicans
Why did only two new robust filamentation phenotypes result out of 4700 potentially homozygosed orf::UAU1/orf::URA3 mutants? First, only about 50% of the insertions were directly in an ORF or in a putative promoter region (see Supporting information). For our analysis, we restricted in-depth characterization and deletion mutant construction of orf::UAU1/orf::URA3 mutants to ORF insertions, because these showed the most consistent and pronounced phenotypes. Second, many insertion mutants were aneuploid, consistent with previous findings when directed UAU1 plasmids were used (Enloe et al., 2000), or showed major chromosomal rearrangements on chromosomes other than where the initial UAU1 marker had inserted (Figs S2 and S3). Finally, we cannot exclude multiple insertions per transformant, which might explain why we could not map some insertions. As well, insertions into repeated regions can be difficult to map. For instance, we had several interesting phenotypes mapped to genes of the TLO (TeLOmere-associated gene) family, but because these genes are known to contain 80% or more similarity (van het Hoog et al., 2007) we could not conveniently map the insertions to a single gene.
Taken together, based on random selection and verification of the WT band, we speculate that out of the 4700 potentially homozygosed orf::UAU1/orf::URA3 mutants, about 5% or 200 would be simple homozygous gene insertion mutants with no aneuploidy at the site of insertion. This would make the overall isolation rate of hyphal defects close to one in 100 genes inactivated.
Exploiting the non-essential function of the Arp2/3 complex in C. albicans
The actin cytoskeleton plays a crucial role in cell polarity across most eukaryotes and hyphal growth in several fungal species in particular (Pruyne and Bretscher, 2000; Steinberg, 2007). In C. albicans, for instance, the importance of actin cytoskeleton structure and dynamics to morphogenesis was shown by experiments where C. albicans cells treated with Cytochalasin A, an actin monomer sequestering drug, led to hyphal formation defects while preserving other cellular growth processes (Akashi et al., 1994). Subsequent work demonstrated a link between actin cytoskeleton stability and hyphae-specific gene expression, observations that corroborate our findings in that disrupting the Arp2/3 complex prevented morphological switching and that many hyphal-specific genes are not properly induced. This is consistent with the idea that besides a complex transcriptional regulatory system the actin cytoskeleton itself could influence hyphal gene activation (Hazan and Liu, 2002; Wolyniak and Sundstrom, 2007), potentially through cAMP signalling (Zou et al., 2009). However, our data also show that partial derepression of the hyphal programme does not restore hyphae formation in the absence of a functional Arp2/3 complex, suggesting that in Arp2/3 complex mutants, additional factors are involved.
Several actin-patch phenotypes previously described for C. albicans myo5Δ/Δ, wal1Δ/Δ, sla2Δ/Δ or RAM pathway mutants could be confirmed with arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants. While a link between the Arp2/3 complex and the two Arp2/3 complex activators, WAL1 and MYO5, or SLA2, an actin binding protein, are more obvious, it remains speculative how the RAM pathway could influence actin dynamics in general and the Arp2/3 complex in particular. Of all phenotypes displayed by Arp2/3 complex mutants, the most surprising was the ability to endocytose as it is now widely accepted that actin patches are the major sites of endocytosis from yeast to mammals (Kaksonen et al., 2006; Moseley and Goode, 2006; Smythe and Ayscough, 2006; Galletta and Cooper, 2009). In general, although delayed, arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ cells were clearly able to endocytose LY and FM4-64, which was not the case for the Arp2/3 complex activator Myo5p (compare movie 2 with movie 3, Supporting information), suggesting that in myo5Δ/Δ cells Arp2/3 complex-independent, MYO5-dependent pathways exist that contribute to plasma membrane component uptake. Together with our microarray studies that showed that the degree of similarity of the transcriptional response of arp2Δ/Δ and myo5Δ/Δ mutants was condition-dependent, this supports previous conclusions (Oberholzer et al., 2006) that some myo5Δ/Δ phenotypes are Arp2/3 complex-independent.
The process of endocytosis can be divided into four phases (Robertson et al., 2009a). During the earliest phase, assembly of the endocytic coat complex takes place and involves cargo recruitment, initial membrane curvature to facilitate the invagination process, as well as recruitment of proteins that will trigger actin polarization. This initial process seems unaffected in Arp2/3 complex mutants as bright FM4-64 dots still accumulate at the plasma membrane prior to membrane internalization in both WT and arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ deletion mutants. These observations are consistent with the model that this first step of endocytosis is actin-independent (Robertson et al., 2009a). During the second step of endocytosis, WT cells invaginate the FM4-64-loaded vesicles, while in Arp2/3 complex mutants a distinct invagination was not observed, again in good agreement with current models in that Arp2/3 complex-mediated actin polymerization is a central driving force during this step of membrane invagination. Instead, in arp2Δ/Δ and arp2Δ/Δarp3Δ/Δ mutants bright distinct dots appeared to be stuck in the membrane with little movement within the membrane and no appearance of invagination even after 3 h. Finally, during later steps of endocytosis, vesicle scission and movement away from the plasma membrane was readily detected in WT cells showing internalization of rapidly moving vesicles with subsequent dye accumulation in the vacuole. While although vacuolar dye accumulation was observed somewhat delayed and appeared overall weaker in Arp2/3 complex mutants, vacuolar accumulation of FM4-64 gradually occurred concomitantly with fading of the plasma membrane staining. Curiously, this vacuolar staining appeared independent of vesicle movement, as no clear, distinct vesicles emanating from the plasma membrane could be detected. This observation is lending support to recent reports in S. cerevisiae that Arp2/3-independent routes for actin-driven polymerization could be involved in endocytosis (Robertson et al., 2009b), an idea that is particularly attractive in more distantly related eukaryotes like red algae where BLAST searches did not detect Arp2/3 complex subunits (Galletta and Cooper, 2009).
While more work is needed to refine the interplay of various endocytotic machinery components and the Arp2/3 complex in order to explore their spatiotemporal involvement in processes such as endocytosis or filamentous growth in C. albicans, the Arp2/3 complex mutants in this molecularly manipulatible model organism provide a powerful tool to probe this process in greater depth.
Experimental procedures
Strains, plasmids, primers and media
Strains, plasmids and primers (oligonucleotides) used in this study are listed in Tables S6–S8. YPD media consisted of 1% yeast extract, 2% peptone, 2% dextrose and 2% agar supplemented with 50 mg l−1 of uridine. YPD + 10% FBS and Spider media (1% Difco nutrient broth, 1% mannitol, 0.2% K2HPO4, 1.35% agar, pH 7.2), two media known to potently induce hyphal formation, were used for the hyphae screen (Liu et al., 1994; Uhl et al., 2003). SD media contained 2% dextrose, 6.7% yeast nitrogen base without amino acids, 2% agar and was supplemented with the appropriate amino acids.
In vivo random mutagenesis and mapping the mutation
The Escherichia coli-based UAU1 plasmid pool (Davis et al., 2002) was first amplified on Luria broth (LB) plates supplemented with 50 μg ml−1 kanamycin and 50 μg ml−1 ampicillin at 37°C for 2 days followed by a 2 h liquid incubation at 37°C. For each transformation round into C. albicans, roughly 5 μg of a maxi-kit (Quiagen) extracted plasmid mix was NotI-digested and transformed (Chen et al., 1992) into BWP17 (relevant genotype: ura3/ura3 his1/his1 arg4/arg4) (Wilson et al., 1999). The orf::UAU1/ORF heterozygotes were selected on SD-Arg + Uri plates, patched on YPD media, grown at 30°C for 1–2 days and serially replica-plated up to eight times onto SD-Arg-Ura plates selecting for orf::UAU1/orf::URA3 homozygotes. Generally, ~94% of the orf::UAU1/ORF heterozygotes gave rise to one or more orf::UAU1/orf::URA3 homozygote colonies when replica-plated to SD-Arg-Ura plates from YPD plates compared with ~30% of heterozygotes that generated homozygotes when replica-plating the transformation plates directly to SD-Arg-Ura plates. For phenotypic analysis, potential orf::UAU1/orf::URA3 insertion mutants were screened for a desired phenotype and simultaneously for Arg+/Ura+ prototrophy. If a desired phenotype was detected, a single colony of that mutant was selected, retested for growth on SD-Arg and SD-Ura media to verify segregation of the markers and at the same time retested for robustness of the desired phenotype. Only when each colony of this second testing showed a consistent phenotype, i.e. growth on SD-Arg, SD-Ura media together with a stable phenotype, was the insertion mapped. If the insertion was mapped directly in an ORF and no WT band was found by PCR analysis, a deletion mutant was constructed of this gene.
To map the transposon insertions inverse PCR was performed (Ochman et al., 1988). For all mutants where we used inverse PCR or PCR to verify the absence of a WT band in orf::UAU1/orf::URA3 mutants, a single colony was isolated and genomic DNA was extracted. Briefly, 20 μg of extracted DNA (Rose et al., 1990) was purified using an ice-cold 2.5 M NH4AcO solution (Maniatis et al., 1982). MboI (New England Biolabs, NEB) was added at 2 U μg−1 of DNA and incubated at 37 C for 2 h. After enzyme inactivation and dilution of the DNA, T4 DNA ligase (NEB) was added to circularize the fragments that were then PCR-amplified with Taq polymerase (NEB) and oligonucleotides (primers) oEE5/oEE6. PCR products were sequenced with primers oEE9 and oEE23 at the Genome Sequencing Centre at McGill (http://www.genomequebecplatforms.com/mcgill). Sequences obtained after inverse PCR were mapped using the BLAST tools on the CDG homepage (http://www.candidagenome.org/).
Deletion mutant construction and phenotypic analysis
Strain SN95 (Noble and Johnson, 2005) was used for deletion mutant constructions. For each deletion mutant, at least two homozygotes derived from two different heterozygotes were constructed. One hundred-mer or 120-mer oligos flanking the coding sequence of genes ARP2 or VPS52 were used to amplify the Arginine and Histidine auxotrophic marker cassettes (Wilson et al., 1999) for sequential disruption of both alleles. For the arp2Δ/Δarp3Δ/Δ double mutant, a fusion PCR approach and strain SN148 were used (Noble and Johnson, 2005). Correct integration of the marker cassettes and the absence of the WT gene were verified for each mutant by standard PCR analysis (Schaub et al., 2006) (Fig. S2). In order to reintegrate the WT gene in each mutant at the WT locus, the nourseothricin marker was used (Reuss et al., 2004). Briefly, the WT gene was amplified with primers oEE166/oEE228, oEE174/oEE232, oEE170/oEE229 for genes ARP2, VPS52 and ORF19.875, respectively, and cloned into the KpnI/XhoI-digested pSFS2A plasmid (Reuss et al., 2004). The resulting plasmids, designated pEE22, pEE18 and pEE23, were sequenced. Downstream flanking sequences for ARP2, VPS52 and ORF19.875 were amplified with primers oEE167/oEE169, oEE233/oEE234 and oEE230/oEE231 and cloned into the NotI/SacII-digested plasmids pEE22, pEE18 and pEE23 respectively. This resulted in plasmids pEE33, pEE20 and pEE30, which were then KpnI/SacII-digested and directly transformed into the corresponding deletion mutant. Selection was done on 200 μg ml−1 of nourseothricin, and correct integration was verified by PCR analysis. Finally, before revertant phenotypes were compared with mutant and WT phenotypes, the nourseothricin marker cassette was looped out by incubating the revertants for 6 h in YP media supplemented with 2% maltose. Loop out of the SAT1 flipper cassette was confirmed by nourseothricin sensitivity, Histidine or Arginine auxotrophy and finally by PCR analysis (not shown).
The nrg1Δ/Δarp2Δ/Δ mutant was created in the nrg1Δ/Δ deletion strain (Murad et al., 2001). Briefly, the upstream flanking sequence of ARP2 was first amplified with primers oEE166/oEE382 and cloned into the KpnI/NotI-digested plasmid pEE33, which resulted in plasmid pEE59. This plasmid was then used to sequentially delete both alleles of the ARP2 gene in the nrg1Δ/Δ mutant.
Microscopy and time-lapse
For microscopic analysis, an upright Leitz Aristoplan or an inverted Leica DMIRE2 microscope with a 100× immersion oil objective, and a 10× projection lense was used. Characterization of Arp2/3 complex mutant phenotypes was done as previously described for CFW, rhodamine/phalloidin and FM4-64 staining in (Vida and Emr, 1995; Oberholzer et al., 2002) and LY visualization in (Baggett et al., 2003).
Time-lapse movies (Supporting information) were performed with TEF1-GFP labelled WT cells that were co-incubated with mutant cells in a one-to-one ratio. Cells were grown to exponential phase and washed with SD media. Agar slides were prepared as follows: 0.75 ml of half strength YPD media was mixed with 0.75 ml of 3.4% Agarose (preheated) after which 1 μl of FM4-64 (200 μg ml−1, in dimethylsulphoxide) was added. A total of 100 μl of this mixture was then transferred to the deep well slides and covered with a coverslip without sealing. After the medium had solidified, the coverslip was removed; 1 μl of mixed WT/mutant culture was applied and covered with a fresh coverslip on top. Differential interference contrast (DIC) and GFP images were taken prior to time-lapse microscopy to distinguish between WT and mutant cells (Fig. S4). Time-lapse image acquisition started 5 min after application of the cells and was followed for 3 h with one image per minute. Images were taken with a Zeiss Axio Imager M1 microscope with a Photometrics Coolsnap HQ camera. Image acquisition and movie file assembly was performed with Metamorph 7 software (Molecular Devices).
Comparative genome hybridization and microarray studies
Comparative genome hybridization analysis was done as previously described (Znaidi et al., 2007) with the following modifications. Genomic DNA was extracted from a C. albicans culture grown to saturation with the Qiagen Genomic DNA Extraction kit according to manufacturer’s instructions. DNA hybridization was done with the Advalytix SlideBooster for 16 h at 42°C according to manufacturer’s instructions.
For the transcriptional profiling experiment, total RNA was extracted using the RNeasy Mini kit (Qiagen). Total RNA quantification was measured by nanodrop (ND-1000 spectrophotometer, NanoDrop Technologies). The quality of the mRNA was verified with a RNA lab-on-chip assay using a 2100 expert mRNA nano BIO analyser (Agilent). cDNA labelling, DNA microarray hybridization, washing and scanning was adapted from a standardized protocol (Nantel et al., 2006). Each condition was covered by a minimum of three DNA microarrays, and results were analysed using Gene-Spring software (Silicon Genetics, Redwood City, CA). In order to compare significantly overlapping gene lists, the P-value was calculated using hypergeometric distribution as described in the GO Term Finder Tool web site (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Gene lists can be found in Tables S2–S5. The entire data set for all microarray experiments has also been deposited at GEO under the accession number GSE19583. (http://www.ncbi.nlm.nih.gov/geo). For calculating the Pearson correlation, GraphPad Prism 5 was used.
Virulence studies
Virulence testing of C. albicans, including survival experiments, tissue fungal burden counting and biochemical analysis of the host blood, was done as previously described (Mullick et al., 2004; 2006). Briefly, 8- to 12-week-old A/J mice (Jackson Laboratories, Bar Harbor, ME) were inoculated via the tail vein with 200 μl of a suspension containing 3 × 105 C. albicans in PBS. Three male and three female mice were used for each experimental group. Mice were closely monitored over a period of maximal 4 days for clinical signs of disease such as lethargy, ruffled fur or hunched back. Mice showing extreme lethargy were considered moribund and were euthanized. All experimental procedures involving animals were approved by the Biotechnology Research Institute Animal Care Committee, which operates under the guidelines of the Canadian Council of Animal Care.
Supplementary Material
Acknowledgments
We thank Aaron P. Mitchell for providing the UAU1 plasmid library, Cynthia Hélie and Mario Mercier for excellent technical assistance in animal handling, Jean-Sébastien Deneault and André Nantel for help with microarray studies and Lichun Liang (a summer student from John Abbott College, Montréal) for help in screening random orf::UAU1/orf::URA3 mutants. This work was supported by a CIHR team grant in fungal pathogenesis to A.M., M.R. and M.W. (CTP-79843). This is NRC publication number 49596.
Footnotes
Additional supporting information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Akashi T, Kanbe T, Tanaka K. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology. 1994;140:271–280. doi: 10.1099/13500872-140-2-271. [DOI] [PubMed] [Google Scholar]
- Arnaud MB, Costanzo MC, Skrzypek MS, Shah P, Binkley G, Lane C, et al. Sequence resources at the Candida Genome Database. Nucleic Acids Res. 2007;35:D452–D456. doi: 10.1093/nar/gkl899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009;5:e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleson CM, Bensen ES, Gale CA, Melms AS, Kurischko C, Berman J. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol Cell Biol. 2001;21:1272–1284. doi: 10.1128/MCB.21.4.1272-1284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggett JJ, Shaw JD, Sciambi CJ, Watson HA, Wendland B. Fluorescent labeling of yeast. Curr Protoc Cell Biol. 2003;Chapter 4(Unit 4):13. doi: 10.1002/0471143030.cb0413s20. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, et al. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- Bernardo SM, Khalique Z, Kot J, Jones JK, Lee SA. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet Biol Mycopathologia. 2008;167:55–63. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, van Het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2006;2:e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci USA. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- Cornet M, Bidard F, Schwarz P, Da Costa G, Blanchin-Roland S, Dromer F, Gaillardin C. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect Immun. 2005;73:7977–7987. doi: 10.1128/IAI.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen L. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- Daugherty KM, Goode BL. Functional surfaces on the p35/ARPC2 subunit of Arp2/3 complex required for cell growth, actin nucleation, and endocytosis. J Biol Chem. 2008;283:16950–16959. doi: 10.1074/jbc.M800783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genet. 2002;162:1573–1581. doi: 10.1093/genetics/162.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Mavor AL, Russell CL, Argimon S, Dennison P, Enjalbert B, Brown AJ. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gow NA. Germ tube growth of Candida albicans. Curr Top Med Mycol. 1997;8:43–55. [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Harris SD, Momany M. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet Biol. 2004;41:391–400. doi: 10.1016/j.fgb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Hazan I, Liu H. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot Cell. 2002;1:856–864. doi: 10.1128/EC.1.6.856-864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol. 2002;156:677–687. doi: 10.1083/jcb.200109065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Kurtz MB, Kirsch DR, Kelly R. The molecular genetics of Candida albicans. Microbiol Sci. 1988;5:58–63. [PubMed] [Google Scholar]
- Laprade L, Boyartchuk VL, Dietrich WF, Winston F. Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genet. 2002;161:509–519. doi: 10.1093/genetics/161.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller JP, Henry G, Helfman DM. Identification of act2, an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc Natl Acad Sci USA. 1992;89:80–83. doi: 10.1073/pnas.89.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy O, Gangneux JP, Montravers P, Mira JP, Gouin F, Sollet JP, et al. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006) Crit Care Med. 2009;37:1612–1618. doi: 10.1097/CCM.0b013e31819efac0. [DOI] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4:728–735. doi: 10.1016/s1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, New York; 1982. [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007a;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Whiteway M. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot Cell. 2007b;6:291–301. doi: 10.1128/EC.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, et al. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winson B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick A, Elias M, Picard S, Bourget L, Jovcevski O, Gauthier S, et al. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect Immun. 2004;72:5868–5876. doi: 10.1128/IAI.72.10.5868-5876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick A, Leon Z, Min-Oo G, Berghout J, Lo R, Daniels E, Gros P. Cardiac failure in C5-deficient A/J mice after Candida albicans infection. Infect Immun. 2006;74:4439–4451. doi: 10.1128/IAI.00159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A. The long hard road to a completed Candida albicans genome. Fungal Genet Biol. 2006;43:311–315. doi: 10.1016/j.fgb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Rigby T, Hogues H, Whiteway M. Microarrays for Studying Pathogenicity in Candida albicans. In: Kavanaugh K, editor. Medical Mycology: Cellular and Molecular Techniques. New York, NJ: Wiley Press; 2006. [Google Scholar]
- Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJ. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell. 2004;3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Marcil A, Leberer E, Thomas DY, Whiteway M. Myosin I is required for hypha formation in Candida albicans. Eukaryot Cell. 2002;1:213–228. doi: 10.1128/EC.1.2.213-228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Iouk TL, Thomas DY, Whiteway M. Functional characterization of myosin I tail regions in Candida albicans. Eukaryot Cell. 2004;3:1272–1286. doi: 10.1128/EC.3.5.1272-1286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Nantel A, Berman J, Whiteway M. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot Cell. 2006;5:1252–1265. doi: 10.1128/EC.00385-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genet. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, Gorman JA. Characterization of the Candida albicans TRP1 gene and construction of a homozygous trp1 mutant by sequential co-transformation. Gene. 1994;148:179–185. doi: 10.1016/0378-1119(94)90687-4. [DOI] [PubMed] [Google Scholar]
- Palmer GE, Cashmore A, Sturtevant J. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot Cell. 2003;2:411–421. doi: 10.1128/EC.2.3.411-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell Mol Life Sci. 2009a;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AS, Allwood EG, Smith AP, Gardiner FC, Costa R, Winder SJ, Ayscough KR. The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol Biol Cell. 2009b;20:1618–1628. doi: 10.1091/mbc.E08-09-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 1990. [Google Scholar]
- Ruhnke M, Maschmeyer G. Management of mycoses in patients with hematologic disease and cancer – review of the literature. Eur J Med Res. 2002;7:227–235. [PubMed] [Google Scholar]
- Santos R, Buisson N, Knight SA, Dancis A, Camadro JM, Lesuisse E. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol Microbiol. 2004;54:507–519. doi: 10.1111/j.1365-2958.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci. 2003;116:1505–1518. doi: 10.1242/jcs.00362. [DOI] [PubMed] [Google Scholar]
- Schaub Y, Dunkler A, Walther A, Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol. 2006;46:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Madhani HD. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- Schwob E, Martin RP. New yeast actin-like gene required late in the cell cycle. Nature. 1992;355:179–182. doi: 10.1038/355179a0. [DOI] [PubMed] [Google Scholar]
- Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc Natl Acad Sci USA. 2008;105:20918–20923. doi: 10.1073/pnas.0809147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E, Ayscough KR. Actin regulation in endocytosis. J Cell Sci. 2006;119:4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- Song Y, Cheon SA, Lee KE, Lee SY, Lee BK, Oh DB, et al. Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol Biol Cell. 2008;19:5456–5477. doi: 10.1091/mbc.E08-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. Hyphal growth: a tale of motors, lipids, and the Spitzenkorper. Eukaryot Cell. 2007;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Tuite A, Elias M, Picard S, Mullick A, Gros P. Genetic control of susceptibility to Candida albicans in susceptible A/J and resistant C57BL/6J mice. Genes Immun. 2005;6:672–682. doi: 10.1038/sj.gene.6364254. [DOI] [PubMed] [Google Scholar]
- Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Wendland J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich Syndrome protein homolog Wal1p. Eukaryot Cell. 2004;3:471–482. doi: 10.1128/EC.3.2.471-482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig M, Jansch L, Gross U, De Koster CG, Klis FM, De Groot PW. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology. 2004;150:3129–3144. doi: 10.1099/mic.0.27256-0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Oberholzer U. Candida morphogenesis and host-pathogen interactions. Curr Opin Microbiol. 2004;7:350–357. doi: 10.1016/j.mib.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak MJ, Sundstrom P. Role of actin cytoskeletal dynamics in activation of the cyclic AMP pathway and HWP1 gene expression in Candida albicans. Eukaryot Cell. 2007;6:1824–1840. doi: 10.1128/EC.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, et al. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 2009;9:126–142. doi: 10.1111/j.1567-1364.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- Znaidi S, De Deken X, Weber S, Rigby T, Nantel A, Raymond M. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol Microbiol. 2007;66:440–452. doi: 10.1111/j.1365-2958.2007.05931.x. [DOI] [PubMed] [Google Scholar]
- Zou H, Fang HM, Zhu Y, Wang Y. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06980.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.