Abstract
Background
Previous work examining normal controls from the Alzheimer's Disease Neuroimaging Initiative (ADNI) identified substantial biological heterogeneity. We hypothesized that ADNI mild cognitive impairment (MCI) subjects would also exhibit heterogeneity with possible clinical implications.
Methods
138 ADNI subjects diagnosed with amnestic MCI were clustered based on baseline MRI, cerebrospinal fluid, and serum biomarkers. The clusters were compared with respect to longitudinal atrophy, cognitive trajectory, and time to conversion.
Results
Four clusters emerged with distinct biomarker patterns: The first cluster was biologically similar to normal controls and rarely converted to Alzheimer's disease (AD) during follow-up. The second cluster had characteristics of early Alzheimer’s pathology. The third cluster showed the most severe atrophy but barely abnormal tau levels and a substantial proportion converted to clinical AD. The fourth cluster appeared to be pre-AD and nearly all converted to AD.
Conclusions
Subjects with MCI who were clinically similar showed substantial heterogeneity in biomarkers.
Keywords: ADNI, Alzheimer’s disease, clustering, heterogeneity, amnestic MCI
1. Introduction
The Alzheimer's Disease Neuroimaging Initiative (ADNI) was designed to have three clinically distinct study groups - cognitively normal controls (NC), individuals with mild cognitive impairment (MCI) who had a memory complaint but did not have significant impairment in other domains, and a group of individuals with mild AD who met NINCDS/ADRDA criteria for probable AD. These groups were designed to be reasonably homogeneous within their respective diagnostic categories. Despite attempts to acquire clinically homogeneous groups at baseline, we previously found that normal controls had considerable underlying biological heterogeneity which was associated with cognitive differences [1]. We hypothesized that the MCI group also contains substantial biological heterogeneity, also with possible clinical consequences.
Amnestic MCI is a subset of MCI which is often thought of as prodromal-AD. A substantial proportion of individuals with amnestic MCI will ultimately convert to AD over 2-5 years [2, 3]. The current amyloid cascade hypothesis of AD suggests that the process begins with Amyloid-Beta (Aβ) plaque deposition, followed by measurable changes in CSF tau proteins, then changes in brain volume, and finally clinically detectable cognitive change [4]. However, not all individuals with MCI progress to dementia and there is heterogeneity in the underlying pathology of those that do progress [5]. In an autopsy study, Jicha et al found that a substantial number of amnestic subjects had primary pathologies other than AD [3]. Possible non-AD pathologies that may produce amnestic MCI include vascular dementia, hippocampal sclerosis, and frontotemporal dementia (FTD) [6–9]. It is also common for multiple cognitive pathologies to coexist, which adds to the difficulty of classification, both clinically and pathologically [10].
Understanding the biological and clinical heterogeneity represented by the seemingly specific clinical diagnosis of amnestic MCI is essential to understanding the pathways involved in cognitive decline and ultimately necessary for the development of treatment. The goal of this analysis was to characterize the heterogeneity in the ADNI amnestic MCI group. Cluster analysis allows an unbiased characterization of the data – free from artificial cut-offs or dichotomizations which remove meaningful variability. Subjects in this study were clustered to see if common patterns exist in the biomarker profiles of the MCI participants and these profiles were compared to the anticipated pattern of biomarkers for prodromal AD prescribed by the amyloid cascade hypothesis [11]. Next, clusters were characterized and tested for association with subject characteristics and clinical outcomes.
2. Methods
2.1 Subjects
The data used for this analysis were downloaded from the ADNI database (www.loni.ucla.edu/ADNI) on 14 December 2011. The individuals studied were recruited between 17 August 2005 and 4 September 2007 as ADNI participants. Additional details are given in Petersen et al. [12]. This study was approved by the Institutional Review Boards of all participating institutions. Informed written consent was obtained from all participants at each site. A detailed description of the study design and inclusion criteria is available at clinicaltrials.gov/show/NCT00106899.
All enrolled ADNI MCI subjects were amnestic MCI; the diagnostic classification required MMSE scores between 24-30 (inclusive), a memory complaint, objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, absence of significant impairment in other cognitive domains, essentially preserved activities of daily living, and absence of dementia. The ADNI MCI diagnostic group contained 382 individuals. Of those, 189 individuals consented to cerebrospinal fluid (CSF) testing. Baseline MRI scans failed to meet quality control in 18 subjects, 32 subjects had missing values for white matter hyperintensity, and two subjects were missing homocysteine data. 138 MCI subjects had complete baseline data for the variables chosen for this analysis.
2.2 Measures
The biological focus of the analysis was on eleven variables: total brain volume, hippocampal volume, ventricle volume, entorhinal cortex thickness, CSF amyloid beta (Aβ1-42), CSF total tau (tau), CSF phosphorylized tau (Ptau181P), the ratio of tau to Aβ1-42, the ratio of P-tau181P to Aβ1-42, white matter hyperintensity (WMH), and serum homocysteine. All MRI summary volumes were calculated as fractions of the total intracranial volume, which included the area occupied by the brainstem inside the skull and were calculated using Quarc, a modification of Free-Surfer implemented by the Anders Dale Laboratory at UC San Diego as part of the ADNI shared dataset [13, 14] 4. Individual longitudinal changes were calculated from cross-sectional Quarc summaries at available follow-up times by fitting mixed models [15]. WMH were detected by Imaging of Dementia and Aging (IDeA) laboratory at UC-Davis based on coregistered T1-,T2-, and proton density (PD)-weighted images using an automated protocol [16]. CSF samples were batch processed by the ADNI Biomarker Core at the University of Pennsylvania, School of Medicine [17]. This set of biomarkers was chosen to view the biology underlying cognitive impairment from multiple perspectives by including measures of abnormal protein activity associated with AD, region-specific atrophy, vascular damage, and a blood serum measure associated with dementia. The clinical outcomes used in this analysis were the following cognitive tests: Digit Span forward and backward, Digit Symbol Substitution, Trails B, Category Naming (sum of animal and vegetable scores), the Mini-Mental State Exam (MMSE), ADAS cognitive subscale, the sum of five trials from the Rey's Auditory Vocabulary List Test (RAVLT), and Logical Memory II [18–24].
2.3 Statistical Analysis
Cluster analysis provides a unique opportunity to group individuals based on their biomarkers that is not based on - or biased by - artificial cut-offs or long-term trajectories. The goal of cluster analysis is to separate individuals into groups such that individuals within a group are as similar to each other as possible and groups are as different from each other as possible. To cluster the MCI subjects, we used agglomerative hierarchical clustering with Ward's method of minimum variance and the Euclidean distance metric. This method seeks to minimize the variance of the distances from each individual in a cluster to the cluster center, thereby ensuring similarity of the individuals within a cluster. In preparation for clustering, each clustering variable was standardized by the overall MCI mean and SD for that variable so that variables on different numerical scales could be fairly compared. The number of clusters was not determined a priori. Instead, the number was chosen using the maximum gap statistic, but in a less conservative manner than originally described which showed promising results in simulations [25], along with visual ascertainment of cluster separation and minimum sample size considerations.
ANOVA F-statistics and exact tests were used to test for baseline differences in demographics, genetics, and baseline cognitive scores among the four clusters. Comparisons of cognitive test scores at baseline did not control for education or age. Baseline biomarkers are reported with 95% confidence intervals, as significant differences between the clusters are to be expected. Linear regression models with random slopes and intercepts were used to estimate adjusted baseline measures and test for longitudinal differences in anatomical and cognitive change. Models of anatomical and cognitive outcomes controlled for age (centered) and the interaction of age with time. Models of cognitive outcomes also controlled for years of education (centered). Time to conversion was assessed with an accelerated failure time (AFT) model assuming a Weibull distribution and controlling for age. The interval censored accelerated failure time model was parameterized in two different ways. First, the healthiest looking cluster (MCI 1) was made the reference and all others were compared to it. Next, the model was parameterized in the following way in order to achieve sequential comparisons (i.e. MCI 2:MCI 1, MCI 3:MCI 2, MCI 4:MCI 3). Individuals who were classified as MCI at baseline were included in these analyses, even if they later were reported as normal. P-values for cluster comparisons have not been adjusted for multiple comparisons. All analysis was performed in R 2.10.0. and R 2.11.0 [26].
3. Results
3.1 Baseline differences
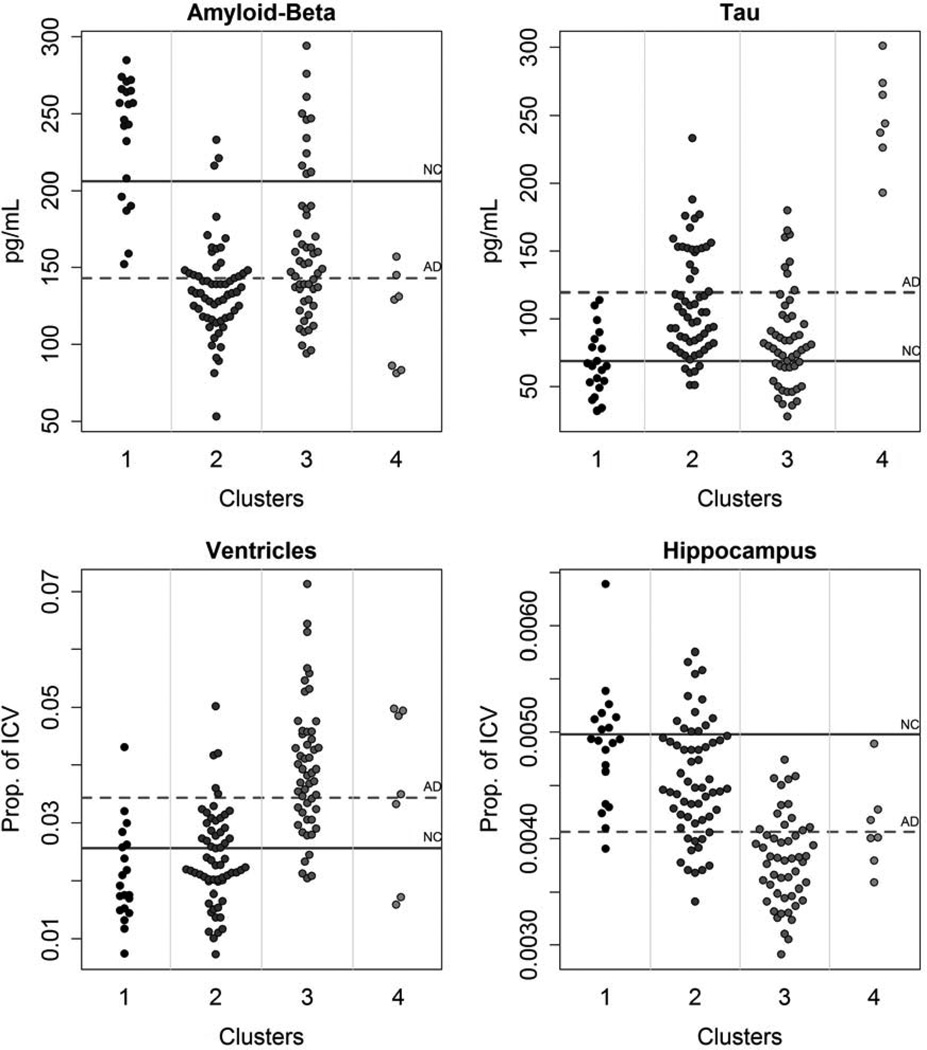
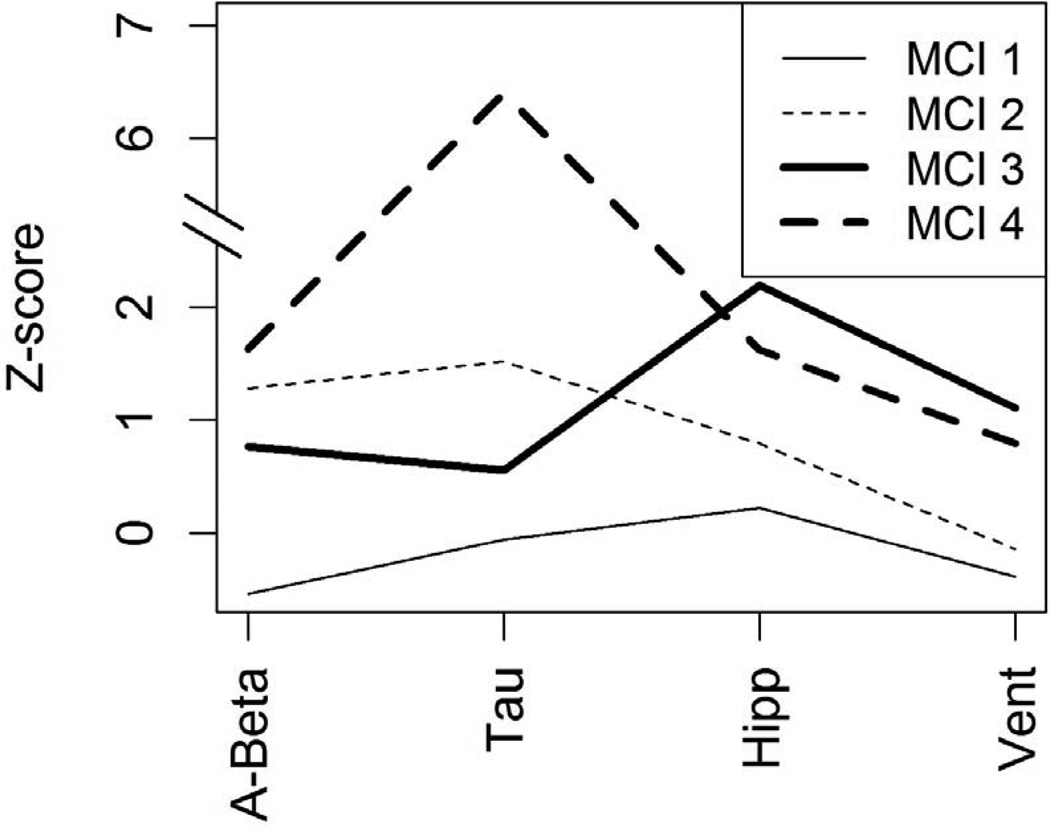
Four clusters were found; Figure 1 displays some of the biomarker characteristics in what are essentially small bin histograms showing each subject (means from the normal controls and AD subjects in ADNI are provided as reference, more information on the ADNI control and AD subjects can be found in the supplemental table). Biologically, the clusters can be summarized as follows: MCI 1 (n=20) had the healthiest biomarker profile, often centered around the normal control (NC) mean and sometimes less abnormal than the NC mean (Figure 1). MCI 4 (n=7) had the least healthy biomarker profile, often near or beyond the averages in the AD group. MCI 2 (n = 60) was between these and had characteristics suggestive of the early stages of AD, as described in the Jack et al. model [4]. MCI 3 (n=51) was similar in many ways to MCI 2, but with substantially lower tau and much worse MRI measures. The clusters differed on age (Table 1); post hoc testing using Tukey’s Honest Significant Difference method indicated that MCI 1 and 2 were significantly younger than MCI 3 and 4, but MCI 1 and 2 did not differ significantly from each other and MCI 3 and 4 did not differ significantly from each other. Table 2 shows means and 95% confidence intervals in each cluster for all of the clustering biomarkers, in which the extent of cluster separation on each biomarker can be seen. There were significant differences among the clusters on the Digit Symbol Substitution test and Trails B, as well as all memory tests (Table 1). In general, MCI 1 had the best cognitive scores, MCI 2 and 3 were in the middle and often very close to one another, and MCI 4 had the most severe impairment. This cognitive continuum might suggest that all of the clusters are on the same path to AD, with some simply farther along the path. However, the plot of Aβ1-42, tau, hippocampal volume, and ventricle volume in Figure 2 suggests this is not the case. For this figure, group z-scores have been scaled by the cognitively normal group's mean and standard deviation, so zero represents true cognitive normality, not the average of MCI subjects. If all of the individuals were on the same biological path, one would expect that the clusters could be ordered with increasingly abnormality. Instead, the plot demonstrates a lack of consistent ordering, with more abnormal CSF measures in MCI 2 than MCI 3.
Figure 1.
Distributions by cluster for a subset of clustering biomarkers. The solid gray line represents the mean from the ADNI normal controls and the dashed line represents the AD mean.
Table 1.
Cluster comparison on baseline demographic, genetic, clinical, and biomarker measures. Means and standard deviations are reported unless otherwise noted. Confidence intervals (95%, two-sided) have been reported for the clustering biomarkers as significant differences are expected. P-values come from ANOVA F-statistics and exact tests for categorical variables.
| MCI 1 (N = 20) |
MCI 2 (N = 60) |
MCI 3 (N = 51) |
MCI 4 (N = 7) |
|||
|---|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | p-val | ||
| Demographic | ||||||
| Age (years) | 71.7 ± 8 | 72.2 ± 7.2 | 76.7 ± 7.1 | 76.3 ± 3.6 | 0.001 | |
| Gender (male) | 60% | 55% | 73% | 57% | 0.27 | |
| Education (years) | 15.5 ± 2.7 | 15.9 ± 3.1 | 16.2 ± 3 | 15.7 ± 1.8 | 0.50 | |
| ApoE4 (+)a | 40% | 60% | 51% | 71% | 0.44 | |
| Parent with AD | 25% | 38% | 20% | 29% | 0.34 | |
| Follow-up (years) | 2.8 ± 0.5 | 2.7 ± 0.6 | 2.5 ± 0.6 | 2.6 ± 0.6 | 0.08 | |
| Clinical | ||||||
| FAQ | 1.8 ± 3.2 | 4.3 ± 4.7 | 3.8 ± 4.4 | 3.1 ± 3.9 | 0.38 | |
| Geriatric Depression | 1.8 ± 1.1 | 1.5 ± 1.4 | 1.6 ± 1.1 | 1.1 ± 0.7 | 0.56 | |
| Attention | ||||||
| Digit Span Forward | 8 ± 2.2 | 8.2 ± 2.3 | 8.5 ± 2.1 | 8 ± 1.4 | 0.50 | |
| Digit Span Backward | 7.2 ± 2 | 6.2 ± 1.9 | 6.1 ± 2 | 7.3 ± 1.3 | 0.38 | |
| Executive | ||||||
| Digit Symbol | 45 ± 11.4 | 36.8 ± 11.6 | 34.6 ± 9.4 | 39.6 ± 9.4 | 0.01 | |
| Trails B | 85.9 ± 51.9 | 132 ± 76.7 | 141.4 ± 66.2 | 148.7 ± 61.3 | 0.01 | |
| Category Naming | 29.3 ± 6.1 | 26.9 ± 7.4 | 27.8 ± 8.1 | 21.7 ± 6.4 | 0.18 | |
| Memory | ||||||
| MMSE | 27.6 ± 1.7 | 27 ± 1.8 | 26.4 ± 1.8 | 26.9 ± 2 | 0.02 | |
| ADAS-cog | 9.8 ± 4.4 | 11.8 ± 4.4 | 12.3 ± 4.8 | 12.8 ± 4.2 | 0.046 | |
| RAVLT (sum of 5 trials) | 34.5 ± 10.3 | 30.3 ± 8.3 | 28.5 ± 8.1 | 29.1 ± 6.9 | 0.02 | |
| RAVLT 30 min | 4.3 ± 3.6 | 2.8 ± 3.6 | 1.8 ± 2.2 | 0.9 ± 1.6 | 0.001 | |
| RAVLT (% savings) | 0.5 ± 0.3 | 0.3 ± 0.4 | 0.2 ± 0.2 | 0.1 ± 0.3 | 0.001 | |
| Logical Memory II | 5.3 ± 2.4 | 3.4 ± 2.8 | 3.5 ± 2.5 | 2.3 ± 2.1 | 0.01 | |
Proportion with at least one E4 allele
Table 2.
Means and associated 95% confidence intervals for clustering biomarkers by cluster.
| MCI 1 | MCI 2 | MCI 3 | MCI 4 | |
|---|---|---|---|---|
| Whole Brain Vol.a | 0.6982 | 0.6801 | 0.6586 | 0.659 |
| (0.6888, 0.7076) | (0.6747, 0.6855) | (0.6527, 0.6645) | (0.6431, 0.6748) | |
| Hippocampal Vol.a | 0.0049 | 0.0046 | 0.0038 | 0.0041 |
| (0.0046, 0.0051) | (0.0044, 0.0047) | (0.0037, 0.0039) | (0.0037, 0.0045) | |
| Ventricle Vol.a | 0.0209 | 0.0239 | 0.0395 | 0.0356 |
| (0.0165, 0.0252) | (0.0214, 0.0264) | (0.0368, 0.0422) | (0.0282, 0.0429) | |
| Entorhinal Thick.b | 3.206 | 3.0501 | 2.5764 | 2.3871 |
| (3.0327, 3.3792) | (2.95, 3.1501) | (2.4679, 2.6849) | (2.0943, 2.6800) | |
| WMHc | 1.433 | 2.557 | 2.52 | 3.624 |
| (0.558, 2.308) | (2.051, 3.062) | (1.972, 3.068) | (2.145, 5.103) | |
| Amyloid Beta1-42d | 236.1 | 135.3 | 163.7 | 116 |
| (218.5, 253.7) | (125.1, 145.5) | (152.7, 174.8) | (86.2, 145.8) | |
| Taud | 67.2 | 111.5 | 84.5 | 248.6 |
| (51.2, 83.1) | (102.3, 120.7) | (74.5, 94.4) | (221.7, 275.5) | |
| P-Tau181d | 21.1 | 42.7 | 29.8 | 68.3 |
| (15, 27.1) | (39.3, 46.2) | (26, 33.6) | (58.1, 78.4) | |
| P-Tau/A-Beta | 0.3 | 0.86 | 0.57 | 2.3 |
| (0.16, 0.44) | (0.78, 0.94) | (0.48, 0.66) | (2.06, 2.54) | |
| Tau/A-Beta | 0.1 | 0.33 | 0.2 | 0.62 |
| (0.04, 0.15) | (0.3, 0.37) | (0.17, 0.23) | (0.53, 0.7) | |
| Homocysteinee | 10.31 | 9.64 | 10.81 | 10.11 |
| (9.07, 11.56) | (8.92, 10.36) | (10.03, 11.59) | (8, 12.22) |
Presented as fraction of ICV
Average of right and left in mm
cm3
pg/mL
μmol/L
Figure 2.
The Z-scores shown were created by subtracting the ADNI normal mean and dividing by the ADNI normal standard deviation for each biomarker shown, so that zero represents actual normality, not normality within the MCI subgroup. Signs have been reversed so that large, positive values always indicate more severe damage. Hipp = hippocampal volume, Vent = ventricle volume.
3.2 Atrophy
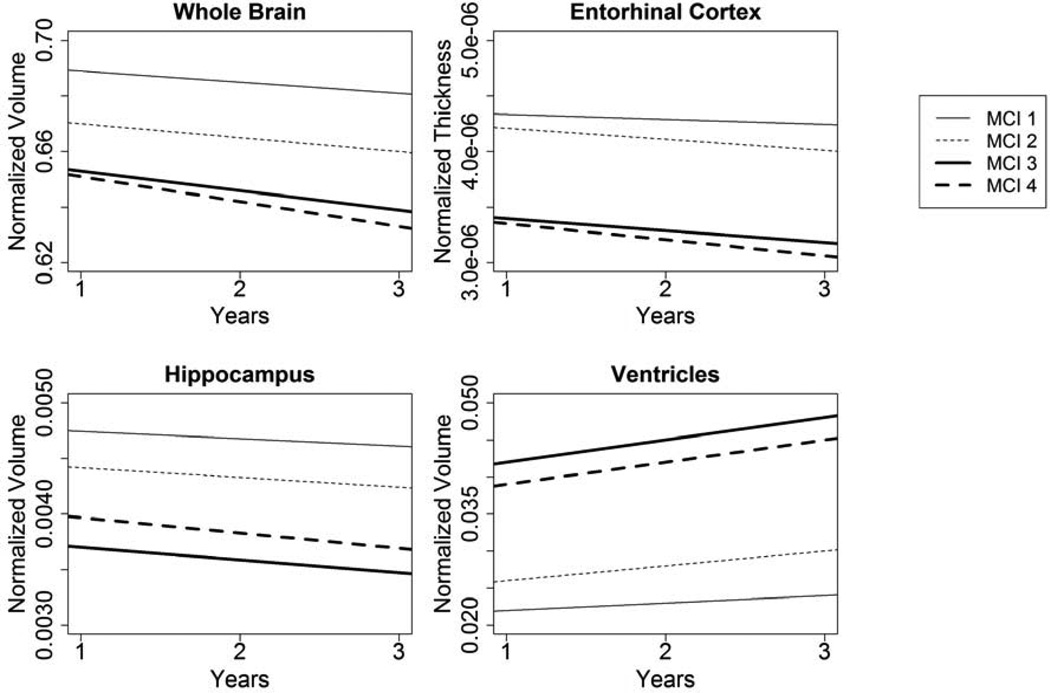
The clusters were created using baseline MRI measurements and consequently showed significant differences in atrophy at baseline, but the clusters also had significantly different rates of atrophy over time (Figure 3). MCI 1 (reference group) showed atrophic change for brain (−0.004, p < 0.001), hippocampus (−6.58×10−5, p < 0.001), entorhinal cortex (−4.44×10−8, p = 0.01), and ventricular volume (0.001, p = 0.04). Compared to the reference group, MCI 2 had a significantly worse atrophic change in the entorhinal cortex (−5.66×10−8 additional decline per year, p = 0.002) and ventricular volume (0.001 additional increase per year, p = 0.01), but not for hippocampal or total brain volumes (p > 0.15). MCI 3 and 4 experienced significantly greater annual change than MCI 1 on every measure, with rates of change approximately two to four times greater than MCI 1 (all p ≤ 0.01).
Figure 3.
Estimated atrophy in all four MCI clusters over a 3 year period. Estimates come from linear mixed effects regression models with random effects for slope and intercept and control for age and the interaction of age with time. There were significant differences between MCI 1 and MCI 3 and 4 for all measures. MCI 1 differed from MCI 2 in ventricles and entorhinal cortex at baseline and brain and hippocampus atrophy over time. Secondary analysis excluding MCI 1 and using MCI 3 as the referent found no significant differences in intercept between MCI 3 and MCI 4 for any of the MRI measures. MCI 4 experienced significantly more rapid deterioration in the entorhinal cortex than MCI 3. MCI 2 and MCI 3 differed significantly in both slope and intercept for all MRI measures.
3.3 Cognitive decline
Groups also differed significantly in both baseline and longitudinal cognitive performance (Table 3). Baseline performance differed on a number of memory and executive function tests, but not on the Mini Mental State Exam (MMSE). Over time, the models showed significant differences between groups in the areas of memory and executive function. For example, MCI 1 generally showed no change in cognitive performance and even showed significant improvement on the Logical Memory II test (0.75 points per year, p <0.001). MCI 2, in contrast, showed significant decline on every test except Digit Span (forward and backward) and Logical Memory II, although Logical Memory II was nearly significant (95%CI: −0.46, 0.06). MCI 3 and MCI 4 also showed global declines on nearly all measures. For the few cognitive measures in which MCI 3 and 4 did not have significant decline, the baseline values for MCI 3 and 4 were substantially worse, suggesting possible plateauing.
Table 3.
Adjusted baseline and annual change results (coefficients, standard errors, and p-values) from longitudinal linear regression models with random effects for slope and intercept. Cluster was used as a predictor in models with cognitive scores as outcomes. All models were adjusted for age, age*time, and education. Age and education have been centered; results are for individuals of average age and education. The intercept is the average baseline level of MCI 1. The rest of the baseline values show how the other clusters differ from MCI 1. The reference slope is the slope for MCI 1. The rest of the annual change values show the difference in slope between the rest of the clusters and MCI 1. The significant p-values for the baseline referent are for the trivial test that the intercept equals zero.
| MC1 (referent) | MCI 2 (vs MCI 1) | MCI 3 (vs MCI 1) | MCI 4 (vs MCI 1) | |||||
|---|---|---|---|---|---|---|---|---|
| Baseline | Est ± SE | p-value | Est ± SE | p-value | Est ± SE | p-value | Est ± SE | p-value |
| Digit Span | ||||||||
| Forward | 8.28 ± 0.43 | <0.0001 | −0.05 ± 0.49 | 0.91 | −0.13 ± 0.51 | 0.80 | −0.65 ± 0.83 | 0.44 |
| Digit Span | ||||||||
| Backward | 7.37 ± 0.41 | <0.0001 | −1.31 ± 0.47 | 0.01 | −1.29 ± 0.49 | 0.01 | −0.72 ± 0.80 | 0.37 |
| Digit Symbol | 46.50 ± 2.47 | <0.0001 | −8.92 ± 2.8 | 0.002 | −12.10 ± 2.95 | 0.0001 | −5.12 ± 4.81 | 0.29 |
| Trails B | 84.76 ± 15.21 | <0.0001 | 44.4 ± 17.3 | 0.01 | 61.26 ± 18.18 | 0.001 | 73.61 ± 29.58 | 0.01 |
| Category Naming | 29.31 ± 1.53 | <0.0001 | −2.42 ± 1.74 | 0.17 | −2.10 ± 1.83 | 0.25 | −5.73 ± 2.98 | 0.06 |
| ADAS-cog | 9.22 ± 0.97 | <0.0001 | 2.07 ± 1.11 | 0.06 | 3.05 ± 1.16 | 0.001 | 2.74 ± 1.90 | 0.15 |
| RAVLT (sum of 5) | 34.00 ± 1.83 | <0.0001 | −4.21 ± 2.08 | 0.045 | −6.29 ± 2.19 | 0.005 | −6.87 ± 3.57 | 0.06 |
| Logical Memory II | 5.83 ± 0.72 | <0.0001 | −2.21 ± 0.82 | 0.01 | −2.55 ± 0.86 | 0.004 | −3.83 ± 1.41 | 0.008 |
| MMSE | 27.72 ± 0.63 | <0.0001 | −1.04 ± 0.72 | 0.15 | −1.33 ± 0.76 | 0.08 | −0.79 ± 1.24 | 0.52 |
| MC1 (referent) | MCI 2 (vs MCI 1) | MCI 3 (vs MCI 1) | MCI 4 (vs MCI 1) | |||||
| Annual Change | Est ± SE | p-value | Est ± SE | p-value | Est ± SE | p-value | Est ± SE | p-value |
| Digit Span | ||||||||
| Forward | 0.03 ± 0.12 | 0.79 | −0.09 ± 0.14 | 0.51 | −0.44 ± 0.14 | 0.002 | −0.40 ± 0.24 | 0.09 |
| Digit Span | ||||||||
| Backward | −0.24 ± 0.16 | 0.13 | 0.10 ± 0.18 | 0.56 | −0.05 ± 0.19 | 0.80 | −0.50 ± 0.33 | 0.13 |
| Digit Symbol | 0.53 ± 0.71 | 0.45 | −2.82 ± 0.81 | <0.0001 | −2.75 ± 0.86 | 0.001 | −6.24 ± 1.44 | <0.0001 |
| Trails B | 3.97 ± 5.48 | 0.47 | 10.88 ± 6.32 | 0.09 | 8.01 ± 6.68 | 0.23 | 3.21 ± 10.92 | 0.77 |
| Category Naming | −0.08 ± 0.58 | 0.89 | −1.32 ± 0.67 | 0.05 | −2.00 ± 0.71 | 0.005 | −3.61 ± 1.17 | 0.002 |
| ADAS-cog | −0.26 ± 0.59 | 0.66 | 1.72 ± 0.68 | 0.01 | 2.74 ± 0.71 | 0.0001 | 6.33 ± 1.19 | <0.0001 |
| RAVLT (sum of 5) | −0.30 ± 0.46 | 0.51 | −0.80 ± 0.54 | 0.14 | −1.61 ± 0.56 | 0.005 | −4.10 ± 0.97 | <0.0001 |
| Logical Memory II | 0.75 ± 0.22 | <0.0001 | −0.95 ± 0.26 | 0.0002 | −1.37 ± 0.27 | <0.0001 | −1.56 ± 0.45 | <0.0001 |
| MMSE | 0.09 ± 0.21 | 0.68 | −0.78 ± 0.24 | 0.001 | −1.43 ± 0.25 | <0.0001 | −2.56 ± 0.41 | <0.0001 |
3.4 Conversion to AD
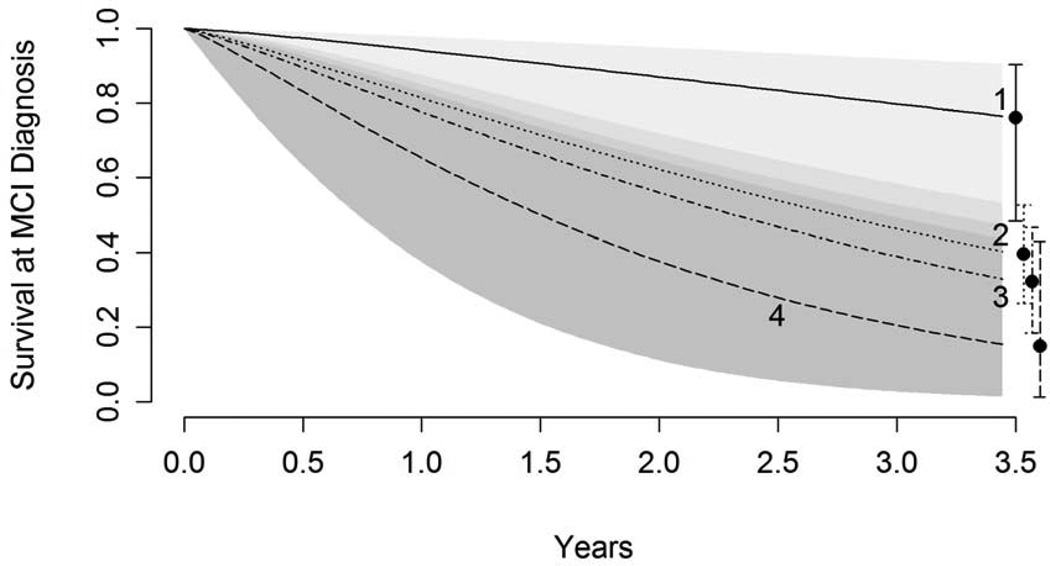
After an average of 2.6 years of follow-up, 70 subjects (51%) converted to AD; four conversions in MCI 1(20%), 32 conversions in MCI 2 (53%), 28 conversions in MCI 3 (55%), and 6 conversions in MCI 4 (85%). Accelerated failure time models were used to compare time until diagnosis of AD. MCI 2, MCI 3, and MCI 4 were all significantly different from MCI 1 in time to conversion but not from each other (Figure 4). There were three individuals who reverted back to the normal control diagnostic category at some time during follow up (two from MCI 2 and one from MCI 1).
Figure 4.
‘Survival’ estimates from interval censored accelerated failure time models, where survival indicates maintaining a diagnosis of MCI rather than converting to AD. Model results indicate that MCI 2, 3, and 4 all differ significantly from MCI 1 but do not differ significantly among each other in time to conversion.
3.5 Secondary analyses
MCI 1 was clearly distinguishable from the rest of the MCI group, with a healthier biomarker profile, a lack of cognitive decline, and minimal conversions. This motivated a secondary analysis comparing MCI 1 to the ADNI normal controls. MCI 1 differed from the normal group at baseline on tests of memory (as expected due to the criteria for the MCI diagnostic category) but not on tests of executive function (Supplemental Table 1). The rate of annual change on cognitive tests in MCI 1 did not differ from the normal group (all p > 0.10), with the exception of Digit Span backward in which MCI 1 performed worse (−0.32 additional points per year in MCI 1, p = 0.02). MCI 1 did not differ from normal controls in baseline values or annual change for total brain, ventricles, and entorhinal cortex (all p > 0.25). There were significant differences in baseline and annual change measures for the hippocampus, where MCI 1 began with smaller hippocampal volumes (approximately 5% smaller, p = 0.02), and experienced an atrophy rate approximately 2.5 times that of the normal controls (p = 0.048).
In order to better understand how MCI 2, MCI 3, and MCI 4 relate to one another, a secondary analysis was undertaken that excluded MCI 1 and took MCI 3 as the reference category. In general, the decrease in cognitive function in MCI 3 was significantly greater than MCI 2, but significantly smaller than MCI 4. This analysis was also performed on the longitudinal MRI measures in which MCI 3 and MCI 4 were not significantly different from each other for any of the measures except entorhinal cortex, where MCI 4 showed more rapid atrophy. MCI 2 showed significantly less atrophy than MCI 3 in all regions.
4. Discussion
This study resulted in three key findings. First, there is biological heterogeneity at baseline among the ADNI aMCI subjects, despite being intentionally selected as a clinical phenotype often presumed to be the precursor to Alzeimer’s disease [27]. Second, a substantial portion of the individuals in this group (MCI 1) showed no clinical decline and was characterized by a remarkably healthy looking biomarker profile. Third, a large cluster (MCI 3) failed to conform to the amyloid cascade hypothesis despite ultimately experiencing a large number of conversions to AD.
The amyloid cascade hypothesis of AD suggests that the process begins with amyloid plaque deposition, followed in sequence by measurable changes in CSF tau proteins, then changes in brain volume, and finally clinically detectable cognitive change [4]. The goal of this study was to characterize heterogeneity within the ADNI MCI population, not to specifically test the veracity of the amyloid hypothesis [11]. Nonetheless, our findings are not entirely consistent with the proposed cascade of biological changes within this phenotypically refined cohort. Specifically, a large proportion of the subjects demonstrated biomarker abnormalities that appeared out of sequence relative to the model and another group of subjects showed clinical memory deficits but lacked substantive biomarker abnormalities. Differences between the hypothesized sequence of events and our observations could happen for multiple reasons. First, it is likely that AD is not the sole cause of memory deficits in all subjects, despite attempting to recruit a homogenous group. For example, previous reports have identified a vascular subgroup of amnestic MCI subjects [28]. Furthermore, individuals may suffer with multiple pathologies that could modify the biomarkers or hypothesized sequence of pathological events [10]. It may also be possible that the amyloid cascade hypothesis describes the transition from memory loss to AD in some groups and the data in aggregate, but substantial variation in the sequence of biomarkers may be present among certain subgroups of amnestic individuals [29]. In fact, originators of the amyloid cascade hypothesis have since identified similar heterogeneity [30, 31].
We found that MCI 1 had pronounced memory deficits that distinguished them from normal controls but had significantly less severe memory deficits than the rest of the MCI group. There was limited evidence to suggest that their memory impairment and smaller hippocampal volume was due to AD pathology. For example, CSF protein levels in this group were within normal limits and there was a slow or nonexistent rate of change in clinical function. These findings suggest that the individuals in MCI 1 are not in a temporary transition state to Alzheimer’s dementia as presumed by the presence of impaired memory performance [27]. Whitwell et al studied a similar group of stable amnestic MCI subjects (defined as no conversion to AD in three year follow-up after aMCI diagnosis), comparing them both to cognitively normal subjects and amnestic MCI subjects who rapidly converted to AD. The stable MCI group had significantly smaller hippocampal volume than the normal controls although there were not significant differences in gray matter loss [32]. Similarly, Wolk et al. found that 42% (8 of 19) of single domain amnestic MCI subjects were amyloid-negative on PiB amyloid imaging supporting the notion of heterogeneity [33] These findings may suggest that a non-AD process such as hippocampal sclerosis may be involved in the pathology of this subgroup of MCI [34], although other non-identified causes—including congenital factors—may be explain these findings as well. Further studies of these subjects are clearly indicated.
MCI 2 and 4 seemed to follow the amyloid cascade hypothesis, with MCI 4 further along the trajectory than MCI 2. They had similarly low levels of Aβ1-42 and had elevated tau proteins (Figure 2). MCI 4 had higher CSF tau levels, and correspondingly, had more severe atrophy and cognitive deficits. Despite entering the study with already severe levels of brain injury, MCI 4 continued to experience rapid change in regional brain volumes and cognitive abilities, and six out of seven of the subjects in MCI 4 converted to AD within three years. While this hypothesis should be interpreted with some caution because MCI 4 is a very small group, the clinical and biological differences between MCI 4 and the other clusters were substantial and favored a more severe state of neurodegeneration.
The biological profile of MCI 3 is the most inconsistent with the presumed sequence of biological markers leading to AD. For example, The Aβ1-42 levels in MCI 3 were clearly abnormal, but in a range intermediate between normal and pathological levels commonly associated with Alzheimer’s dementia [35]. In addition, average tau levels were essentially normal in MCI 3. This is in stark contrast to MCI 2 and 4 where total tau means were approximately 1.5 and 6.5 normal standard deviations, respectively, above the normal controls. Moreover, there was no evidence for a second pathological process such as cerebrovascular brain injury to explain the biomarker findings. The hypothesized sequence of events prescribed by Jack et al suggests that there should be substantial changes in tau before there are drastic changes in brain volume or cognitive function if the changes are due to AD [4]. MCI 3 had substantial brain atrophy, which is similar to the levels seen in the AD group and exhibited significantly lower cognitive function than MCI 1 and 2 both at baseline and longitudinally.
Although it is not prominently discussed in the AD literature, there are other documented AD subgroups with minimal tau elevation. A recent CSF clustering paper separated AD subjects into three clusters, one of which had substantially lower tau and p-tau, despite exhibiting a wide range of Aβ1-42, similar to the pattern seen in MCI 3 [36]. While the CSF assays in the Wallin et al study differed from ADNI and therefore cannot be directly compared, the same group has performed other studies which included normal control subjects using the same assay. The total tau levels in their low-tau AD cluster (397 ± 113, n= 87) hardly differed from the total tau levels in aged normal controls in another study using the same assay (412 ± 232, n = 34) [37]. The normal controls in the aforementioned study likely contained some individuals who are in the early stages of AD and did not yet exhibit cognitive deficits, but this possibility was reduced by including in the analysis only controls which showed no substantive cognitive decline over a four year follow-up period. Another study examining subgroups of AD subjects using latent profile analysis found a similar group characterized by low tau [38]. This group was the oldest, approximately 76 years, similar to our study.
Further research is necessary to determine whether the lack of congruence between the cascade model and the pathology in MCI 1 and 3 exists because the model is not suited to all cases of AD or because the cognitive deficits seen in MCI 1 and 3 are not due to AD. This question may be answerable in the future by examining longitudinal biomarker and cognitive data, as well as neuropathological evidence from ADNI subjects who consented to post-mortem examination.
Our data can also be viewed in light of the recently revised NIA/AA diagnostic criteria for MCI due to AD [39]. According to this criteria, individuals with evidence of amyloid pathology (based on amyloid PET imaging or CSF Aβ) and neuronal injury (based on FDG-PET, CSF tau, or atrophy) have the greatest likelihood of MCI due to AD, while for those with conflicting biomarker information (e.g. low CSF Aβ but high CSF tau), the biomarkers are considered uninformative and the default clinical criteria hold. It is interesting to note that the smallest MCI clusters (1 and 4) are the ones that seem to correspond most consistently to this diagnostic criteria, since MCI 1 is generally negative for both categories of biomarkers, while MCI 4 is generally positive. For a large proportion of the individuals in MCI 2 and 3, however, the biomarker measurements would be considered conflicting (and therefore uninformative) when, for example, CSF Aβ was abnormal but CSF tau (or atrophy) was normal. Even more troublesome are cases for which markers of neuronal injury conflict, for example, the extensive atrophy and relatively normal CSF tau seen in MCI 3. This observation provides further support for the notion that the clinical phenotype of MCI is biologically heterogeneous [40].
The primary strength of this study is the use of unsupervised clustering without regard to cutoffs for dichotomous biomarker status, clinical outcomes or longitudinal trajectories of biomarkers. Thus, any longitudinal patterns or clinical associations with cluster membership were not manufactured by the clustering process. The methodology is also a benefit in that it allows an examination of multivariate structure in the data which thrives on correlation between variables, as opposed to being hindered by such correlation as is the case with many regression methods. The primary weakness of this study is the limited number of subjects overall, and the limited number with CSF fluid samples, which reduced the sample size available to study CSF and imaging biomarkers simultaneously. Another weakness is the fact the clusters found were not compact and well-separated, but instead show overlap. While this is to be expected in a biological system where individuals are “moving" from cluster to cluster and may be exhibiting multiple pathologies simultaneously, it does leave the membership of individuals on the boundaries in some question. We view the use of cluster analysis, in this case, not as a definitive classification method where we sought to develop new categorical phenotypes, but as a tool for simultaneously using multiple biomarkers to understand the biological heterogeneity apparent with MCI subjects within the well-defined clinical phenotype of amnestic MCI. This analysis was exploratory in nature and would benefit from replication in other similar populations where both CSF and MRI measures are available to determine the extent to which the patterns found here are representative. Another limitation is the possibility that atrophy was non-linear and yet was modeled as linear; however, we believe that such non-linearity is likely to be minimal over the short time frame.
The most important finding to come out of this analysis is the identification of biological and cognitive heterogeneity within the presumed homogenous clinical phenotype of amnestic MCI. Furthermore these findings are relevant not only to the understanding of biological processes leading to memory loss, but to clinical trials methodology. For example, subjects such as those in MCI 1 (14% of the subjects) whose memory deficits placed them in the MCI diagnostic group but who exhibited very little change over time. Their slow rate of change and the likelihood that their deficits were not clearly related to AD would make them poor candidates for inclusion in clinical trials. Similarly, individuals with profiles matching MCI 3, which made up 37% of the subjects, may not be suitable for inclusion in treatment trials that would emphasize reductions in tau as a treatment outcome. Importantly, these two groups combined made up over 50% of the subjects in this analysis. If ADNI had been running a clinical trial specific to tau-mediated brain injury in the MCI diagnostic group, the inclusion of MCI 1 and MCI 3 could have resulted in a substantial loss of power to detect beneficial effects of treatment.
In conclusion, our findings indicate that the clinical phenotype of amnestic MCI is biologically and behaviorally heterogeneous and a more complete understanding of this heterogeneity will not only improve our understanding of transition phases from normal cognitive aging, but will also likely benefit the design and implementation of clinical trials aimed at treatment of the earliest pathological changes associated with AD.
Supplementary Material
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants R01 AG021028 and UL1 TR0002.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Conflict of interest
None reported for JN. Dr. DeCarli is a member of the MRI core and Dr. Beckett leads the biostatistics core of ADNI. Each receives research support from ADNI for these services. Dr. Landau works with the ADNI PET core, receives ADNI research support, and has also received consulting fees from Avid Radiopharmaceuticals and Biogen Idec.
References
- 1.Nettiksimmons J, et al. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiology of aging. 2010;31(8):1419–1428. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubois B, Albert ML. Amnestic MCI or prodromal Alzheimer's disease? Lancet neurology. 2004;3(4):246–248. doi: 10.1016/S1474-4422(04)00710-0. [DOI] [PubMed] [Google Scholar]
- 3.Jicha GA, et al. Neuropathologic outcome of mild cognitive impairment following progression to clinical dementia. Archives of neurology. 2006;63(5):674–681. doi: 10.1001/archneur.63.5.674. [DOI] [PubMed] [Google Scholar]
- 4.Jack CR, Jr, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet neurology. 2010;9(1):119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganguli M, et al. Mild cognitive impairment, amnestic type: an epidemiologic study. Neurology. 2004;63(1):115–121. doi: 10.1212/01.wnl.0000132523.27540.81. [DOI] [PubMed] [Google Scholar]
- 6.Zanetti M, et al. Mild cognitive impairment subtypes and vascular dementia in community-dwelling elderly people: A 3-year follow-up study. Journal of the American Geriatrics Society. 2006;54(4):580–586. doi: 10.1111/j.1532-5415.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- 7.Yaffe K, et al. Subtype of mild cognitive impairment and progression to dementia and death. Dementia and geriatric cognitive disorders. 2006;22(4):312–319. doi: 10.1159/000095427. [DOI] [PubMed] [Google Scholar]
- 8.Caroli A, et al. Cerebral perfusion correlates of conversion to Alzheimer's disease in amnestic mild cognitive impairment. Journal of neurology. 2007;254(12):1698–1707. doi: 10.1007/s00415-007-0631-7. [DOI] [PubMed] [Google Scholar]
- 9.Dickson DW, et al. Hippocampal sclerosis: a common pathological feature of dementia in very old (>or = 80 years of age) humans. Acta neuropathologica. 1994;88(3):212–221. doi: 10.1007/BF00293396. [DOI] [PubMed] [Google Scholar]
- 10.Schneider JA, et al. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69(24):2197–2204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- 11.Jack CR, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet neurology. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen RC, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jack CR, Jr, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of magnetic resonance imaging : JMRI. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland D, Dale AM. Nonlinear registration of longitudinal images and measurement of change in regions of interest. Medical image analysis. 2011;15:489–497. doi: 10.1016/j.media.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 16.Schwarz C, et al. Fully-automated white matter hyperintensity detection with anatomical prior knowledge and without FLAIR. Information processing in medical imaging : proceedings of the ... conference. 2009;21:239–251. doi: 10.1007/978-3-642-02498-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw LM. PENN biomarker core of the Alzheimer's disease Neuroimaging Initiative. Neuro-Signals. 2008;16(1):19–23. doi: 10.1159/000109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wechsler D. In: Wechsler Memory Scale-Revised. Corporation P, editor. San Antonio, Texas: 1987. [Google Scholar]
- 19.Wechsler D. In: Wechsler Adult Intelligence Scale - Revised. Corporation TP, editor. 1981. [Google Scholar]
- 20.Partington JE. In: Partington's Pathway Test, in The Psychological Service Center Bulletin. Center TPS, editor. 1949. pp. 9–20. [Google Scholar]
- 21.Morris JC, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer's disease. Neurology. 1989;39(9):1159–1165. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- 22.Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. Journal of psychiatric research. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 23.Mohs RC. Administration and Scoring Manual for the Alzheimer's Disease Assessment Scale, 1994 Revised Edition. In: Medicine TMSSo., editor. The Mount Sinai School of Medicine.p. Test manual. 1994. [Google Scholar]
- 24.Rey a. In: L'Examen Clinique en Psychologie (Psychological Clinical Examination) Paris PUdF., editor. 1964. [Google Scholar]
- 25.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2001;63:411–423. [Google Scholar]
- 26.R Development Core Team, R. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 27.Morris JC, Cummings J. Mild cognitive impairment (MCI) represents early-stage Alzheimer's disease. J Alzheimers Dis. 2005;7(3):235–239. doi: 10.3233/jad-2005-7306. discussion 255-62. [DOI] [PubMed] [Google Scholar]
- 28.Nordahl CW, et al. Different mechanisms of episodic memory failure in mild cognitive impairment. Neuropsychologia. 2005;43(11):1688–1697. doi: 10.1016/j.neuropsychologia.2005.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nettiksimmons J, et al. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiology of aging. 2010;31:1419–1428. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack CR, Jr, et al. An operational approach to National Institute on Aging-Alzheimer's Association criteria for preclinical Alzheimer disease. Ann Neurol. 2012;71(6):765–775. doi: 10.1002/ana.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knopman DS, et al. Brain injury biomarkers are not dependent on beta-amyloid in normal elderly. Ann Neurol. 2013 doi: 10.1002/ana.23816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitwell JL, et al. MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology. 2008;70(7):512–520. doi: 10.1212/01.wnl.0000280575.77437.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolk DA, et al. Amyloid Imaging in Mild Cognitive Impairment Subtypes. Annals of neurology. 2009;65(5):557–568. doi: 10.1002/ana.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarow C, Sitzer TE, Chui HC. Chui, Understanding hippocampal sclerosis in the elderly: Epidemiology, characterization, and diagnostic issues. Current Neurology and Neuroscience Reports. 2008;8(5):363–370. doi: 10.1007/s11910-008-0057-3. [DOI] [PubMed] [Google Scholar]
- 35.Petersen RC, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallin AK, et al. CSF biomarkers predict a more malignant outcome in Alzheimer disease. Neurology. 2010;74(19):1531–1537. doi: 10.1212/WNL.0b013e3181dd4dd8. [DOI] [PubMed] [Google Scholar]
- 37.Buchhave P, et al. Longitudinal study of CSF biomarkers in patients with Alzheimer's disease. PloS one. 2009;4(7):e6294. doi: 10.1371/journal.pone.0006294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iqbal K, et al. Subgroups of Alzheimer's disease based on cerebrospinal fluid molecular markers. Annals of neurology. 2005;58(5):748–757. doi: 10.1002/ana.20639. [DOI] [PubMed] [Google Scholar]
- 39.Albert MS, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeCarli C. Mild cognitive impairment: prevalence, prognosis, aetiology, and treatment. Lancet Neurol. 2003;2(1):15–21. doi: 10.1016/s1474-4422(03)00262-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.