Abstract
Energy failure from mitochondrial dysfunction is proposed to be a central mechanism leading to neuronal death in a range of neurodegenerative diseases. However, energy failure has never been directly demonstrated in affected neurons in these diseases, nor has it been proved to produce degeneration in disease models. Therefore, despite considerable indirect evidence, it is not known whether energy failure truly occurs in susceptible neurons, and whether this failure is responsible for their death. This limited understanding results primarily from a lack of sensitivity and resolution of available tools and assays and the inherent limitations of in vitro model systems. Major advances in these methodologies and approaches should greatly enhance our understanding of the relationship between energy failure, neuronal dysfunction, and death, and help us to determine whether boosting bioenergetic function would be an effective therapeutic approach. Here we review the current evidence that energy failure occurs in and contributes to neurodegenerative disease, and consider new approaches that may allow us to better address this central issue.
Bioenergetic failure has been suggested to cause neuronal death in a range of neurodegenerative diseases, including Parkinson disease (PD),1 Alzheimer disease (AD),2,3 and Huntington disease (HD).4 However, energy failure has never been directly demonstrated to occur in dying neurons in these diseases or even in intact neurons in genetic models of these diseases. Why, then, is bioenergetic dysfunction considered by many to be a central mechanism that produces neurodegeneration? This assertion is supported by the nearly overwhelming evidence—from human, genetic, and animal studies—that mitochondria are altered in multiple respects in all of these conditions, and because many of these mitochondrial changes have the potential to cause bioenergetic failure. However, whether this actually occurs in affected neurons is almost always unknown. Furthermore, in addition to producing adenosine triphosphate (ATP), mitochondria have other functions, including the production of reactive oxygen species (ROS), calcium buffering, and the regulation of apoptotic pathways, lipid biosynthesis, and neurotransmitter metabolism,5,6 and changes in these processes could also contribute to neurodegeneration. Therefore, although suggestive, altered mitochondrial function per se cannot be automatically equated with energy failure.
How, then, can we establish whether bioenergetic dysfunction is a central mechanism that produces neurodegeneration? A major issue is that most of the available tools, techniques, and model systems lack sufficient resolution to establish a direct relationship. The development of new and improved methods that overcome some of these challenges could provide new and important insight into the initial mitochondrial changes that occur in neurodegeneration, and how these affect bioenergetic function.
Before reviewing the evidence for bioenergetic dysfunction in neurodegeneration, we must first define energy failure. For the purposes of this review, we define energy failure as insufficient ATP for a cell to maintain its cellular functions and/or defenses against stresses. If relatively mild, energy failure will impair neuronal function–for instance, by blocking synaptic transmission7–but may still be compatible with neuronal survival. However, more profound energy failure will trigger active forms of cell death, including apoptosis and/or pathways that involve the release of mitochondrial factors from damaged mitochondria. When particularly severe, there may be insufficient energy for classic apoptotic pathways to proceed, and energy failure may trigger other cell death pathways.8,9 Energy failure might result either from impaired energy production, increased energy consumption, or both. In addition, energy failure might result from defects in either aerobic or anaerobic respiration, but this review focuses on bioenergetic dysfunction caused by mitochondrial failure.
Evidence for Energy Failure from Mitochondrial Dysfunction in Neurodegenerative Disease
Mitochondrial Changes Identified at Autopsy
The enzymatic function of specific respiratory-chain complexes are decreased in the brain tissue of patients with AD,10 HD,11 and PD.12 With the essential role of these complexes for aerobic respiration, these changes might certainly produce—or at least reflect—bioenergetic stress. However, further mechanistic interpretation is difficult for several reasons. First, many of these changes were assayed from total brain tissue, and likely reflect changes in glia rather than neurons,13,14 making it impossible to discern whether the same enzymatic changes also occurred in affected neurons. Some studies have specifically targeted individual affected neurons and identified defects in the expression of genes that regulate bioenergetic function.1,15 However, such changes may still not translate into decreased ATP production,16 depending on the specific complex inhibited and the extent to which enzymatic activity is compromised. In yet other cases, decreased ATP production might be a compensatory response to decreased ATP demand and, therefore, may not contribute to the degeneration.
A number of other intriguing changes in mitochondrial function have been observed in postmortem tissue, although their effects on bioenergetic function are similarly difficult to predict. For instance, mitochondrial DNA mutations accumulate with age and appear to accumulate at a faster rate in individuals with neurodegenerative diseases,17,18 although this is controversial in some cases.19,20 In many cases, these mutations are found in the affected neuronal subtypes (eg, through laser capture of dopaminergic [DA] neurons17). These mutations likely produce respiratory defects, considering that many of the genes encoded by mitochondrial DNA contribute to bioenergetic function, and because the percentage of mitochondrial DNA with deletions is increased in those neurons with decreased cytochrome c oxidase activity,17,21 although this remains to be formally proved. Alternatively, these mutations might affect other functions, such as ROS production, and/or make little contribution to disease progression.
Other studies revealed intriguing changes in mitochondrial biogenesis. For example, decreased expression of PGC1-α and PGC1-α-regulated genes was observed in DA neurons in patients with PD,1 in striatal tissue in HD patients,22 and in the hippocampus of AD patients.23 Once again, these changes would likely result in bioenergetic dysfunction, but this is also not yet proved.
Nuclear Magnetic Resonance Spectroscopy
Magnetic resonance spectroscopy (MRS) provides one of the only methods to directly visualize energy metabolites in the brains of living patients, and it has provided strong evidence for energy deficits in neurodegenerative disease. For instance, levels of lactate are increased in the basal ganglia and occipital cortex in patients with HD.24 In addition, MRS studies have revealed decreased resting levels of ATP/(phosphocreatine + inorganic phosphate) in the muscles of symptomatic and presymptomatic HD patients and a decreased maximal rate of ATP production and phosphocreatine recovery after exercise.25,26 In early HD, ATP levels in the brain also fail to upregulate normally when energy demands are increased.27 Levels of ATP are also decreased in the midbrain and putamen of patients with early and advanced PD,28 and levels of high-energy phosphates (ATP and phosphorylated creatine), but not low-energy phosphates (ADP and unphosphorylated creatine), are decreased in the basal ganglia and frontal lobes of patients with progressive supranuclear palsy.29 MRS approaches, thus, provide strong evidence that energy failure occurs in neurodegeneration. However, at present, they lack the sensitivity to discriminate changes between adjacent neurons and their surrounding glia, and hence are unable to prove that the energy failure occurs within affected neurons, or to provide insight into how any changes may differ in susceptible versus resistant cell types.
Evidence from Mitochondrial Neurotoxins
The susceptibility of vulnerable neurons to inhibitors of the mitochondrial respiratory chain also suggests a role for bioenergetic failure in neurodegenerative disease. For instance, the complex I inhibitors 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and rotenone are selectively toxic to DA neurons,30,31 and striatal interneurons are selectively susceptible to the complex II inhibitors 3-nitropropionic acid32,33 and malonate.34 However, 1 methyl-4-phenylpyridinium (MPP1) may produce death through mechanisms that are at least partly independent of its effects on complex I,35–37 and even rotenone, a prototypical complex I inhibitor, may exert toxicity through other mechanisms.36,37 It is thus unclear whether the susceptibility to these toxins truly reflects intrinsic differences in the respiratory chain function between neuronal subtypes, even for those toxins, such as rotenone, that are imported into all cells equally (in contrast to MPTP, whose metabolite MPP+ is selectively concentrated within DA neurons by the dopamine transporter38). Furthermore, even if they do kill susceptible neurons by blocking energy production, this still would not prove that neuronal death actually occurs through bioenergetic failure in the human disease. There is no conclusive evidence that any of these toxins produce the idiopathic conditions, although several epidemiologic studies have associated certain pesticides (such as rotenone and paraquat) with a somewhat higher disease prevalence.39 Thus, although suggestive, susceptibility of a cell type to a mitochondrial toxin does not directly prove that neuronal death occurs through bioenergetic failure.
Genetic Forms of Neurodegenerative Disease Implicate Mitochondria
The finding that mutations in the mitochondrial protein PINK1 produce an autosomal-recessive form of PD40 established that specific defects in mitochondria can cause PD. Subsequent studies in Drosophila41,42 and cell-culture models43,44 linking PINK1 and parkin, another autosomal-recessive PD protein, strongly suggest that mutations in parkin also cause PD through effects on mitochondria. However, the specific mechanism(s) are unclear. A number of functions have been ascribed to PINK1 and parkin, including roles in mitophagy,43,45 mitochondrial dynamics,46,47 mitochondrial mobility,48 and mitochondrial biogenesis.49 All of these changes might affect mitochondrial bioenergetics (Fig 1), but could also influence other mitochondrial functions. In the case of PINK1, subtle impairments in basal mitochondrial membrane potential and a paradoxical depolarization follow exposure to oligomycin.50,51 This latter finding has generally been interpreted to arise from reverse transport of protons through the ATP synthase to maintain membrane potential, in the context of a dysfunctional respiratory chain.51 In agreement with this, a subsequent study using luciferase-based approaches has demonstrated decreased basal ATP levels in the striatum of living PINK1 knockout animals, and an impaired ability of mouse embryonic fibroblasts lacking PINK1 to upregulate ATP synthesis in response to stress.52 Although these studies still do not establish that ATP levels are actually decreased and limiting within individual neurons or provide insight as to why DA neurons are selectively susceptible, they nonetheless provide some of the strongest evidence to date that mutations in PD proteins produce bioenergetic failure in neurons.
FIGURE 1.
Potential mitochondria-based mechanisms by which Parkinson disease proteins may produce energy failure and neuronal death. The schematic illustrates the known effects of loss of parkin or PINK1 (mitochondrial biogenesis,49 mitochondrial transport,48 mitochondrial turnover,44,45 dynamics,46,58 and respiration50,51,124) or increased synuclein (mitochondrial turnover,125 dynamics,54,55 and respiration126,127). These primary changes may result in disruptions of the normal mitochondrial distribution and/or function, and this in turn could lead to energy failure and neuronal death. Other mitochondrial functions, such as reactive oxygen species (ROS) production, calcium buffering, and roles in apoptotic pathways could also be altered and contribute to cell-death pathways. DJ-1 and LRRK2 can also affect mitochondrial function,128–130 but are not depicted in this schematic.
A number of other genes implicated in AD and PD can impact various mitochondrial functions, although it is unknown whether these effects lead to neuronal dysfunction or death. Of particular interest is the relationship between mitochondrial dynamics, the balance between fusion and fission, and bioenergetics. A number of PD-associated proteins, including α-synuclein, PINK1, parkin, and LRRK2, as well as amyloid-beta, tau, and huntingtin, disrupt normal dynamics across a range of paradigms. In some cases, normalizing mitochondrial morphology protected against these insults.46,53–59
The mechanism by which changes in mitochondrial dynamics might produce degeneration is unknown, but has been hypothesized to ultimately involve bioenergetic failure.55,60–62 This may occur through changes in the intrinsic function of mitochondria55,60 and/or changes in the mass or distribution of mitochondria within neuronal processes, for instance, through decreased transit of excessively tubulated mitochondria61,63 or changes in the rate of mitochondrial degradation.58 Understanding the relationship between mitochondrial dynamics and bioenergetics, versus other nonenergetic consequences of altered mitochondrial morphology, will help to clarify how changes in dynamics may contribute to disease.
Mitochondrial DNA Disorders
The neurologic diseases that provide the most compelling evidence for bioenergetic failure are the mitochondrial myopathies. Patients suffering from mitochondrial myopathies show elevated lactate levels,64 suggesting that these diseases cause impaired aerobic respiration with a compensatory shift toward increased anaerobic respiration. But is this compensation sufficient, or does energy failure still occur? It is somewhat surprising that mice with severely compromised complex I function exhibit broad neurologic and systemic deficits, but fail to show significant changes in ATP or phosphocreatine levels in muscle.65 In another study, deletion of a mitochondrial transcription factor (Tfam) in skeletal muscle also resulted in only a small decrease in ATP production when standardized to overall muscle mass.66 However, further analysis revealed a substantial increase in mitochondrial mass, suggesting that dysfunctional mitochondria proliferated to compensate for their impaired bioenergetic function.66 In yet another study, McKenzie et al showed that mitochondrial mutations producing the syndrome MELAS (mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes) resulted in impaired mitochondrial function. Although ATP levels were largely maintained at baseline through compensation by glycolysis, severe bioenergetic deficits were revealed when glycolysis was blocked.67 These results thus highlight the importance of assay design to detect mitochondrial failure, and provide evidence that bioenergetic dysfunction can occur with certain mitochondrial mutations. Future experiments will be required to confirm that this energy failure is ultimately responsible for neuronal dysfunction and death.
Contribution of Glucose and Overall Metabolic Changes to Bioenergetic Failure in Neurodegenerative Disease
Although this review focuses on energy failure from mitochondrial dysfunction, primary changes in glucose uptake or metabolism could also produce bioenergetic dysfunction (eg, by impairing glycolysis). Changes in glucose metabolism occur early in the pathogenesis of AD and HD sufferers as well as in those susceptible to these diseases.68–72 In some cases, this may represent a shift in the extent of glucose allocated to glycolysis versus the pentose–phosphate pathway.73 However, it is unknown whether changes in glucose metabolism produce energy failure in dying neurons, nor is it clear whether they are the cause or the result of mitochondrial dysfunction in these diseases.
On a broader level, patients and animal models of several neurodegenerative diseases, including AD,74, HD,75 and likely PD,76 exhibit early weight loss that cannot be explained by changes in caloric intake or activity. Although the etiology of this weight loss is unknown, it could reflect underlying changes in mitochondrial and other bioenergetic functions at the cellular level.
Obstacles to Measuring Energy Failure in Neurodegenerative Disease
Considering that bioenergetic failure is a feature common to many neurodegenerative diseases, it is remarkable that our understanding of its role in neurodegeneration is so limited and largely speculative. However, progress in this area has been hampered by a lack of appropriate tools, as well as limitations in the existing approaches and methods.
Model Systems
The limitations of existing model systems greatly impede our understanding of bioenergetics in neurodegeneration. Although neurodegenerative diseases are characterized by the selective vulnerability of specific neuronal populations, many studies rely on cell lines or study neuronal types that are not affected by the disease. This approach can yield important insights. For instance, energy failure might be equally likely to occur in all cell types that express a given protein of interest, and selective degeneration may simply reflect a greater susceptibility to the toxicity of these effects and/or selectively increased expression of the toxic proteins in affected versus unaffected cells. However, in other cases, bioenergetic changes may occur only in certain neuronal subtypes, depending on the presence of discrete characteristics. For instance, susceptible neurons may contain relevant compounds (eg, dopamine) that contribute to mitochondrial impairment, or have mitochondria with subtype-specific functions or properties. Clearly, there are fundamental differences between neurons grown in culture and in vivo, including the extent of myelination,77,78 axonal arbor size,79 and oxygen tension,80,81 that are likely to affect bioenergetic function. We do not know whether the critical mechanisms producing degeneration are recapitulated in culture.
In vivo model systems have similar limitations, and rodent models in particular have consistently failed to replicate important features of the human condition. For PD, no faithful disease-based genetic model has produced the selective loss of DA neurons, with the possible exception of PINK1 knockout loss in rats.82 This may result partly from the shorter lifespan of mice, but rodent neurons also have intrinsic differences in susceptibility to mitochondrial stressors. For instance, rats are more sensitive than mice to loss of PINK1,82,83 whereas primates are far more sensitive to the mitochondrial toxin MPTP than mice, which in turn are more susceptible than rats.84 In this respect, human-derived neurons might provide valuable information, although the lack of in vivo context remains an important limitation.
Subcellular Compartment
Synaptic terminals degenerate early in a range of neurodegenerative diseases associated with impaired energy metabolism, including AD,85 HD,86 and PD,87,88 and in animal models of these diseases,30 indicating that there may be significant changes in bioenergetic function that are either restricted to—or occur first in—neuronal processes. The regulation and maintenance of energy metabolism in axons versus the cell body almost certainly differ in multiple ways, including the reliance on oligodendrocytes to support the energy requirements of myelinated axons.78 In axons, loss of myelin results in increased mitochondrial content as well as mitochondrial redistribution,89,90 and unmyelinated axons may be more susceptible to energy failure.91 In contrast to the cell body, many synaptic boutons also lack mitochondria (for instance, only about ½ of hippocampal boutons contain mitochondria92), raising questions about how energy is dispersed at nerve terminals as a function of their presence or absence, and whether there are regional gradients of ATP in synapses that may impact neuronal function and/or axonal health (Fig 2). It will be interesting to learn whether regional energy failure can be influenced by changes in the regional distribution or function of mitochondria.
FIGURE 2.
Regulation of mitochondrial bioenergetics in axons. In most non-neural cells, the capacity of mitochondria to produce energy depends on the mitochondrial mass and function, but they may be less dependent on subcellular distribution due to adenosine triphosphate (ATP) diffusion. In axons, however, a normal distribution of mitochondria may also be required to minimize energy gradients. This is illustrated here by the hypothetical gradations in color in the neuron, in which lighter blue reflects higher ATP levels, whereas darker reflects lower levels. Therefore, factors that disrupt the normal distribution of mitochondria, such as mitochondrial dynamics (the balance between mitochondrial fusion and fission) and mitochondrial motility may have more prominent effects on energy levels in axons. Notably, mitochondria in axons are also smaller and more mobile94 and may have different lifespans than those in the cell body, suggesting that the intrinsic function of mitochondria in the cell body versus axons may also differ.
However, despite these important differences, for technical reasons, most studies in intact neurons have focused on the cell body rather than processes. It is unknown whether the bioenergetic status at the cell body provides an accurate reflection of the energy status at the nerve terminal, but there are several reasons why this may not be the case. First, the mitochondria in these compartments may be different, despite coexisting within the same cell. Mitochondrial heterogeneity within the same cell occurs even in nonpolarized cells,93 the mitochondria in axons are generally much smaller and may be more mobile,94 and their ability to produce ATP is possibly more susceptible to complex I inhibition.95 Axonal mitochondria may also be much older, on average, than somatic mitochondria, although very little is known about mitochondrial turnover in neuronal processes. In addition to intrinsic function and mitochondrial mass, regional bioenergetics are also likely to depend on how mitochondria are distributed92 (see Fig 2). Factors that affect mitochondrial distribution, such as the machinery that moves mitochondria and mitochondrial shape—which is, in turn, regulated by mitochondrial dynamics—are likely to affect bioenergetic function to a far greater extent in the axon versus the cell body (see Fig 2). Differences in local energy requirements, such as the energy requirements for synaptic transmission in neuronal processes, are also likely to differentially impact bioenergetic function at the nerve terminal versus the cell body. It is thus likely that the bioenergetic status is quite different between the cell body and processes, suggesting the need to directly examine bioenergetic function in axons and dendrites.
Tools to Measure Bioenergetic Function Specifically in Neurons
Our ability to study bioenergetic function specifically in neurons is limited by a lack of appropriate tools. At present, there are a number of important approaches to study bioenergetic function in brain tissue as a whole, such as: (1) measurements of regional brain activity and metabolism by positron emission tomography and functional magnetic resonance imaging72,73,96; (2) measurements of key energy metabolites, including lactate, creatine, phosphocreatine, and ATP, by MRS27,64,97; or by (3) high-performance liquid chromatography after microwave fixation of animal tissues.98 These approaches have provided clear evidence for bioenergetic changes in specific brain regions, forming much of the rationale for ongoing studies in the area. However, they generally lack cell-type specificity and thus cannot provide conclusive evidence for energy failure within specific neuronal populations and their subcompartments. In certain cases, some cell-type specificity has been attained by combining MRS and measurements of energy metabolites with transgenic approaches targeting specific glial cell types,78,99 and it may be possible to apply similar methods to study neurons.
There is a growing number of methods to assay bioenergetic function within cultured neurons. Among these approaches, new technical advances allow for the sensitive analysis of respiration, glycolysis, and other metabolic functions in adherent cells, including cultured neuron–based models of neurodegenerative disease.100,101 In addition, assays with radiolabeled glucose can monitor bioenergetic flux through glycolysis and the tricarboxylic acid cycle.102,103
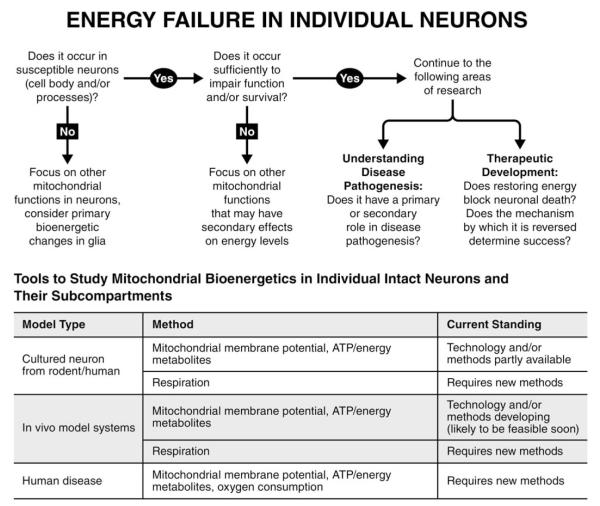
Of particular importance, however, may be new tools that allow measurements within individual neurons, thus factoring out the oftentimes underestimated contribution of glia and allowing the study of specific neuronal subpopulations and subcompartments, such as the nerve terminal (Fig 3). There are now arrays of fluorescence-based sensors that measure bioenergetic function, including those targeting ATP104–106 and glucose.107,108 Although there is no way to directly assay respiration at a subcellular level in mammalian neurons, approaches have been developed to allow for measurements of energy production on a single-neuron level.109,110 These methods complement longstanding approaches, such as the measurement of mitochondrial membrane potential (ΦΨm) with voltage sensitive dyes,111,112 and the use of synaptosomes to study the respiratory function of isolated synaptic boutons.16,113 Thus, these provide new opportunities to understand whether and how energy failure contributes to neuronal dysfunction and death. Although these single-cell approaches are applied primarily to in vitro cell-culture paradigms (see Fig 3), future in vivo applications with 2-photon–based approaches have tremendous potential.
FIGURE 3.
Energy failure in individual neurons. The schematic illustrates an algorithm of critical but challenging questions to determine whether energy failure occurs in individual neurons, including their processes, whether and how it contributes to degeneration, and how it might be targeted therapeutically. The table summarizes the availability of tools and methods to assess the bioenergetic function of mitochondria in individual neurons in model systems and human disease. ATP = adenosine triphosphate.
Assay Design to Target Mitochondrial Bioenergetics
Optimizing assay sensitivity is likely critical for models of neurodegenerative disease where the phenotypes may be subtle, given that these diseases can take years to develop in humans. It may be necessary to perform assays under conditions that minimize alternative energy sources (namely glycolysis) and thus force reliance on mitochondria for energy. For instance, cell lines are commonly grown in the presence of galactose, which is primarily metabolized through the pentose–phosphate pathway, thus forcing reliance on oxidative phosphorylation for ATP.114,115 In this way, the effects of mitochondrial dysfunction or loss on ATP levels and survival can be revealed.45,105 However, galactose is rarely used in studies of neurodegeneration in primary neurons, and glucose levels in standard neuronal culture and imaging buffers are roughly 20-fold higher than in rat brain (1–1.5 mM116,117). Under these nonphysiological conditions, glycolysis compensates for deficits in aerobic respiration, allowing cells to maintain nearly normal levels of ATP and support key functions including synaptic transmission,105,118,119 and this markedly decreases the sensitivity to observe and detect mitochondrial dysfunction.67
Conclusions and Directions
Considering the many unknowns about whether and how energy failure contributes to neurodegeneration, it is not surprising that most clinical trials targeting bioenergetics have had little success. In some cases, the limited success may have resulted from limitations in study design or execution, and hopefully ongoing studies will have better results. Importantly, the conditions where restoring bioenergetics has been most successful, such as replenishing CoQ10 in some individuals with primary CoQ10 deficiency120,121 or creatine in guanidinoacetate methyltransferase deficiency,122,123 are diseases where we have significant insights into the defective element and how to replace it. Clearly, we require a far deeper understanding of how mitochondrial changes lead to energy failure. Understanding these mechanisms will require that we disentangle the effects of mitochondrial morphology, transport, turnover, and ROS formation on bioenergetic function. In addition, in many cases, energy failure may not be an initiating insult, but may occur downstream within cell-death pathways. In those instances, we must understand whether modulating energy failure is still potentially useful as a therapeutic intervention, or whether the affected cells are past the point of no return. Understanding how, when, and in which cells or subcellular compartments energy failure occurs, as well as the threshold levels required to produce neuronal dysfunction and death, would greatly clarify the therapeutic potential of bioenergetic restoration for neurodegenerative disease.
Acknowledgment
This work was supported by a Burroughs-Wellcome Medical Scientist Fund Career Award (K.N.) and grants 1KO8NS062954-01A1 (K.N.) and P30NS069496 from the NIH National Institute of Neurological Disorders and Stroke.
We thank L. Goss for administrative assistance, G. Maki for assistance with illustrations, and A. L. Lucido and G. Howard for assistance editing the manuscript.
Footnotes
Potential Conflicts of Interest Nothing to report.
References
- 1.Zheng B, Liao Z, Locascio JJ, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parihar MS, Brewer GJ. Mitoenergetic failure in Alzheimer disease. Am J Physiol Cell Physiol. 2007;292:C8–C23. doi: 10.1152/ajpcell.00232.2006. [DOI] [PubMed] [Google Scholar]
- 3.Kapogiannis D, Mattson MP. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer’s disease. Lancet Neurol. 2011;10:187–198. doi: 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waagepetersen HS, Sonnewald U, Schousboe A. Compartmentation of glutamine, glutamate, and GABA metabolism in neurons and astrocytes: functional implications. Neuroscientist. 2003;9:398–403. doi: 10.1177/1073858403254006. [DOI] [PubMed] [Google Scholar]
- 7.Vos M, Lauwers E, Verstreken P. Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front Synaptic Neurosci. 2010;2:139. doi: 10.3389/fnsyn.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberthal W, Menza SA, Levine JS. Graded ATP depletion can cause necrosis or apoptosis of cultured mouse proximal tubular cells. Am J Physiol. 1998;274(2 pt 2):F315–F327. doi: 10.1152/ajprenal.1998.274.2.F315. [DOI] [PubMed] [Google Scholar]
- 9.Chiarugi A. “Simple but not simpler”: toward a unified picture of energy requirements in cell death. FASEB J. 2005;19:1783–1788. doi: 10.1096/fj.05-4200rev. [DOI] [PubMed] [Google Scholar]
- 10.Valla J, Berndt JD, Gonzalez-Lima F. Energy hypometabolism in posterior cingulate cortex of Alzheimer’s patients: superficial laminar cytochrome oxidase associated with disease duration. J Neurosci. 2001;21:4923–4930. doi: 10.1523/JNEUROSCI.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabrizi SJ, Cleeter MW, Xuereb J, et al. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Schapira AHV, Mann VM, Cooper JM, et al. Anatomic and disease specificity of NADH CoQ1 reductase (complex I) deficiency in Parkinson’s disease. J Neurochem. 1990;55:2142–2145. doi: 10.1111/j.1471-4159.1990.tb05809.x. [DOI] [PubMed] [Google Scholar]
- 13.Azevedo FA, Carvalho LR, Grinberg LT, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 14.Vives-Bauza C, de Vries RL, Tocilescu MA, Przedborski S. Is there a pathogenic role for mitochondria in Parkinson’s disease? Parkinsonism Relat Disord. 2009;15(suppl 3):S241–S244. doi: 10.1016/S1353-8020(09)70823-5. [DOI] [PubMed] [Google Scholar]
- 15.Liang WS, Reiman EM, Valla J, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telford JE, Kilbride SM, Davey GP. Complex I is rate-limiting for oxygen consumption in the nerve terminal. J Biol Chem. 2009;284:9109–9114. doi: 10.1074/jbc.M809101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 18.Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancuso M, Nardini M, Micheli D, et al. Lack of association between mtDNA haplogroups and Alzheimer’s disease in Tuscany. Neurol Sci. 2007;28:142–147. doi: 10.1007/s10072-007-0807-z. [DOI] [PubMed] [Google Scholar]
- 20.Elson JL, Herrnstadt C, Preston G, et al. Does the mitochondrial genome play a role in the etiology of Alzheimer’s disease? Hum Genet. 2006;119:241–254. doi: 10.1007/s00439-005-0123-8. [DOI] [PubMed] [Google Scholar]
- 21.Moraes CT, Sciacco M, Ricci E, et al. Phenotype-genotype correlations in skeletal muscle of patients with mtDNA deletions. Muscle Nerve. 1995;3:S150–S153. doi: 10.1002/mus.880181429. [DOI] [PubMed] [Google Scholar]
- 22.Cui L, Jeong H, Borovecki F, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Qin W, Haroutunian V, Katsel P, et al. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch Neurol. 2009;66:352–361. doi: 10.1001/archneurol.2008.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins BG, Rosas HD, Chen YC, et al. 1H NMR spectroscopy studies of Huntington’s disease: correlations with CAG repeat numbers. Neurology. 1998;50:1357–1365. doi: 10.1212/wnl.50.5.1357. [DOI] [PubMed] [Google Scholar]
- 25.Lodi R, Schapira AH, Manners D, et al. Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann Neurol. 2000;48:72–76. [PubMed] [Google Scholar]
- 26.Saft C, Zange J, Andrich J, et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov Disord. 2005;20:674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- 27.Mochel F, N’Guyen TM, Deelchand D, et al. Abnormal response to cortical activation in early stages of Huntington disease. Mov Disord. 2012;27:907–910. doi: 10.1002/mds.25009. [DOI] [PubMed] [Google Scholar]
- 28.Hattingen E, Magerkurth J, Pilatus U, et al. Phosphorus and proton magnetic resonance spectroscopy demonstrates mitochondrial dysfunction in early and advanced Parkinson’s disease. Brain. 2009;132(pt 12):3285–3297. doi: 10.1093/brain/awp293. [DOI] [PubMed] [Google Scholar]
- 29.Stamelou M, Pilatus U, Reuss A, et al. In vivo evidence for cerebral depletion in high-energy phosphates in progressive supranu-clear palsy. J Cereb Blood Flow Metab. 2009;29:861–870. doi: 10.1038/jcbfm.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Betarbet R, Sherer TB, MacKenzie G, et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 31.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 32.Massieu L, Del Rio P, Montiel T. Neurotoxicity of glutamate uptake inhibition in vivo: correlation with succinate dehydrogenase activity and prevention by energy substrates. Neuroscience. 2001;106:669–677. doi: 10.1016/s0306-4522(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 33.Brouillet E, Jacquard C, Bizat N, Blum D. 3-Nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington’s disease. J Neurochem. 2005;95:1521–1540. doi: 10.1111/j.1471-4159.2005.03515.x. [DOI] [PubMed] [Google Scholar]
- 34.Greene JG, Greenamyre JT. Characterization of the excitotoxic potential of the reversible succinate dehydrogenase inhibitor malonate. J Neurochem. 1995;64:430–436. doi: 10.1046/j.1471-4159.1995.64010430.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Xu Z, Fang H, et al. Gene expression profiling of MPP+-treated MN9D cells: a mechanism of toxicity study. Neurotoxicology. 2007;28:979–987. doi: 10.1016/j.neuro.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Choi WS, Kruse SE, Palmiter RD, Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc Natl Acad Sci U S A. 2008;105:15136–15141. doi: 10.1073/pnas.0807581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi WS, Palmiter RD, Xia Z. Loss of mitochondrial complex I activity potentiates dopamine neuron death induced by microtubule dysfunction in a Parkinson’s disease model. J Cell Biol. 2011;192:873–882. doi: 10.1083/jcb.201009132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Javitch JA, D’Amato RJ, Strittmatter SM, Snyder SH. Parkinsonism-induced neurotoxin, N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine: uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc Natl Acad Sci U S A. 1985;82:2173–2177. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freire C, Koifman S. Pesticide exposure and Parkinson’s disease: epidemiological evidence of association. Neurotoxicology. 2012;33:947–971. doi: 10.1016/j.neuro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 41.Clark IE, Dodson MW, Jiang C, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 42.Park J, Lee SB, Lee S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 43.Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole AC, Thomas RE, Andrews LA, et al. The PINK1/parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Ouyang Y, Yang L, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Winter D, Ashrafi G, et al. PINK1 and parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin JH, Ko HS, Kang H, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morais VA, Verstreken P, Roethig A, et al. Parkinson’s disease mutations in PINK1 result in decreased complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gandhi S, Wood-Kaczmar A, Yao Z, et al. PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heeman B, Van den Haute C, Aelvoet SA, et al. Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. J Cell Sci. 2011;124(pt 7):1115–1125. doi: 10.1242/jcs.078303. [DOI] [PubMed] [Google Scholar]
- 53.Cho DH, Nakamura T, Fang J, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamp F, Exner N, Lutz AK, et al. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, parkin and DJ-1. EMBO J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura K, Nemani VM, Azarbal F, et al. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J Biol Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song W, Chen J, Petrilli A, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Su B, Lee HG, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka A, Cleland MM, Xu S, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duboff B, Gotz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 61.Misko AL, Sasaki Y, Tuck E, et al. Mitofusin2 mutations disrupt axonal mitochondrial positioning and promote axon degeneration. J Neurosci. 2012;32:4145–4155. doi: 10.1523/JNEUROSCI.6338-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Laar VS, Berman SB. The interplay of neuronal mitochondrial dynamics and bioenergetics: implications for Parkinson’s disease. Neurobiol Dis. 2013;51:43–55. doi: 10.1016/j.nbd.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verstreken P, Ly CV, Venken KJ, et al. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 64.Kaufmann P, Engelstad K, Wei Y, et al. Dichloroacetate causes toxic neuropathy in MELAS: a randomized, controlled clinical trial. Neurology. 2006;66:324–330. doi: 10.1212/01.wnl.0000196641.05913.27. [DOI] [PubMed] [Google Scholar]
- 65.Kruse SE, Watt WC, Marcinek DJ, et al. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wredenberg A, Wibom R, Wilhelmsson H, et al. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKenzie M, Liolitsa D, Akinshina N, et al. Mitochondrial ND5 gene variation associated with encephalomyopathy and mitochondrial ATP consumption. J Biol Chem. 2007;282:36845–36852. doi: 10.1074/jbc.M704158200. [DOI] [PubMed] [Google Scholar]
- 68.Powers WJ, Videen TO, Markham J, et al. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ciarmiello A, Cannella M, Lastoria S, et al. Brain white-matter volume loss and glucose hypometabolism precede the clinical symptoms of Huntington’s disease. J Nucl Med. 2006;47:215–222. [PubMed] [Google Scholar]
- 70.Kadir A, Almkvist O, Forsberg A, et al. Dynamic changes in PET amyloid and FDG imaging at different stages of Alzheimer’s disease. Neurobiol Aging. 2012;33:198 e1–198.e14. doi: 10.1016/j.neurobiolaging.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 71.Forster S, Grimmer T, Miederer I, et al. Regional expansion of hypometabolism in Alzheimer’s disease follows amyloid deposition with temporal delay. Biol Psychiatry. 2012;71:792–797. doi: 10.1016/j.biopsych.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 72.Reiman EM, Chen K, Alexander GE, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vlassenko AG, Vaishnavi SN, Couture L, et al. Spatial correlation between brain aerobic glycolysis and amyloid-beta (Abeta) deposition. Proc Natl Acad Sci U S A. 2010;107:17763–17767. doi: 10.1073/pnas.1010461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchman AS, Wilson RS, Bienias JL, et al. Change in body mass index and risk of incident Alzheimer disease. Neurology. 2005;65:892–897. doi: 10.1212/01.wnl.0000176061.33817.90. [DOI] [PubMed] [Google Scholar]
- 75.Djousse L, Knowlton B, Cupples LA, et al. Weight loss in early stage of Huntington’s disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 76.Chen H, Zhang SM, Hernan MA, et al. Weight loss in Parkinson’s disease. Ann Neurol. 2003;53:676–679. doi: 10.1002/ana.10577. [DOI] [PubMed] [Google Scholar]
- 77.Fex Svenningsen A, Shan WS, Colman DR, Pedraza L. Rapid method for culturing embryonic neuron-glial cell cocultures. J Neurosci Res. 2003;72:565–573. doi: 10.1002/jnr.10610. [DOI] [PubMed] [Google Scholar]
- 78.Funfschilling U, Supplie LM, Mahad D, et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuda W, Furuta T, Nakamura KC, et al. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci. 2009;29:444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tiede LM, Cook EA, Morsey B, Fox HS. Oxygen matters: tissue culture oxygen levels affect mitochondrial function and structure as well as responses to HIV viroproteins. Cell Death Dis. 2011;2:e246. doi: 10.1038/cddis.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu J, Aja S, Kim EK, et al. Physiological oxygen level is critical for modeling neuronal metabolism in vitro. J Neurosci Res. 2012;90:422–434. doi: 10.1002/jnr.22765. [DOI] [PubMed] [Google Scholar]
- 82.Dave KD, De Silva S, Ramboz S, et al. MJFF animal models 2: phenotypic characterization of the autosomal recessive (parkin, Pink-1 and DJ-1) gene knockout rat models of Parkinson’s disease. Soc Neurosci. 2012 Abstract #854.23. [Google Scholar]
- 83.Kitada T, Pisani A, Porter DR, et al. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci U S A. 2007;104:11441–11446. doi: 10.1073/pnas.0702717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Przedborski S, Jackson-Lewis V, Naini AB, et al. The parkinsonian toxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP): a technical review of its utility and safety. J Neurochem. 2001;76:1265–1274. doi: 10.1046/j.1471-4159.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- 85.Scheff SW, Price DA, Schmitt FA, et al. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology. 2007;68:1501–1508. doi: 10.1212/01.wnl.0000260698.46517.8f. [DOI] [PubMed] [Google Scholar]
- 86.Li H, Li SH, Yu ZX, et al. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng HC, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chu Y, Morfini GA, Langhamer LB, et al. Alterations in axonal transport motor proteins in sporadic and experimental Parkinson’s disease. Brain. 2012;135(pt 7):2058–2073. doi: 10.1093/brain/aws133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zambonin JL, Zhao C, Ohno N, et al. Increased mitochondrial content in remyelinated axons: implications for multiple sclerosis. Brain. 2011;134(pt 7):1901–1913. doi: 10.1093/brain/awr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohno N, Kidd GJ, Mahad D, et al. Myelination and axonal electrical activity modulate the distribution and motility of mitochondria at CNS nodes of Ranvier. J Neurosci. 2011;31:7249–7258. doi: 10.1523/JNEUROSCI.0095-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braak H, Del Tredici K. Poor and protracted myelination as a contributory factor to neurodegenerative disorders. Neurobiol Aging. 2004;25:19–23. doi: 10.1016/j.neurobiolaging.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 92.Shepherd GM, Harris KM. Three-dimensional structure and composition of CA3-->CA1 axons in rat hippocampal slices: implications for presynaptic connectivity and compartmentalization. J Neurosci. 1998;18:8300–8310. doi: 10.1523/JNEUROSCI.18-20-08300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuznetsov AV, Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci. 2009;10:1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang DT, Honick AS, Reynolds IJ. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J Neurosci. 2006;26:7035–7045. doi: 10.1523/JNEUROSCI.1012-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davey GP, Peuchen S, Clark JB. Energy thresholds in brain mitochondria. Potential involvement in neurodegeneration. J Biol Chem. 1998;273:12753–12757. doi: 10.1074/jbc.273.21.12753. [DOI] [PubMed] [Google Scholar]
- 96.Musiek ES, Chen Y, Korczykowski M, et al. Direct comparison of fluorodeoxyglucose positron emission tomography and arterial spin labeling magnetic resonance imaging in Alzheimer’s disease. Alzheimers Dement. 2012;8:51–59. doi: 10.1016/j.jalz.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mandal PK. Magnetic resonance spectroscopy (MRS) and its application in Alzheimer’s disease. Concepts Magn Reson Part A. 2007;30:40–65. [Google Scholar]
- 98.Mochel F, Durant B, Meng X, et al. Early alterations of brain cellular energy homeostasis in Huntington disease models. J Biol Chem. 2012;287:1361–1370. doi: 10.1074/jbc.M111.309849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Viader A, Sasaki Y, Kim S, et al. Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron. 2013;77:886–898. doi: 10.1016/j.neuron.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu M, Neilson A, Swift AL, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 101.Yao J, Irwin RW, Zhao L, et al. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herrero-Mendez A, Almeida A, Fernandez E, et al. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- 103.Bak LK, Obel LF, Walls AB, et al. Novel model of neuronal bioenergetics: postsynaptic utilization of glucose but not lactate correlates positively with Ca2+ signalling in cultured mouse glutamatergic neurons. ASN Neuro. 2012;4(3) doi: 10.1042/AN20120004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Imamura H, Nhat KP, Togawa H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kovac S, Domijan AM, Walker MC, Abramov AY. Prolonged seizure activity impairs mitochondrial bioenergetics and induces cell death. J Cell Sci. 2012;125(pt 7):1796–1806. doi: 10.1242/jcs.099176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takanaga H, Chaudhuri B, Frommer WB. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochim Biophys Acta. 2008;1778:1091–1099. doi: 10.1016/j.bbamem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bittner CX, Loaiza A, Ruminot I, et al. High resolution measurement of the glycolytic rate. Front Neuroenergetics. 2010;2 doi: 10.3389/fnene.2010.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Land SC, Porterfield DM, Sanger RH, Smith PJ. The self-referencing oxygen-selective microelectrode: detection of trans-membrane oxygen flux from single cells. J Exp Biol. 1999;202(pt 2):211–218. doi: 10.1242/jeb.202.2.211. [DOI] [PubMed] [Google Scholar]
- 110.Alavian KN, Li H, Collis L, et al. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat Cell Biol. 2011;13:1224–1233. doi: 10.1038/ncb2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buckman JF, Reynolds IJ. Spontaneous changes in mitochondrial membrane potential in cultured neurons. J Neurosci. 2001;21:5054–5065. doi: 10.1523/JNEUROSCI.21-14-05054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ward MW, Rego AC, Frenguelli BG, Nicholls DG. Mitochondrial membrane potential and glutamate excitotoxicity in cultured cerebellar granule cells. J Neurosci. 2000;20:7208–7219. doi: 10.1523/JNEUROSCI.20-19-07208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 115.Rossignol R, Gilkerson R, Aggeler R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 116.McNay EC, Fries TM, Gold PE. Decreases in rat extracellular hippocampal glucose concentration associated with cognitive demand during a spatial task. Proc Natl Acad Sci U S A. 2000;97:2881–2885. doi: 10.1073/pnas.050583697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rex A, Bert B, Fink H, Voigt JP. Stimulus-dependent changes of extracellular glucose in the rat hippocampus determined by in vivo microdialysis. Physiol Behav. 2009;98:467–473. doi: 10.1016/j.physbeh.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 118.Ames A, III, Li YY, Heher EC, Kimble CR. Energy metabolism of rabbit retina as related to function: high cost of Na+ transport. J Neurosci. 1992;12:840–853. doi: 10.1523/JNEUROSCI.12-03-00840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Winkler BS, Arnold MJ, Brassell MA, Sliter DR. Glucose dependence of glycolysis, hexose monophosphate shunt activity, energy status, and the polyol pathway in retinas isolated from normal (nondiabetic) rats. Invest Ophthalmol Vis Sci. 1997;38:62–71. [PubMed] [Google Scholar]
- 120.Rotig A, Appelkvist EL, Geromel V, et al. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 121.Duncan AJ, Bitner-Glindzicz M, Meunier B, et al. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am J Hum Genet. 2009;84:558–566. doi: 10.1016/j.ajhg.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tarnopolsky MA, Beal MF. Potential for creatine and other therapies targeting cellular energy dysfunction in neurological disorders. Ann Neurol. 2001;49:561–574. [PubMed] [Google Scholar]
- 123.Mercimek-Mahmutoglu S, Sinclair G, van Dooren SJ, et al. Guanidinoacetate methyltransferase deficiency: first steps to newborn screening for a treatable neurometabolic disease. Mol Genet Metab. 2012;107:433–437. doi: 10.1016/j.ymgme.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 124.Vos M, Esposito G, Edirisinghe JN, et al. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336:1306–1310. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- 125.Choubey V, Safiulina D, Vaarmann A, et al. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem. 2011;286:10814–10824. doi: 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu G, Zhang C, Yin J, et al. alpha-Synuclein is differentially expressed in mitochondria from different rat brain regions and dose-dependently down-regulates complex I activity. Neurosci Lett. 2009;454:187–192. doi: 10.1016/j.neulet.2009.02.056. [DOI] [PubMed] [Google Scholar]
- 127.Loeb V, Yakunin E, Saada A, Sharon R. The transgenic over expression of alpha-synuclein and not its related pathology associates with complex I inhibition. J Biol Chem. 2010;285:7334–7343. doi: 10.1074/jbc.M109.061051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guzman JN, Sanchez-Padilla J, Wokosin D, et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang X, Yan MH, Fujioka H, et al. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]