Abstract
Over the last decade transcriptome studies of postmortem tissue from subjects with schizophrenia revealed that synaptic, mitochondrial, immune system, GABA-ergic and oligodendrocytic changes are all integral parts of the disease process. The combined genetic and transcriptomics studies argue that the molecular underpinnings of the disease are even more varied than the symptomatic diversity of schizophrenia. Ultimately, to decipher the pathophysiology of human disorders in general, we will need to understand the function of hundreds of genes and regulatory elements in our genome, and the consequences of their overexpression and reduced expression in a developmental context. Furthermore, integration of knowledge from various data sources remains a monumental challenge that has to be systematically addressed in the upcoming decades. In the end, our success in interpreting the molecular changes in schizophrenia will depend on our ability to understand the biology using innovative ideas and cannot depend on the hope of developing novel, more powerful technologies.
Keywords: schizophrenia, gene expression, genetics, gene network, convergence, postmortem
1) Gene expression changes in schizophrenia
Gene expression changes in the postmortem brains of subjects with schizophrenia have been studied for many decades. These studies initially included low throughput methodologies, encompassing northern blotting and in situ hybridization, and were expanded more recently to qPCR, DNA microarrays, and RNAseq (1). Over the last half century a tremendous amount of data has been generated, yet our understanding of gene expression changes in schizophrenia is still limited. Reasons for this incomplete knowledge are complex and include both disease-related factors and technical limitation. Schizophrenia is a spectrum disorder rather than a single diagnosis (2) and the etiology of the disease is complex, encompassing both environmental and genetic factors (3). Substance abuse is quite common in the patient population (4), and various comorbidities and lifestyle differences have also a strong effect on the findings. The picture is further complicated by limited availability of postmortem material, postmortem interval, medication history, circumstances of death and the disease progression between disease onset and the brain harvest (5). In addition, considering the potential cohort biases and very small sample sizes, differences in experimental methodology, and the diverse data analytical methods used, it is perhaps not surprising that transcriptome findings often do not replicate across the investigated cohorts (6).
Still, while most of the single-gene expression changes in schizophrenia have poor reproducibility across studies, data-driven approaches were able to provide us with a more reproducible list of gene expression network disruptions that are related to schizophrenia. While the cascade of causality remains uncertain, it appears that synaptic (7, 8), mitochondrial (9, 10), immune system (6, 11), GABA-ergic (12, 13), and oligodendrocytic (14, 15) mRNA changes are all integral parts of the disease process (1, 16). However, it is important to point out that not all patients show deficits in all molecular domains: there is a clear molecular sub-stratification of patients (6, 11), and that synaptic, immune, oligodendrocytic, GABA-ergic or other, etiologically diverse processes might give rise to the same behavioral disturbance. Thus, schizophrenia is not a disease of a single molecular pathway – rather, transcriptome changes argue for the existence of predominantly “synaptic”, oligodendrocytic” and multiple other molecular subtypes of schizophrenia (17) that sort along a continuum in a complex, partially overlapping pattern: each subject with schizophrenia might show a dominant deficit in one of the molecular domains, yet the overall molecular deficit might also encompass elements from other molecular pathways.
The current review is focused on mRNA changes – however, it is clear that other, non-coding RNA species also play a critical role in regulating gene expression, and appear to significantly contribute to the disease process of schizophrenia (18, 19).
2) Small signals in genetics vs. strong signals in transcriptome
Postmortem gene expression studies are typically performed on dozens of brains, while genome-wide association studies (GWAS) include thousands of patient samples. To date, GWA studies identified a number of genetic elements that predispose to schizophrenia (20–23). It appears that two different, but interrelated mechanisms are at work: common alleles conferring small, cumulative risk to the disease through single nucleotide polymorphisms (SNPs), and low-frequency large effect structural chromosomal abnormalities known as copy-number variants (CNVs). Yet, common SNPs with relatively small effect sizes that can only partially explain the strong heritability of the disease (20–24), and defining a CNV as causal to the disease is even more challenging. In contrast, postmortem gene expression studies reveal much stronger disease-associated signals: even with small sample sizes, there are well-replicated gene expression disturbances that are present in a significant subpopulation of subjects with schizophrenia. For example, GAD1/GAD67 underexpression appears to be a hallmark of the illness (25), and present in the majority of the subjects with the disease – yet, this cannot be explained by genetic susceptibility in the GAD1 gene itself, lifestyle, medication history or other confounds. Similarly, immune system disturbances can be identified in >20% of the postmortem brains of diseased subjects (6, 11, 26), but these changes cannot be traced back to a specific genetic predisposition. Thus, this strongly suggest that gene expression changes are a cumulative readout of different genetic susceptibilities and environmental insults, which converge onto common molecular (and ultimately functional cellular) pathways (5, 27).
Expression quantitative trait loci (eQTLs) studies of schizophrenia also support the notion of common, converging expression readout of genetic vulnerabilities. eQTLs are genomic loci that regulate expression levels of mRNAs or proteins. In context of schizophrenia, one would predict that schizophrenia susceptibility alleles are enriched for those that can affect gene expression, and that eQTLs should carry more true association signals. Recent studies provide strong support for this prediction: schizophrenia susceptibility alleles are enriched for SNPs that affect gene expression in adult human brain. Furthermore, higher probability eQTLs predict schizophrenia better than those with a lower probability for being a eQTL (21, 28).
Further evidence for genome-transcriptome convergence comes from an interesting relationship between genetic susceptibility found in patient DNA and gene expression changes seen in postmortem brains of subjects. The vast majority of the schizophrenia susceptibility genes also show altered transcript expression in the postmortem brain, even in subjects that do not harbor these particular disease-predisposing variants (5). This is true for both the previously identified candidate gene studies and the GWAS uncovered susceptibility genes. For example, the major histocompatibility complex locus confers genetic susceptibility to schizophrenia (29), and transcripts originating from this cytogenetic region also show dysregulation in postmortem transcriptome studies (30, 31) – even in the brains with no apparent genetic susceptibility in the same locus. Similarly, risk-associated SNP of the alpha-1C subunit of the L-type voltage-gated calcium channel (CACNA1C) shows mRNA expression changes in the brain of subjects with schizophrenia (32), and a similar relationship is observed for transcription factor 4 (TCF4) (33), and possibly other GWAS-uncovered schizophrenia-predisposing genes. This can be explained by the above-discussed pathway view of schizophrenia: there might be various, patient-specific disease-predisposing genetic elements in the pathways upstream of the changed transcript. Yet, the different genetic susceptibilities are likely to give rise to a common expression change at points of molecular convergences.
3) Environmental influences and genetic vulnerability converge on the transcriptome
As mentioned above, genetic susceptibility can be strongly potentiated by environmental factors. Increased incidence of schizophrenia has been associated with urban lifestyle, prenatal infections, malnutrition, adolescent cannabis abuse, perinatal hypoxia, and other factors (34). These adverse events act in concert with genetic predisposition and the transcriptome changes represent a sum of gene*environment interactions that jointly tip the balance of the transcriptome. Ultimately, the transcriptome changes will affect growth, axonal pathfinding, neuronal arborization, synapse formation and pruning, energy metabolism and many other developmental and cellular processes. Once the compensatory mechanisms are exceeded, these changes manifest themselves as a specific behavioral phenotype, which we define as the symptoms of the disease.
Many human and transgenic mouse studies demonstrate this very eloquently. However, while human evidence comes from epidemiological studies (34) and can be considered only indirect proof, the animal studies provide clear evidence that the effect of putative schizophrenia susceptibility genes is greatly influenced by environment. DISC1 mutant mice, when exposed to early immune activation or social paradigms, show more pronounced behavioral and molecular deficits not observed in the unchallenged mutants (35). In addition, a mild isolation stress affects the mesocortical projection of dopaminergic neurons, but only when combined with a relevant genetic risk for neuropsychiatric disorders (36). Furthermore, ifitm3−/− mice do not develop the characteristic cellular-molecular-behavioral phenotype after maternal immune activation (e.g. impaired neurite outgrowth and dendritic spine formation, diminished MAP2 expression, altered object recognition and exploratory behavior), underscoring that the gene*environment interaction can act both as detrimental and as protective factors (37).
It is important to note that the environmental modification of genetic disease predisposition is not only related to schizophrenia, but appears to be a universal theme across many neurological and psychiatric disorders. For example, when coupled with adverse life experiences, individuals with one or two copies of the short allele of the 5-HTT promoter polymorphism show more depressive symptoms, diagnosable depression, and suicidality than individuals homozygous for the long allele (38, 39). In contrast, Alzheimer’s Disease mutant mice models, when exposed to enriched environment, show significantly reduced amyloid deposition in the brain and remarkable sparing of cognitive functions (40).
4) Environment predisposes, genetic susceptibility specifies disease
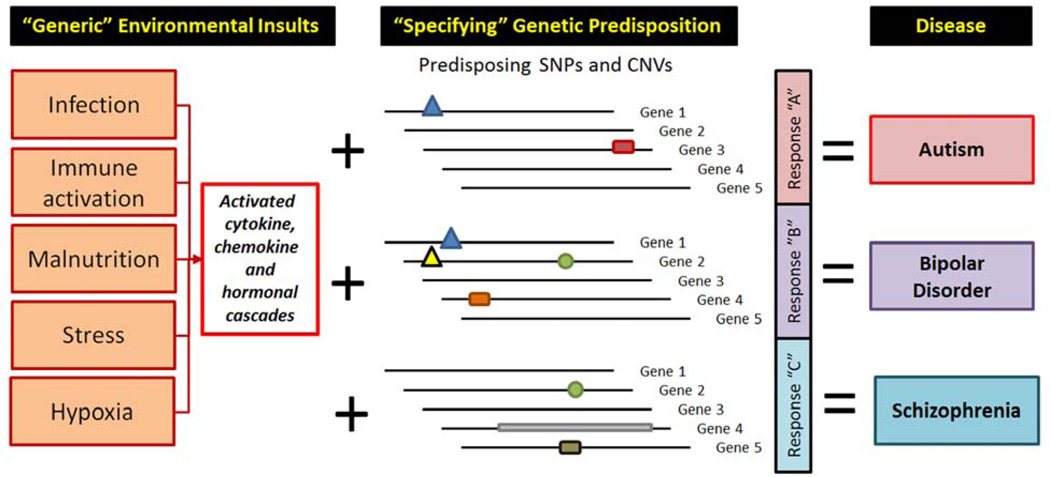
It is well established that environmental influences protect or predispose to disease (34). Yet, environmental factors appear to be quite non-disease specific in their actions. For example, physical exercise slows the progression of Alzheimer’s Disease (41), Parkinson’s Disease (42), Huntington’s Disease (43), and many other brain disorders. Similarly, prenatal immune activation by various agents appears to predispose both to schizophrenia and autism (3). Thus, most of the environmental influences could be viewed as common predisposers/protectors across various brain disorders. As a result, we hypothesize that the disease specificity and phenotypic symptoms are defined by the genetic predisposition: the same environmental insult could have no effect at all, have only a minor effect, or result in well-defined behavioral end-phenotype, often giving rise to a full-blown clinical diagnosis (Figure 1). Thus, the specific behavioral end-results are likely to be defined by the genetic susceptibility. The recent findings of the Cross Disorders PGC group, showing that some disorders have considerable genetic co-variation between disorders (21), actually strengthen our hypothesis. This study basically states that there are common building blocks (genes) of multiple mental disorders. Knowing that the GWAS uncovered disease-predisposing genetic elements are quite distinct for schizophrenia and major depression (22, 44), it is likely that specific disease predisposition is a result of a combination of various genetic elements within the individual’s genome. Thus, the same genetic elements, in different arrangement, will predispose to different disorders.
Figure 1. Environmental influences predispose to disease, genetic susceptibility specifies phenotypic manifestations of brain disorders.
Various environmental influences might predispose to brain disease, but they do this is a non-disease-specific fashion. The disease specificity arises from the genetic makeup of the individual (predisposing mutations, SNPs, or CNVs), and the gene*environment interactions together give rise to diverse symptoms. The combination of various phenotypic manifestations we classify as a disease, and group them together into diagnoses.
Furthermore, it is also noteworthy that the exact timing, duration, frequency and magnitude of the deleterious environmental influences have a huge influence on the ultimate outcome, as genetic predisposition and environmental influences interact on a developmental timeline (45, 46). Although we know that developmental insults during prenatal development predispose to schizophrenia (e.g. prenatal maternal infection), it appears that there are multiple vulnerability periods that extend all the way to adolescence, as cannabis abuse during adolescence has been associated with a greater incidence of the disease (34). Unfortunately, at the current time we do not understand how the behavioral phenotypes are related to the transcriptome changes produced by the gene*environment interaction in the human brain – these processes can only be partially modeled in animals.
5) Genetic diversity and phenotypic similarity
The diagnosis of schizophrenia is established based on phenotypic-symptomatic manifestations of the disease (2). While there is a tremendous genetic diversity across the genome of patients, the manifestations of the disease (established by the DSM diagnostic criteria) are relatively common. The treatment of schizophrenia today is also quite similar, regardless of the underlying genetic diversity (47). Personalized treatment based on genetic makeup of patients, although showing considerable promise, remains elusive (48). However, the phenotypic similarity and use of similar therapy across patients with schizophrenia suggests that there is a strong molecular convergence at work: while the genetic*environmental pathophysiology might be unique for each patient, they feed into common disease-related pathways, disrupting fundamental processes that give rise to altered information processing, emotion, and behavior. This concept is strongly supported by the microanatomical disturbances and transcriptome signature in the postmortem brain from subjects with schizophrenia, where the disturbances are characteristic of a relatively large subgroup of the diseased subjects (6) – even when the studied cohort is too small for the detection of the genetic predisposition to the disease. In this view, schizophrenia can be considered a disease of converging molecular pathways.
6) Hub genes and converging pathways
From the perspective of disease pathophysiology, not all genes and transcripts are created equal. Some genes encode proteins that have hundreds of interacting partners, while others serve a single purpose. Nonsense mutations or deletion of some genes lead to death or a well-defined disease, while others can be removed from the genome with no apparent deleterious effects on overall health or behavior. Thus, gene transcripts can be compared to a set of complexly arranged domino tiles with many converging lines, where a fall of a single domino can have an enormous effect or quite small impact on the overall network.
To address this, the main foci of brain transcriptome studies today are the molecular pathways, rather than single genes. This approach may group genes together based on known functions (e.g. synaptic release genes, transcription factors), gene-centered pathways (e.g. TrKB, AKT, BDNF or DISC1 signaling) (49), disease pathophysiology (e.g. ApoE or PS1 function), metabolic pathways (e.g. oxidative phosphorylation, pentose cycle), substrate activity (e.g. receptor tyrosine kinases, GPCRs), intracellular location of their encoded protein (cell membrane, cytosol, ER), chromosomal location (22q11-13 genes), or any other principle that is appropriate for a particular study. Then, after the pathway analysis is performed, the investigator can make conclusions about the preferential involvement or enrichment of a particular group of transcripts.
These molecular pathway analyses can be performed by a wide variety of commercial or custom-made tools. Over the last ten years several customizable, complementary, and free software packages and approaches were developed. They were instrumental for shifting our attention from “single-gene analysis” to “pathways assessments”. In Supplement 1 we will review only a few of the commonly used bioinformatics toolsets for comprehensive assessment of molecular differences. However, it should be pointed out that our goal was only to highlight the various strategies and the remaining challenges, and not striving for a comprehensive review of the various analytical approaches and methods.
While these analytical tools contribute greatly to our understanding of the disturbed transcriptome and altered cellular function, several significant challenges remain. First, as already mentioned, the relative weight of any gene in a particular network is poorly understood. Should the so called “hub genes” (points of molecular convergence between various pathways) receive special consideration in these analyses, based on their participation in a variety of molecular networks? Should transcription factors be more heavily weighted, as they often control the expression of multiple genes and extensive downstream molecular cascades?
Second, including many members of a single gene family in a pathway can overwhelm the studied pathway, leading to both false negative and false positive findings. For example, including the >15 synaptotagmin genes (50) in a synaptic release pathway could potentially mask the effect on the molecular cascade - especially if the synaptic release pathway is narrowly defined and includes relatively few gene transcripts beyond the synaptotagmin family. Assessing the expression of such a group of genes would represent a “synaptotagmin family” analysis rather than a “synaptic pathway” assessment, potentially leading to false conclusions about the involvement of a synaptic pathway.
Third, how inclusive should any molecular pathway be? Transcripts encoding multifunctional proteins (e.g. early-immediate genes, 14-3-3 gene family) (51) could potentially influence hundreds of molecular pathways, with various effect sizes on each of those. Including them into all the pathways they might participate in might compromises pathway specificity, yet how can we decide when should they be included or left out?
Fourth, most of the pathway analyses do not respect the microanatomical compartmentalization and functional diversity of the brain cells. Yet, the different cell types often use different transcript networks. Most transcriptome experiments are performed on RNA originating from diverse cell populations (e.g. brain region or cortical layer), which contains multiple cell type specific transcriptome profiles (glia, principal neurons, interneurons) and restricted, cell-type specific changes are washed out and blended into an artificial average for the entire sample. For example, while GABA receptors and GAD67 transcripts can be both classified as part of the GABA-ergic pathway, they might not be expressed on the same cell type. Furthermore, underexpression of the same receptor can have vastly different effects on the neural network and behavior if it occurs in the inhibitory interneurons or in principal, glutamatergic excitatory neurons. Our analysis tools are not well suited to respect this anatomical compartmentalization. By finding biologically meaningful correlations, WGCNA is perhaps the best suited analytical method to alleviate some of these concerns (52, 53), yet even this approach cannot assign transcriptome changes to specific cell types of anatomical substructures. As a result, cell type specific enrichment of starting material using laser dissection microscopy (54–57) and flow sorting (58, 59) can take transcriptome profiling to a whole new level of resolution. However, these harvesting methods are labor intensive, time and resource consuming, and low throughput and have seen only limited use in transcriptome profiling experiments to date.
The above-mentioned four challenges still significantly limit our understanding of transcriptome data (60). We must be aware of them in performing our analyses, and as a result, we should always tailor the analytical strategy to the experimental design and the scientific question pursued (1).
7) Methods evolve, the main challenges remain
Transcriptome profiling of human brain disorders is still evolving, and we have an ever-expanding arsenal of tools available to us (61). Northern hybridizations gave way to in situ hybridization and qPCR while gene expression microarrays and SAGE are rapidly losing ground to RNA sequencing. While all of these methods generate valuable, interesting, and technically correct data, significant challenges remain.
First, it appears that our main challenge is biological and not technical. Schizophrenia is a very broad diagnosis, as it describes a spectrum rather than a well-defined disease entity (2). The combined genetic and transcriptomics studies argue that the molecular underpinnings might be as heterogeneous as the clinical presentations among patients, but this will have to be further validated on large-scale studies. Thus, it is not surprising that gene expression findings of schizophrenia often do not replicate across different cohorts of subjects (6). Categorizing schizophrenia into more homogenous biological and genetic constructs has been a major challenge: the clinical sub-classification efforts have not yet resulted in the discovery of clearly predictive (central or peripheral) biomarkers of the disease (62).
The second challenge of molecular profiling is access to meaningful biological material, and postmortem studies are typically performed on dozens of samples, potentially biasing the outcome of such studies. Postmortem brain tissue is a limited, non-renewable resource, obtained several decades after the onset of the disease. The obtained transcriptome profiles reflect a combination of the disease process, progress of illness, postmortem interval, lifestyle changes, comorbidity, treatment of disease, individual variability, and many other factors (5). In contrast, peripheral biomarkers, while readily accessible and better controlled, do not clearly capture the disease process that primarily occurs in the brain. Induced pluripotent stem cells (IPSCs) and olfactory cells from patients are much better suited for understanding the disease mechanisms than assessing biochemical analyses, and the initial studies suggest that they hold a great promise for deciphering disease pathophysiology (63, 64). Yet, at the current time this is a low throughput technology with considerable limitations: schizophrenia is also about connectivity between cells, various cell types, lamination patterns, and many other factors that are extremely challenging to study using in vitro systems.
8) What does the future hold?
At the end, understanding the biology and integration of data are as critical as developing a novel, more powerful technologies. While novel technologies clearly open the door to new, exciting discoveries, diseases are about disturbed function, superimposed on individual variability. We can find enrichment in CNVs in patients with schizophrenia, identify predisposing SNPs, sequence the DNA of all the individuals on the planet, describe transcriptome disturbances associated with the disease, but all this knowledge might not sufficient: such knowledge alone will provide us very limited understanding of the underlying pathophysiology. Our ultimate understanding of the disease will partially come from novel analytical approaches (such as mapping de novo mutations on human brain transcriptome networks throughout development (65)), combined with targeted, functional and mechanistic experiments. Ultimately, to decipher the disease process of schizophrenia, we will need to understand the function of hundreds of genes and regulatory elements in our genome and the consequences of their overexpression and underexpression in a developmental context. Furthermore, we will need to integrate the genetic-transcriptomic-epigenetic-proteomic knowledge with each other and with all other data sources, including (but not limited to) imaging, anatomical, electrophysiological, and epidemiological information. This is the only way that we can understand the phenotypic diversity that characterizes schizophrenia and only this approach will lead to true personalized diagnosis and treatment.
These are monumental tasks and will take many years to complete, yet there is reason for optimism. The CommonMind Consortium is a multi-institutional academic-industrial-non-profit partnership that has as a goal to generate and analyze large scale data from human subjects with neuropsychiatric and neurodevelopmental disorders. The consortium will make this data and the analytical results broadly available to the public as a free resource, and this joint endeavor has a potential to revolutionize our understanding of the molecular pathophysiology of schizophrenia (www.commonmind.org). Importantly, due to a common analysis platform, standardized methodology, uniform processing and large sample size, this approach will eliminate many of the false positive findings and inconsistencies that are found in the literature to date. Thus, the findings might allow a true molecular characterization of postmortem brains into different subgroups of patients, ultimately relating them to the underlying endophenotypes associated with the illness. Furthermore, the NIMH-funded Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative and Human Connectome Project (mapping the neural pathways that underlie human brain function) (66–68), the Neurotherapeutics Network (http://www.neuroscienceblueprint.nih.gov/bpdrugs/- a pipeline between academic and industry drug development research), The Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC - http://www.nitrc.org/), and other large-scale initiatives from the Allen Institute (http://www.alleninstitute.org) have a potential to be even more transformative than the Human Genome Project (69) – but only if the knowledge from the various projects is properly integrated. Where will this all lead? We do not know for certain, and this is the beauty of research. Three decades ago, it was unconceivable that we will ever be able to sequence the whole human genome. To quote Agent K from the Men in Black movie (1997): “1,500 years ago, everybody knew that the Earth was the center of the universe. 500 years ago, everybody knew that the Earth was flat. And 15 minutes ago, you knew that humans were alone on this planet. Imagine what you'll know tomorrow.” So, we just have to keep on working and the new discoveries and breakthroughs will come. We are in it for the long haul.
Supplementary Material
Acknowledgments
The authors are grateful to Martin Schmidt for helpful suggestions and edits of the manuscript. KM’s work is supported by NIMH grants R01MH067234 and R01 MH079299. We are also grateful for the thoughtful and constructive comments of the reviewers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Drs. Mirnics and Horvath reported no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Horvath S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2010;69:157–162. doi: 10.1016/j.biopsych.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhati MT. Defining Psychosis: The Evolution of DSM-5 Schizophrenia Spectrum Disorders. Curr Psychiatry Rep. 2013;15:409. doi: 10.1007/s11920-013-0409-9. [DOI] [PubMed] [Google Scholar]
- 3.Michel M, Schmidt MJ, Mirnics K. Immune system gene dysregulation in autism and schizophrenia. Developmental Neurobiology. 2012;72:1277–1287. doi: 10.1002/dneu.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers RA, Krystal JH, Self DW. A neurobiological basis for substance abuse comorbidity in schizophrenia. Biol Psychiatry. 2001;50:71–83. doi: 10.1016/s0006-3223(01)01134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Horvath S, Mirnics K. Immune System Disturbances in Schizophrenia. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–309. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 9.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2010;29:311–324. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 14.Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V. A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry. 2012;69:1205–1213. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
- 15.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavin DP, Akbarian S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol Dis. 2013;46:255–262. doi: 10.1016/j.nbd.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Mirnics K, Lewis DA. Genes and subtypes of schizophrenia. Trends Mol Med. 2001;7:281–283. doi: 10.1016/s1471-4914(01)02067-6. [DOI] [PubMed] [Google Scholar]
- 18.van Erp TG, Guella I, Vawter MP, Turner J, Brown GG, McCarthy G, et al. Schizophrenia miR-137 Locus Risk Genotype is Associated with Dorsolateral Prefrontal Cortex Hyperactivation. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2011;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 20.Gershon ES, Alliey-Rodriguez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry. 2011;168:253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium PG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aberg KA, Liu Y, Bukszar J, McClay JL, Khachane AN, Andreassen OA, et al. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry. 2013;70:573–581. doi: 10.1001/jamapsychiatry.2013.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan J, Sanders AR, Gejman PV. Genome-wide approaches to schizophrenia. Brain Res Bull. 2010;83:93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2013;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corvin A, Morris DW. Genome-wide Association Studies: Findings at the Major Histocompatibility Complex Locus in Psychosis. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Kano S, Nwulia E, Niwa M, Chen Y, Sawa A, Cascella N. Altered MHC class I expression in dorsolateral prefrontal cortex of nonsmoker patients with schizophrenia. Neurosci Res. 2013;71:289–293. doi: 10.1016/j.neures.2011.07.1818. [DOI] [PubMed] [Google Scholar]
- 31.Sinkus ML, Adams CE, Logel J, Freedman R, Leonard S. Expression of immune genes on chromosome 6p21.3-22.1 in schizophrenia. Brain Behav Immun. 2013;32:51–62. doi: 10.1016/j.bbi.2013.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67:939–945. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S, Cho H, Lee D, Webster MJ. Association between SNPs and gene expression in multiple regions of the human brain. Transl Psychiatry. 2013;2:e113. doi: 10.1038/tp.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2010;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cash-Padgett T, Jaaro-Peled H. DISC1 mouse models as a tool to decipher gene-environment interactions in psychiatric disorders. Front Behav Neurosci. 2013;7:113. doi: 10.3389/fnbeh.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa M, Jaaro-Peled H, Tankou S, Seshadri S, Hikida T, Matsumoto Y, et al. Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science. 2013;339:335–339. doi: 10.1126/science.1226931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibi D, Nagai T, Nakajima A, Mizoguchi H, Kawase T, Tsuboi D, et al. Astroglial IFITM3 mediates neuronal impairments following neonatal immune challenge in mice. Glia. 2013;61:679–693. doi: 10.1002/glia.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonda X, Fountoulakis KN, Harro J, Pompili M, Akiskal HS, Bagdy G, et al. The possible contributory role of the S allele of 5-HTTLPR in the emergence of suicidality. J Psychopharmacol. 2010;25:857–866. doi: 10.1177/0269881110376693. [DOI] [PubMed] [Google Scholar]
- 39.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 40.Lazarov O, Robinson J, Tang YP, Hairston IS, Korade-Mirnics Z, Lee VM, et al. Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell. 2005;120:701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Fang Y. Guiding research and practice: a conceptual model for aerobic exercise training in Alzheimer's disease. Am J Alzheimers Dis Other Demen. 2013;26:184–194. doi: 10.1177/1533317511402317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grazina R, Massano J. Physical exercise and Parkinson's disease: influence on symptoms, disease course and prevention. Rev Neurosci. 2013;24:139–152. doi: 10.1515/revneuro-2012-0087. [DOI] [PubMed] [Google Scholar]
- 43.Kloos AD, Fritz NE, Kostyk SK, Young GS, Kegelmeyer DA. Video game play (Dance Dance Revolution) as a potential exercise therapy in Huntington's disease: a controlled clinical trial. Clin Rehabil. 2013 doi: 10.1177/0269215513487235. [DOI] [PubMed] [Google Scholar]
- 44.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvath S, Mirnics K. Breaking the gene barrier in schizophrenia. Nat Med. 2009;15:488–490. doi: 10.1038/nm0509-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 47.Meltzer HY. Update on typical and atypical antipsychotic drugs. Annu Rev Med. 2013;64:393–406. doi: 10.1146/annurev-med-050911-161504. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JP, Malhotra AK. Pharmacogenetics of antipsychotics: recent progress and methodological issues. Expert Opin Drug Metab Toxicol. 2013;9:183–191. doi: 10.1517/17425255.2013.736964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, Lewis DA, et al. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry. 2006;11:633–648. doi: 10.1038/sj.mp.4001835. [DOI] [PubMed] [Google Scholar]
- 50.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 51.Middleton FA, Peng L, Lewis DA, Levitt P, Mirnics K. Altered expression of 14-3-3 genes in the prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology. 2005;30:974–983. doi: 10.1038/sj.npp.1300674. [DOI] [PubMed] [Google Scholar]
- 52.Mirnics K. What is in the brain soup? Nat Neurosci. 2008;11:1237–1238. doi: 10.1038/nn1108-1237. [DOI] [PubMed] [Google Scholar]
- 53.Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- 55.Sodhi MS, Simmons M, McCullumsmith R, Haroutunian V, Meador-Woodruff JH. Glutamatergic gene expression is specifically reduced in thalamocortical projecting relay neurons in schizophrenia. Biol Psychiatry. 2011;70:646–654. doi: 10.1016/j.biopsych.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arion D, Horvath S, Lewis DA, Mirnics K. Infragranular gene expression disturbances in the prefrontal cortex in schizophrenia: signature of altered neural development? Neurobiol Dis. 2009;37:738–746. doi: 10.1016/j.nbd.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arion D, Unger T, Lewis DA, Mirnics K. Molecular markers distinguishing supragranular and infragranular layers in the human prefrontal cortex. Eur J Neurosci. 2007;25:1843–1854. doi: 10.1111/j.1460-9568.2007.05396.x. [DOI] [PubMed] [Google Scholar]
- 58.Faux C, Rakic S, Andrews W, Yanagawa Y, Obata K, Parnavelas JG. Differential gene expression in migrating cortical interneurons during mouse forebrain development. J Comp Neurol. 2010;518:1232–1248. doi: 10.1002/cne.22271. [DOI] [PubMed] [Google Scholar]
- 59.Nishioka M, Shimada T, Bundo M, Ukai W, Hashimoto E, Saito T, et al. Neuronal cell-type specific DNA methylation patterns of the Cacna1c gene. Int J Dev Neurosci. 2012;31:89–95. doi: 10.1016/j.ijdevneu.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell AC, Mirnics K. Gene expression profiling of the brain: pondering facts and fiction. Neurobiol Dis. 2011;45:3–7. doi: 10.1016/j.nbd.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ginsberg SD, Mirnics K. Functional genomic methodologies. Prog Brain Res. 2006;158:15–40. doi: 10.1016/S0079-6123(06)58002-1. [DOI] [PubMed] [Google Scholar]
- 62.Stober G, Ben-Shachar D, Cardon M, Falkai P, Fonteh AN, Gawlik M, et al. Schizophrenia: from the brain to peripheral markers. A consensus paper of the WFSBP task force on biological markers. World J Biol Psychiatry. 2009;10:127–155. doi: 10.1080/15622970902898980. [DOI] [PubMed] [Google Scholar]
- 63.Yu DX, Marchetto MC, Gage FH. Therapeutic translation of iPSCs for treating neurological disease. Cell Stem Cell. 2013;12:678–688. doi: 10.1016/j.stem.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 64.Kano S, Colantuoni C, Han F, Zhou Z, Yuan Q, Wilson A, et al. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2012;18:740–742. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kandel ER, Markram H, Matthews PM, Yuste R, Koch C. Neuroscience thinks big (and collaboratively) Nat Rev Neurosci. 2013;14:659–664. doi: 10.1038/nrn3578. [DOI] [PubMed] [Google Scholar]
- 67.Lichtman JW, Sanes JR. Ome sweet ome: what can the genome tell us about the connectome? Curr Opin Neurobiol. 2008;18:346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts L, Davenport RJ, Pennisi E, Marshall E. A history of the Human Genome Project. Science. 2001;291:1195. doi: 10.1126/science.291.5507.1195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.