Abstract
Obtaining high-resolution information from a complex system, while maintaining the global perspective needed to understand system function, represents a key challenge in biology. Here we address this challenge with a method (termed CLARITY) for the transformation of intact tissue into a nanoporous hydrogel-hybridized form (crosslinked to a three-dimensional network of hydrophilic polymers) that is fully assembled but optically transparent and macromolecule-permeable. Using mouse brains, we show intact-tissue imaging of long-range projections, local circuit wiring, cellular relationships, subcellular structures, protein complexes, nucleic acids and neurotransmitters. CLARITY also enables intact-tissue in situ hybridization, immunohistochemistry with multiple rounds of staining and de-staining in non-sectioned tissue, and antibody labelling throughout the intact adult mouse brain. Finally, we show that CLARITY enables fine structural analysis of clinical samples, including non-sectioned human tissue from a neuropsychiatric-disease setting, establishing a path for the transmutation of human tissue into a stable, intact and accessible form suitable for probing structural and molecular underpinnings of physiological function and disease.
The extraction of detailed structural and molecular information from intact biological systems has long been a fundamental challenge across fields of investigation, and has spurred considerable technological innovation1–8. The study of brain structure–function relationships in particular may benefit from intact-system tools9–12, and in general, much valuable information on intra-system relationships and joint statistics will be accessible from full structural analysis of intact systems rather than piecemeal reconstruction across preparations. Yet even tissue structure in itself provides only a certain level of insight without detailed molecular phenotyping13,14, which is difficult to achieve within intact tissue.
Current pioneering methods suitable for the mammalian brain either involve sectioning and reconstruction, or are incompatible with molecular phenotyping, or both. Automated sectioning methods have been successfully used to map structure4,5,15–18, in some cases with molecular labelling. However, detailed reconstruction has typically been limited in application to small volumes of tissue. On the other hand, intact-imaging methods that extend the depth of light microscopy by reducing light scattering have emerged19–21, but these preparations are incompatible with intact-tissue molecular phenotyping, and require many weeks of preparation to achieve partial tissue clearing. Studying intact systems with molecular resolution and global scope remains an unmet goal in biology.
We set ourselves the goal of rapidly transforming intact tissue into an optically transparent and macromolecule-permeable construct while simultaneously preserving native molecular information and structure. We took note of the fact that packed lipid bilayers are implicated in rendering tissue poorly accessible—both to molecular probes and to photons—by creating diffusion-barrier properties relevant to chemical penetration, as well as light-scattering properties at the lipid–aqueous interface22,23. If lipid bilayers could be removed non-destructively, light and macromolecules might penetrate deep into the tissue, allowing three-dimensional imaging and immunohistological analysis without disassembly. But removing lipid membranes that provide structural integrity and retain biomolecules would inevitably damage tissue with profound loss of cellular and molecular information. Therefore the provision of a physical framework would first be required to physically support the tissue and secure biological information. We have developed and used such a technology, which we term CLARITY, that addresses these challenges.
Hydrogel-electrophoretic tissue transmutation
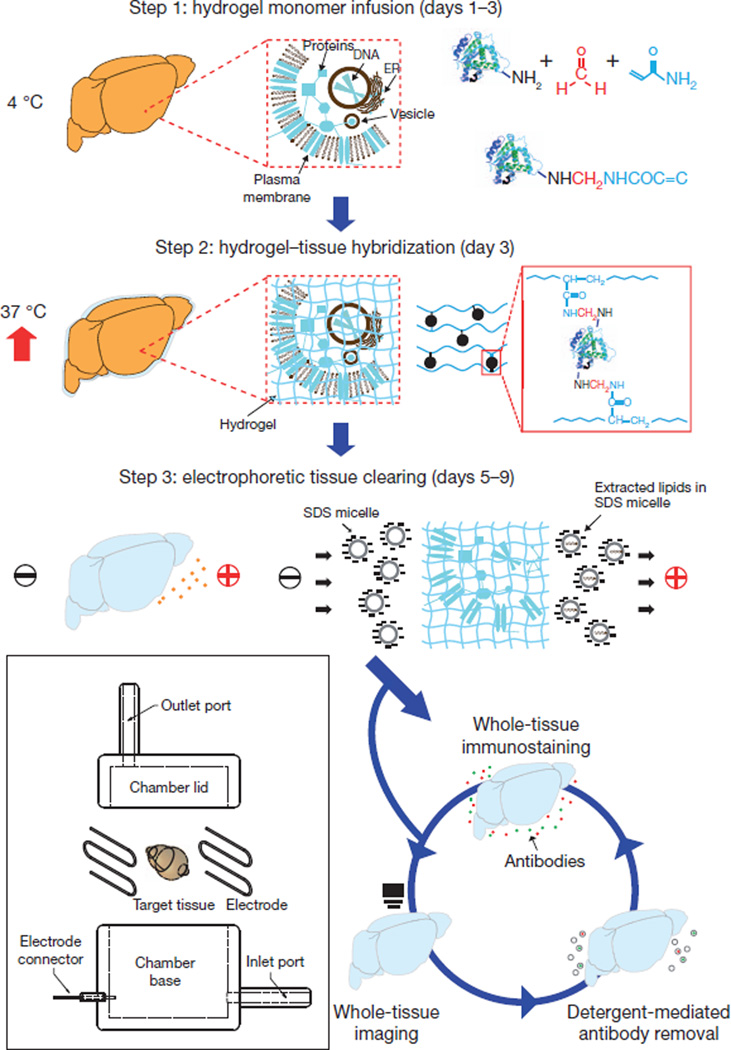
We began by infusing hydrogel monomers (here, acrylamide and bisacrylamide), formaldehyde and thermally triggered initiators into tissue at 4 °C (Fig. 1). In this step, formaldehyde not only crosslinks the tissue, but also covalently links the hydrogel monomers to biomolecules including proteins, nucleic acids and small molecules. Next, polymerization of the biomolecule-conjugated monomers into a hydrogel mesh was thermally initiated by incubating infused tissue at 37 °C for 3 h, at which point the tissue and hydrogel became a hybrid construct. This hydrogel–tissue hybridization physically supports tissue structure and chemically incorporates biomolecules into the hydrogel mesh. Importantly, lipids and biomolecules lacking functional groups for conjugation remain unbound and therefore can be removed from the hybrid. To extract lipids efficiently, we developed an ionic extraction technique rather than using hydrophobic organic solubilization, for two main reasons. First, although organic solvents can extract lipids or reduce refractive-index variations19,20,24, these solvents quench fluorescence, thereby limiting imaging time. Light-sheet microscopy has been used to image benzyl alcohol/benzyl benzoate (BABB)-treated samples while fluorescence persists19,20, but this approach is less compatible with slower high-resolution imaging. Moreover, instability of native fluorescence in BABB constrains imaging of fine neuronal projections or other modest signals that can be easily quenched. Conversely, in the hydrogel–tissue hybrid all fluorescent proteins tested, including green, yellow and red fluorescent proteins (GFP, YFP and RFP, respectively), were robust to ionic detergent extraction (Fig. 2, Supplementary Fig. 1 and Supplementary Video 1). Second, whereas passive diffusion of detergent micelles would take many months to completely extract lipids from the adult mouse brain, we developed an active-transport organ-electrophoresis approach, which we term electrophoretic tissue clearing (ETC), that capitalizes on the highly charged nature of ionic micelles. This method expedites the extraction by orders of magnitude (Fig. 1 and Supplementary Fig. 2). We anticipated that the hydrogel would secure biomolecules and fine structural features such as membrane-localized proteins, synapses and spines in place, whereas membranous lipids that cause light scattering and prevent penetration of macromolecules would be actively removed, leaving behind an otherwise fully assembled biological system suitable for labelling and imaging intact.
Figure 1. CLARITY.
Tissue is crosslinked with formaldehyde (red) in the presence of hydrogel monomers (blue), covalently linking tissue elements to monomers that are then polymerized into a hydrogel mesh (followed by a day-4 wash step; Methods). Electric fields applied across the sample in ionic detergent actively transport micelles into, and lipids out of, the tissue, leaving fine-structure and crosslinked biomolecules in place. The ETC chamber is depicted in the boxed region (Supplementary Fig. 2).
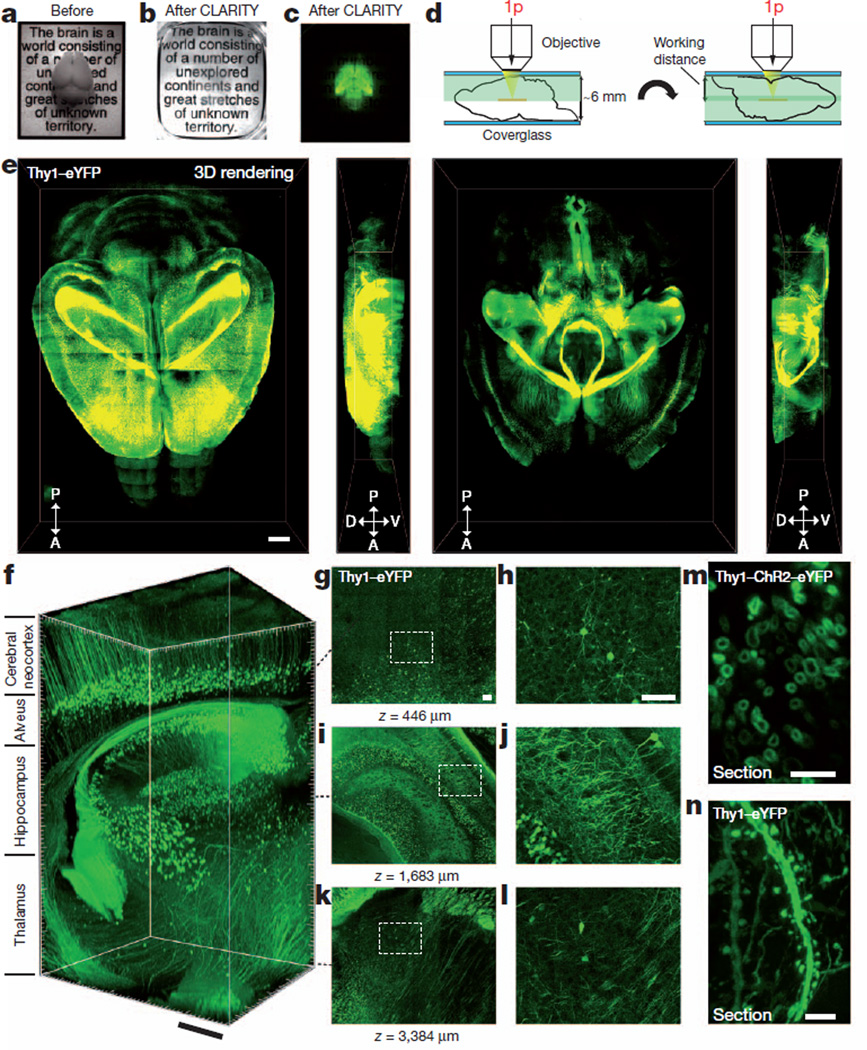
Figure 2. Intact adult mouse brain imaging.
Imaging was performed in adult mouse brains (3 months old). a, Cajal quote before CLARITY. b, Cajal quote after CLARITY: Thy1–eYFP line-H mouse brain after hydrogel–tissue hybridization, ETC and refractive-index matching (Methods). c, Fluorescence image of brain depicted in b. d, Dorsal aspect is imaged (single-photon (1p) microscopy), then brain is inverted and ventral aspect imaged. e, Three-dimensional rendering of clarified brain imaged (×10 water-immersion objective; numerical aperture, 0.3; working distance, 3.6 mm). Left, dorsal half (stack size, 3,100 µm; step size, 20 µm). Right, ventral half (stack size, 3,400 µm; step size, 20 µm). Scale bar, 1 mm (Supplementary Videos 3–5). f, Non-sectioned mouse brain tissue showing cortex, hippocampus and thalamus (×10 objective; stack size, 3,400 µm; step size, 2 µm). Scale bar, 400 µm (Supplementary Video 2). g–l, Optical sections from f showing negligible resolution loss even at ~3,400-µm deep: z=446 µm (g, h), z=1,683 µm (i, j) and z=3,384 µm (k, l). h, j and l, boxed regions in g, i and k, respectively. Scale bars, 100 µm.m, Cross-section of axons in clarified Thy1–channelrhodopsin2 (ChR2)–eYFP striatum: membrane-localized ChR2–eYFP (1-mm-thick coronal block; ×63 glycerol-immersion objective; numerical aperture, 1.3; working distance, 280 µm). Scale bar, 5 µm. n, Dendrites and spines of neurons in clarified Thy1–eYFP line-H cortex (1-mm-thick coronal block; ×63 glycerol objective). Scale bar, 5 µm.
Whole adult mouse brain imaging
To test anticipated features of the technology, we processed with CLARITY (hereafter ‘clarified’) the brain of a 3-month-old line-H mouse, in which the cytosolic fluorescent protein enhanced YFP (eYFP), under control of the Thy1 promoter sequence region, is expressed in a subset of projection neurons25,26. Within 8 days, the intact adult brain was transmuted into a lipid-extracted and structurally stable hydrogel–tissue hybrid. Scattering still occurs owing to heterogeneously distributed protein and nucleic acid complexes in the hybrid; however, after immersion in refractive-index-specified solutions matching the CLARITY hybrid (for example, 85% glycerol or FocusClear, both with a refractive index of ~1.45), the intact brain (including heavily myelinated white matter, thalamus and brainstem) becomes uniformly transparent (Fig. 2a–c and Supplementary Videos 2–5). ETC is crucial: passive refractive-index matching of non-ETC-processed brain provided little clearing (Supplementary Fig. 3), whereas full CLARITY processing enabled imaging of the entire intact mouse brain at cellular resolution even using single-photon microscopy (Fig. 2d–f and Supplementary Videos 2–5); this is important as many dyes and fluorescent proteins are better suited to single-photon rather than two-photon illumination27,28).
Because imaging depth in clarified tissue appeared to be limited only by the working distance of the objective (here 3.6 mm, although 8-mm working distance/0.9 numerical aperture objectives are available), to image the 5–6-mm-thick adult mouse brain, we next imaged the dorsal half of the brain followed by inversion and imaging of the ventral half (Fig. 2d, e). Figure 2f shows a volume of unsectioned mouse brain with visualization through the cortex, hippocampus and thalamus (Fig. 2g–l and Supplementary Video 2). We observed that tissue expanded with ETC and returned to its original size after refractive-index matching; this transient change did not cause net tissue deformation, and remaining secured in place were fine structural details such as membrane-localized proteins (Fig. 2m), dendritic spines (Fig. 2n) and synaptic puncta (Fig. 3e–g). We found that clarified tissue may be compatible with downstream electron microscopy, retaining key ultrastructural features such as synapses and postsynaptic densities, although with current methods not all relevant ultrastructures are apparent, probably owing to lack of membrane lipids (Supplementary Fig. 4). CLARITY thus allows rapid high-resolution optical access to dense intact tissue and, if needed, subsequent electron microscopy analysis. We next sought to determine whether this design enabled intact-tissue molecular phenotyping.
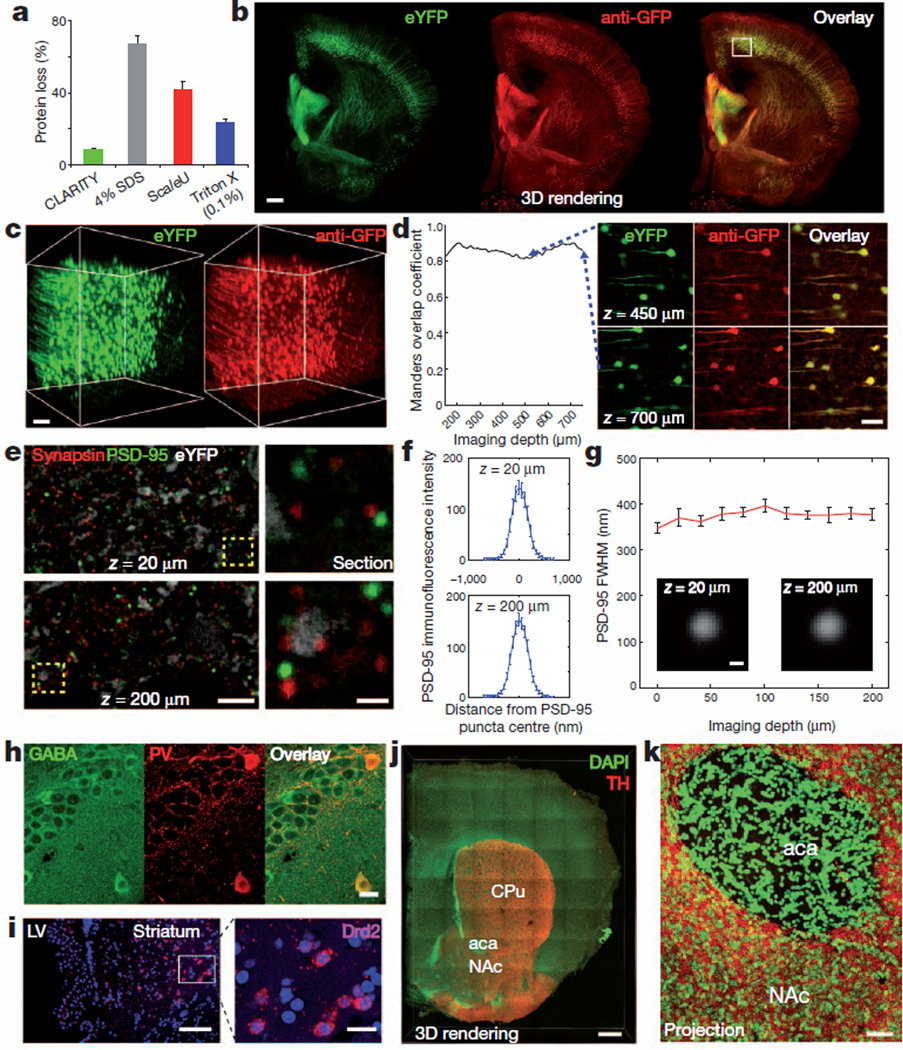
Figure 3. Molecular phenotyping in intact tissue.
a, Protein loss in clarified mouse brain compared to conventional methods (see Supplementary Information for more details); error bars denote s.e.m.; n=4 for each condition. b, Rendering of a 1-mm-thick non-sectioned coronal block of Thy1–eYFP mouse brain immunostained for GFP. The tissue was ETC-cleared (1 day), immunostained (3 days) and imaged (×10 water-immersion objective; single-photon excitation). Left, eYFP (green); middle, anti-GFP (red); right, overlay. Scale bar, 500 µm (Supplementary Video 6). c, Three-dimensional rendering of the boxed region in the cortex in b shows eYFP fluorescence (left) and anti-GFP staining (right). d, Left, co-localization: Manders overlap coefficient plotted versus depth43. Right, optical sections at different depths in three-dimensional rendering. Scale bar, 100 µm. e, f, 500-µm-thick block of line-H mouse brain (2 months old) clarified for 1 day and immunostained for synapsin I (red) and PSD-95 (blue) for 3 days (Methods) (×63 glycerol objective; single-photon excitation). e, Left, optical sections (z=20 µm, z=200 µm). Right, enlarged images of boxed regions on left. Individual synaptic puncta resolved throughout depth. White depicts eYFP staining. f, Average immunofluorescence cross-section of PSD-95 puncta at z=20 µm (top) and z=200 µm (bottom). g, Full width at half maximum (FWHM) of average immunofluorescence cross-section of PSD-95 puncta versus depth. Insets, average puncta at z=20 µm and z=200 µm. h, Hippocampal staining. Left, GABA; middle, parvalbumin (PV); right, overlay. 500-µm-thick block of wild-type mouse brain (3 months) clarified (1 day) and immunostained (3 days) (×25 water-immersion objective; numerical aperture, 0.95; working distance, 2.5 mm; single-photon excitation). Scale bar, 20 µm. i, in situ hybridization. Clarified 500-µm mouse brain block showing dopamine receptor D2 (Drd2) mRNA in the striatum. LV, lateral ventricle. Blue, DAPI. 50-base-pair RNA probes for Drd2 visualized with FastRed (×25 water-immersion objective; single-photon excitation (555 nm) for FastRed, two-photon excitation (720 nm) for DAPI). Scale bars: left, 100 µm; right, 20 µm. j, k, Axonal fibres of tyrosine hydroxylase (TH)-positive neurons in the nucleus accumbens (NAc) and caudate–putamen (CPu). j, Three-dimensional rendering of 1-mm-thick clarified mouse brain block stained for tyrosine hydroxylase (red) and DAPI (green). aca, anterior commissure. Scale bar, 500 µm. k, Maximum projection, NAc/aca volume in j. Scale bar, 50 µm.
Molecular phenotyping of intact-tissue volumes
Interrogating molecular features at subcellular resolution in an intact brain with known global wiring properties may be of value; however, conventional labelling techniques involve: (1) loss of native molecules after the permeabilization required for access to intracellular targets29; (2) time-intensive thin sectioning and reconstruction5; or (3) damage when multiple rounds of labelling are attempted due to harsh proberemoval processes5,30–32. We anticipated that CLARITY could allow intact-tissue and multiple-round molecular phenotyping by overcoming these three main difficulties. First, CLARITY may preserve native antigens with unusual completeness owing to the hydrogel-hybridization process. To quantify molecular preservation associated with tissue–hydrogel fusion, we compared protein loss in clarified mouse brain to loss from conventional methods (Fig. 3a). Of total protein, ~65% was solubilized (lost) when conventionally paraformaldehyde (PFA)-fixed tissue blocks were cleared by 4% SDS for 1 week. Scale, a tissue-clearing method using 4M urea, allowed ~41% protein loss over the same interval. Even PFA-fixed tissue treated only with 0.1% Triton X-100 in PBS buffer, a mild detergent-based permeabilization solution used in conventional histology, allowed ~24% loss of protein. However, when hydrogel-hybridized tissue was cleared with the stringent 4% SDS solution of CLARITY, only ~8% protein loss was seen, indicating that chemical tethering of biomolecules into hydrogel mesh can enhance the preservation of molecular tissue components.
Second, we found that CLARITY, which increases tissue permeability by replacing lipid bilayers with nanoporous hydrogel, enabled rapid diffusion of molecular probes deep into intact tissue, and therefore allows access to preserved biomolecules without sectioning. In a 1-mm-thick clarified coronal block of mouse brain, uniformly antibody stained over 3 days (Fig. 3b and Supplementary Video 6), quantitative co-localization analysis revealed that eYFP fluorescence and anti-GFP staining overlapped throughout the block (Fig. 3c, d). We next wanted to know whether whole-mount mouse brain immunolabelling was possible; although the lipid-cleared brain is highly permeable, it was possible that antibodies would not diffuse throughout the tissue on practical timescales. We conducted immunohistochemistry for tyrosine hydroxylase in clarified intact mouse brain, and found that 2 weeks of incubation each for primary and secondary antibody enabled 2.5 mm of penetration from each surface, enough to stain ~5-mm-thick adult mouse brains (Supplementary Fig. 5 and Supplementary Video 7). Tyrosine-hydroxylase-positive cell bodies and projections were readily visualized throughout the brain, and only at the deepest point was moderate signal attenuation seen, indicating that antibody penetration was not quite maximized and may be further enhanced by tuning conditions.
To investigate axonal projections of the tyrosine hydroxylase neurons further, we clarified, tyrosine-hydroxylase-stained and imaged 1-mm-thick coronal blocks of mouse brain using a high numerical aperture objective (0.95). As shown in Fig. 3j, k, Supplementary Figs 6–8 and Supplementary Video 8, projections of tyrosine-hydroxylase-positive fibres were readily visualized in the neocortex, nucleus accumbens, caudate–putamen and amygdala. Interestingly, certain cells in the infralimbic cortex and amygdala appeared much more heavily invested by tyrosine hydroxylase fibres than neighbouring cells (Supplementary Figs 6f and 8 and Supplementary Video 8). Further supporting specificity of staining in clarified tissue (Fig. 3j, k and Supplementary Fig. 7), visualized tyrosine hydroxylase projections spared anterior commissure while heavily investing the surrounding nucleus accumbens and caudate–putamen as expected.
We next considered that CLARITY could potentially enable analysis of subcellular molecular architecture in large volumes with resolution at the diffraction limit of light microscopy33, in an approach complementary to thin mechanical sectioning and three-dimensional reconstruction5,34,35. Resolving submicron-scale structures deep in tissue poses a challenge as the high numerical aperture objectives used for high-resolution imaging are susceptible to optical aberration caused by refractive-index variations in tissue, which blurs images and impairs differentiation of adjacent fluorescent signals, such as pre- and postsynaptic puncta. But when a clarified 500-µm-thick block of Thy1–eYFP mouse brain was immunolabelled for synapsin I (presynaptic) and PSD-95 (postsynaptic) and imaged with a ×63 glycerol-immersion objective (numerical aperture, 1.3; working distance, 280 µm), throughout the tissue volume we could unequivocally identify individual paired pre- and postsynaptic puncta, molecularly defining identity and position of putative excitatory synapses (Fig. 3e–g). The full-width at half-maximum of the point-spread function was uniform throughout the block, indicating that loss of resolution is negligible even near the diffraction limit of conventional light microscopy (Fig. 3f, g and Supplementary Figs 9 and 10) and demonstrating another feature of the technique: removal of lipid membranes ensures that tissue refractive index remains nearly constant throughout large volumes, allowing high-resolution imaging. We also found that the CLARITY hydrogel-conjugation process preserves small molecules such as the neurotransmitter GABA (γ-aminobutyric acid, Fig. 3h) and (for in situ hybridization) messenger RNAs (Fig. 3i), paving the way for multimodal combinatorial labelling within unsectioned tissue.
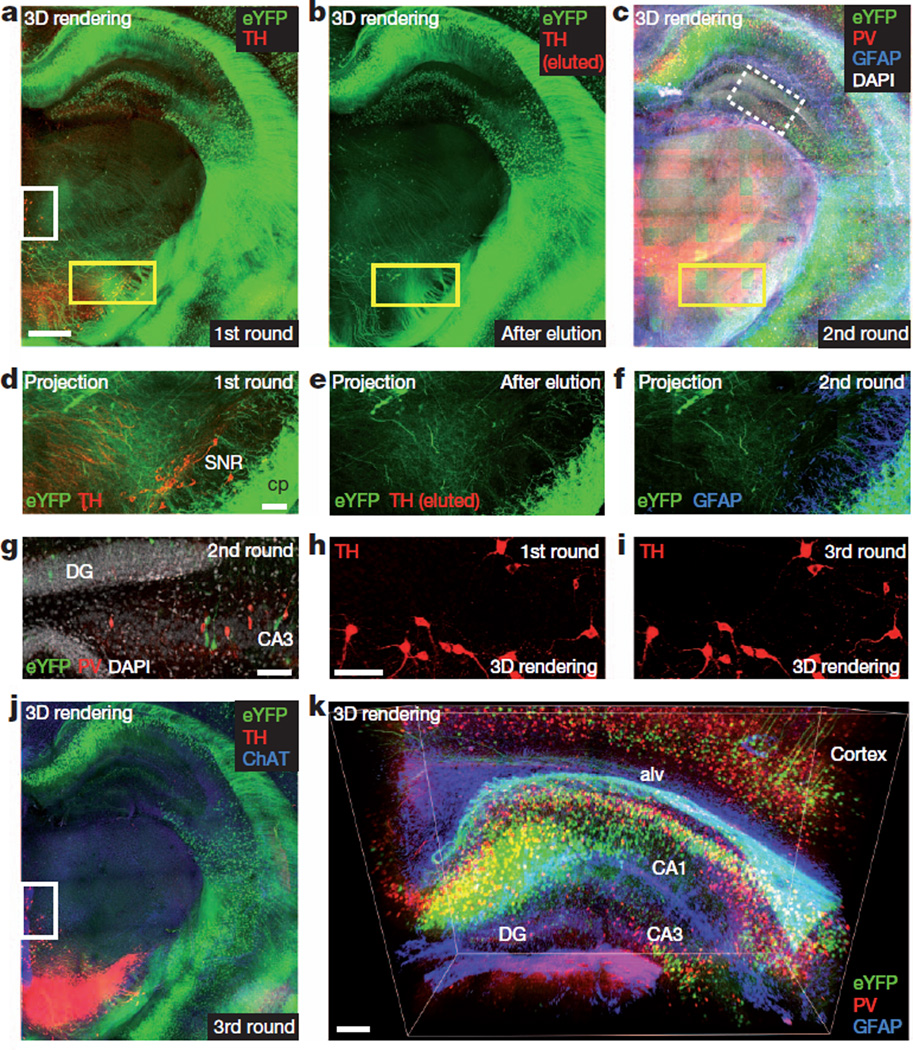
Finally, CLARITY was found to enable multi-round molecular phenotyping; the stable framework allowed effective removal of antibodies without fine structural damage or degraded antigenicity. Traditional elution requires incubation in acidic buffer with potassium permanganate (KMnO4) oxidation or heat, harsh treatments that cause antigen loss and fluorescence quenching5,30–32. But in the hydrogel context of increased structural/chemical stability, the ionic detergent (4% SDS/neutral-pH buffer) used initially to create clarified tissue can be re-used to denature antibodies and disrupt binding, and the permeability of clarified tissue can facilitate rapid antibody exchange. We performed three consecutive rounds of staining in 1-mm-thick coronal blocks from a Thy1–eYFP H-line mouse brain (Fig. 4 and Supplementary Videos 10–14), observing effective antibody removal and preserved eYFP-positive neuronal morphology as well as re-staining capability. Although further quantification would be required to fully map the extent to which CLARITY secures molecular information, this result shows that elution in clarified tissue largely preserves integrity of tissue structure, cellular architecture, fluorescence signals (Fig. 4d–f) and 4′,6-diamidino-2-phenylindole (DAPI) DNA staining (Fig. 4g). Moreover, repeated tyrosine hydroxylase staining in the first and third rounds maintained signal pattern and intensity, confirming that antigenicity is retained throughout multiple rounds of staining and elution.
Figure 4. Multi-round molecular phenotyping of intact tissue.
a, First round. Rendering of 1-mm-thick Thy1–eYFP block immunostained for tyrosine hydroxylase in non-sectioned form. ETC-cleared (1 day) and immunostained (6 days). Scale bar, 500 µm (Supplementary Video 10). b, Antibodies eluted from block in a (4% SDS, 60 °C for 0.5 days). Tyrosine hydroxylase signal was removed and eYFP fluorescence retained (Supplementary Video 11). c, Second round. Three-dimensional rendering of same block now immunostained for parvalbumin (red), glial fribrillary acidic protein (GFAP) (blue) and DAPI (white) (Supplementary Video 12). d–f, Maximum projections of 100 µm volume of yellow-boxed regions in a, b and c, respectively. eYFP-positive neurons preserved. cp, cerebral peduncle; SNR, substantia nigra. Scale bar, 100 µm. g, Optical section of white/dotted-box region in c showing DAPI. CA, cornu ammonis; DG, dentate gyrus. Scale bar, 100 µm. h, i, Tyrosine hydroxylase channel of white box regions in a (h) and j (i). Tyrosine hydroxylase antigenicity preserved through multiple elutions. Scale bar, 100 µm. j, Third round. Block in a–c immunostained for tyrosine hydroxylase (red) and choline acetyltransferase (ChAT) (blue) (Supplementary Video 13). k, Three-dimensional view of hippocampus in c showing eYFP-expressing neurons (green), parvalbumin-positive neurons (red) and GFAP (blue). Alv, alveus. Scale bar, 200 µm (Supplementary Video 14).
Human tissue imaging and molecular phenotyping
CLARITY is, in principle, applicable to any tissue: adult zebrafish imaging and phenotyping are demonstrated36 in Supplementary Fig. 12, and we also tested post-mortem human brains. With important and rare tissue from clinical brain banks residing in formalin, one concern is that these maximally crosslinked samples could be poorly suited to CLARITY, and certainly the mapping of connectivity in the human brain (ref. 37 and http://www.neuroscienceblueprint.nih.gov/connectome/) at cellular resolution has been hampered by lack of methods for labelling individual neurites over distances, as the injection of dyes into living brain or in vivo genetic labelling are feasible only in animals38,39. If extensively fixed human tissues were suitable for CLARITY, new avenues would be opened for the study of clinical samples.
We found that CLARITY was suitable for such long-banked human brain, allowing immunohistological visualization and identification of neurons and projections over large volumes (Fig. 5a–g and Supplementary Videos 15–17). In 0.5-mm-thick blocks of frontal lobe from an autistic patient, stored in formalin for >6 years, we were able to stain for axons with neurofilament protein and myelin basic protein, and trace individual fibres (Fig. 5e and Supplementary Video 15). In addition, by staining for parvalbumin it was possible to visualize the distribution of parvalbumin-positive neurons in the neocortex over large volumes (6.7 × 4.7 × 0.5 mm), and trace individual parvalbumin-labelled processes (Fig. 5g–n and Supplementary Videos 16 and 17).
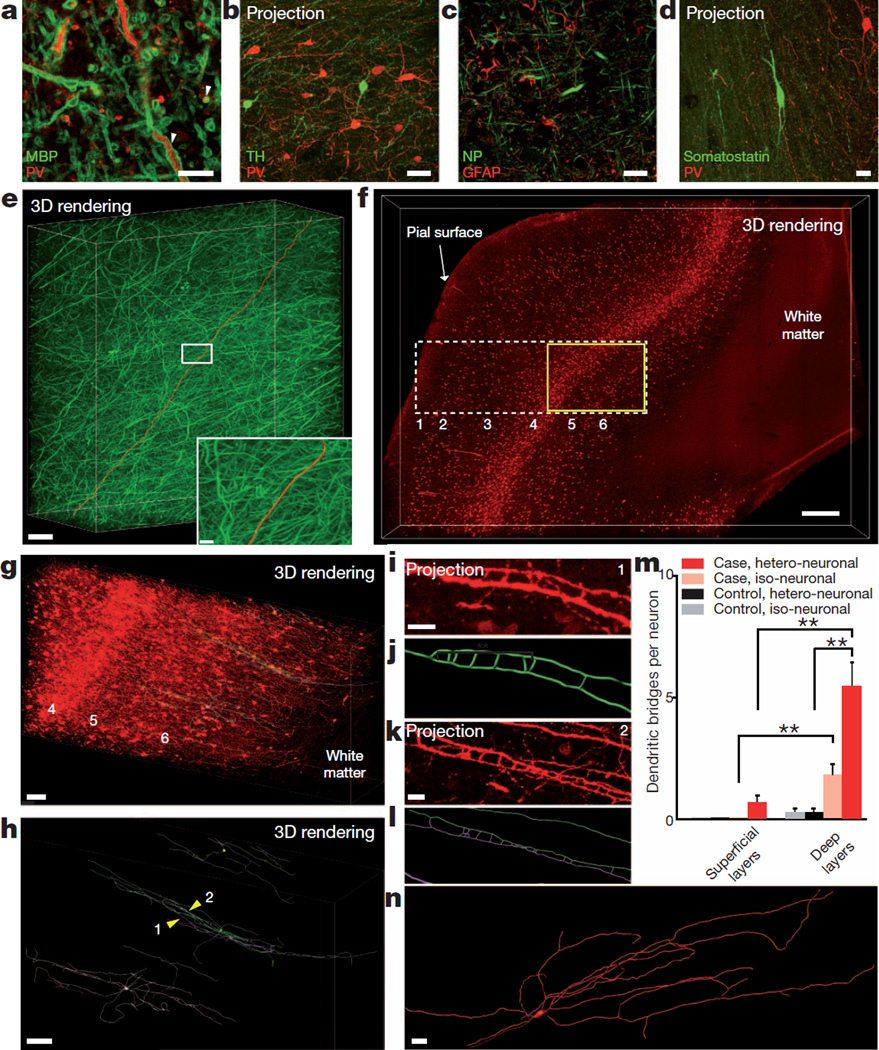
Figure 5. Human brain structural/molecular phenotyping.
Human BA10 500-µm-thick intact blocks clarified (1 day) and immunostained (3 days) (×25 water-immersion objective). a, Optical section: myelin basic protein (MBP) and parvalbumin staining. White arrowheads indicate membrane-localized myelin basic protein around parvalbumin-positive projections. Scale bar, 10 µm. b, Tyrosine hydroxylase and parvalbumin staining (maximum projection; 120 µm volume; step size, 0.5 µm). Scale bar, 50 µm. c, Optical section: neurofilament (NP) and GFAP. Scale bar, 20 µm. d, Somatostatin and parvalbumin staining (maximum projection; 63 µm volume; step size, 0.5 µm). Scale bar, 20 µm. e, Rendering of neurofilament-positive axonal fibres. Red, traced axon across volume. Scale bar, 500 µm. Inset: boxed region. Scale bar, 20 µm (Supplementary Video 15). f, Visualization of parvalbumin-positive neurons in the neocortex of autism case; layers identified as described in ref. 44. Scale bar, 500 µm (Supplementary Video 16). g, Yellow-boxed region in f showing parvalbumin-positive cell bodies and fibres in layers 4, 5 and 6. Three representative parvalbumin-positive interneurons in layer 6 with ladder-shaped hetero- or iso-neuronal connections were traced (green, purple, blue). Scale bar, 100 µm (Supplementary Video 17). h, Three-dimensional rendering of abnormal neurons in g; yellow arrowheads (1, 2) indicate ladder-shaped structures shown below in i and k. Scale bar, 80 µm. i, Zoomed-in maximum projection of 8 µm volume showing morphology of ladder-shaped structure formed by neurites from a single neuron. Scale bar, 10 µm. j, Tracing of structure in i. k, Maximum projection of 18 µmvolume showing ladder-shaped structure formed by neurites from two different neurons. Scale bar, 10 µm. l, Tracing of structure in k. m, Iso- and hetero-neuronal dendritic bridges per neuron. Neurons selected randomly and traced in software (Methods); dendritic bridges were manually counted. **P<0.05; error bars denote s.e.m. n=6 neurons for both superficial and deep layers of autism case and n=4 neurons for both superficial and deep layers of control case. n, Three-dimensional reconstruction of a neuron in layer 2 (superficial) of the autism case. Typical avoidance of iso-dendritic contact was observed.
Unlike mechanical sectioning methods that may involve deformation of tissue and uncertainty in registration across sections, CLARITY preserves continuity of structure, which not only allows tracing of neurites over distances, but also provides a class of distinct information about three-dimensional and topological morphology of traced neurons. As one example, we found that many parvalbum-inpositive interneurons in this human sample, particularly in deep layers, showed isoneuronal and heteroneuronal dendritic bridges (Fig. 5g–m, Supplementary Fig. 13, Supplementary Video 17 and Supplementary Table 2). These ladder-like connections, not typical of age- and sexmatched normal brain (Supplementary Fig. 14), instead resemble abnormalities observed with mutations in the Down syndrome cell-adhesion molecule (Dscam) protein40 or protocadherins41; mutations in the latter family are associated with autism-spectrum disorder42. Although extensive work would be required to define the incidence and implications of abnormal parvalbumin neurons, the observation illustrates the structural and molecular capability that clarified tissue provides by virtue of volumetric continuity, which may help to shed light on structural underpinnings of neuropsychiatric disease.
Discussion
With this hydrogel–tissue fusion and electrophoretic-clearing technology, intact tissue can be rapidly transformed into an optically and chemically accessible form while retaining structural and molecular information, thereby enabling the imaging of entire adult vertebrate brains as well as multiple-round molecular phenotyping without thin sectioning. Currently, the non-corrected water-immersion objectives used, rather than clarified tissue properties, represent the main limiting factor for maximizing imaging depth and resolution; specialized adaptive optics or CLARITY-optimized long-working-distance objectives will further enhance the acquisition of integrated structural and molecular information from intact systems.
Turning immense data sets into useful insights remains a key challenge. Computational approaches to image segmentation, three-dimensional registration and automated tracing require further development. Moreover, for the subset of applications relevant to circuit connectivity, CLARITY does not in itself supplant electron microscopy. However, the potential electron microscopy compatibility of CLARITY (Supplementary Fig. 4) may offer a two-step approach in probing connectivity with synaptic resolution; CLARITY with light microscopy may first be used to visualize and trace fluorescently labelled neurons and projections in the whole brain, followed by electron microscopy analysis of small volumes to define the patterns and rules (for example, postsynaptic cell target type) followed by axon terminals and synaptic contacts. Notably, owing to the absence of lipids, osmium tetroxide/uranyl acetate staining alone with electron microscopy does not provide enough contrast to identify all ultrastructural boundaries, and additional methodological tissue-preparation work is indicated.
Although much remains to be refined, newly enabled access to intact-system properties may dovetail with existing optogenetic-control or activity-imaging methods. For example, cells studied in local and global structural detail with CLARITY could be the same cells previously visualized for activity, or controlled to modulate performance, during behavioural assays in the same organism with proper registration. Moreover, efficient molecular phenotyping applied to immediate early gene products may help to map populations with altered activity downstream of directly modulated populations, across the same intact brain and in the context of global wiring maps. Together with its capability for intersectional definition through multiple rounds of labelling, and broad application domain (including mouse, zebrafish and human), these data suggest that CLARITY provides access to structural and molecular information that may help to support integrative understanding of large-scale intact biological systems.
METHODS
CLARITY
CLARITY refers to transformation of intact biological tissue into a hybrid form in which specific components are replaced with exogenous elements that provide new accessibility or functionality. Initially, the term was an acronym to describe the Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/In situ hybridization-compatible Tissue-hYdrogel. However, CLARITY is more inclusively defined because infused elements need not be exclusively hydrogel monomers or acrylamide-based, and the properties of infused elements may be adjusted for varying degrees of clarity, rigidity, macromolecule-permeability or other functionality.
Immunostaining of CLARITY-processed mouse brain tissue
For GFP staining shown in Fig. 3b–d, the clarified 1-mm-thick block of Thy1–eYFP line-H mouse (2 months old) brain was incubated at 37 °C for 2 days in 0.1% Triton X-100 (wt/vol), anti-GFP antibody conjugated with Alexa Fluor 594 (Invitrogen, 1:50 dilution), 1M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 1 day with 0.1% (wt/vol) Triton X-100 and 1 M sodium borate buffer, pH 8.5. For tyrosine hydroxylase staining of whole mouse brain in Supplementary Fig. 4, the clarified C57BL/6 mouse (4 weeks old) brain was incubated at 37 °C for 2 weeks in 0.1% (wt/vol) Triton X-100, primary antibody (dilution, 1:50), 0.5M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 1 week with 0.1% (wt/vol) Triton X-100 and 0.5M sodium borate buffer, pH 8.5. The tissue was then incubated at 37 °C for 2 weeks in 0.1% (wt/vol) Triton X-100, secondary antibody (dilution, 1:50) and 0.5M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 1 week with 0.1% (wt/vol) Triton X-100 and 0.5M sodium borate buffer, pH 8.5. For other staining, the clarified 1-mm-thick block of Thy1–eYFP H-line mouse (2 months old) brain was incubated at 37 °C for 2 days in 0.1% (wt/vol) Triton X-100, primary antibody (dilution, 1:50–1:100) and 1M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 1 day with 0.1% (wt/vol) Triton X-100 and 1 M sodium borate buffer, pH 8.5. The tissue was then incubated at 37 °C for 1 day in 0.1% (wt/vol) Triton X-100, secondary antibody (dilution, 1:50 – 1:100) and 1M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 1 day with 0.1% (wt/vol) Triton X-100 and 1M sodium borate buffer, pH 8.5.
Immunostaining of CLARITY-processed post-mortem human brain tissue
Clarified 500-µm-thick blocks were incubated at 37 °C for 1 day in 0.1% (wt/vol) Triton X-100, primary antibody (dilution, 1:50–1:100) and 1M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 12 h with 0.1% (wt/vol) Triton X-100 and 1M sodium borate buffer, pH 8.5. The tissue was then incubated at 37 °C for 1 day in 0.1% (wt/vol) Triton X-100, secondary antibody (dilution, 1:50–1:100) and 1M sodium borate buffer solution, pH 8.5, followed by wash at 37 °C for 12 h with 0.1% (wt/vol) Triton X-100 and 1 M sodium borate buffer, pH 8.5.
Imaging of CLARITY-processed mouse brain tissue
For imaging the intact Thy1–eYFP H-line mouse brain (Fig. 2e), the clarified brain (3 months old) was incubated in FocusClear, a water-based immersion medium, for 2 days. The brain was then enclosed between two coverglass-bottom Petri dishes. The dorsal half of the brain was first imaged (stack size, 3.1 mm; step size, 20 µm) using a Leica SP5 system equipped with the ×10 water-immersion objective (Leica HCX AOP L; numerical aperture, 0.30; working distance, 3.6) and 514-nm excitation. The mounted brain was then inverted and the ventral half was imaged in the same way. To obtain the 3.4-mm-thick volume image visualizing from the cortex to the thalamus (Fig. 2f–l), the intact line-H mouse brain was mounted as described and imaged using the ×10 objective and 514-nm excitation (stack size, 3.4 mm; step size, 2 µm). The immunostained 1-mm-thick coronal blocks were incubated in FocusClear for 1 day and enclosed between the coverglass-bottom Petri dish and slide glass. The mounted samples were imaged using the ×10 objective and single-photon excitation (514 nm, 591 nm and 654 nm).
Imaging of CLARITY-processed post-mortem human brain tissue
Clarified and immunostained tissues were incubated in FocusClear for 1 day and mounted as described above. The tissues were then imaged using the Leica SP5 system equipped with the ×25 water-immersion objective (Leica HCX IRAPOL; numerical aperture, 0.95; working distance, 2.4 mm; stack size, 500 µm; step size, 0.5 µm).
Protein-loss measurement
Six PFA-fixed adult mouse (4 weeks old) brains (for 4% SDS, Scale and Triton X-100 treatments) and two (4 weeks old) PFA-fixed, hydrogel-embedded brains (for CLARITY) were cut into 1-mm-thick coronal blocks. One half of each PFA-fixed brain was weighed and placed in 2.5 ml of 4% SDS, ScaleU2 (a mixture of 4M urea and 30% glycerol) or 0.1% Triton X-100 solution. One half of each PFA-fixed, hydrogel-embedded brain was placed in 2.5 ml of 4% SDS solution. The tissues were allowed to clear for 1 week at 37 °C in the respective solutions and quantity of protein lost from tissue by diffusing into solution was measured using the BCA (bicinchoninic acid) protein assay; total protein inmouse brain was estimated at 10%(wt) on the basis of previous work45,46.
Experimental subjects
Animal husbandry and all aspects of animal care and euthanization as described were in accordance with guidelines from the National Institutes of Health and have been approved by members of the Stanford Institutional Animal Care and Use Committee. Use of post-mortem human tissue was in accordance with guidelines from the National Institutes of Health and approved by the Stanford Institutional Review Board.
Neurite tracing
Manual tracing of individual neurons was performed using Imaris software (Bitplane). Neurons with cell bodies located in the middle 150 µm of the z-stack were randomly sampled and chosen for tracing. The cell body was reconstructed semi-automatically through a user-defined threshold that included as much of the cell body as possible but less than 5 µm of any dendrite. All neurites originating from the cell body were traced manually in short segments in the ‘Surpass’ mode, and each segment was automatically centred (with the opportunity for user corrections) before being connected. The number of interconnections between filaments of the same cell, and the number of interconnections between a filament from the traced cell and that of any other cell, was counted manually. Separately, for Fig. 5h–n neurons with interesting structures were chosen non-randomly and traced using the same method described above.
Basic CLARITY protocol
Hydrogel solution preparation: (1) Combine and mix 40 ml of acrylamide (40%), 10 ml of bis-acrylamide (2%), 1g of VA-044 initiator (10% wt), 40 ml of ×10 PBS, 100 ml of 16% PFA and 210 ml of dH2O with special attention to temperature and safety precautions. Keep all components on ice at all times to prevent polymerization. Caution: PFA is an irritant, sensitizer, carcinogen and toxin. Acrylamide is a potent neurotoxin, a respiratory and skin sensitizer, carcinogen, irritant, mutagen, teratogen and reproductive hazard. Many of the chemical constituents of hydrogels that could be used for CLARITY would fall into one or more of these categories. Therefore, to avoid skin contact or inhalation of monomers and/or crosslinkers (for example, acrylamide or PFA), solution preparation and all subsequent handling of hydrogel solution and polymer must be conducted in a fume hood with personal protective equipment including face shield, laboratory coat, gloves and closed-toe shoes.
Saponin is a widely used mild non-ionic surfactant often used to permeabilize cellular membranes in conventional immunohistochemistry. In CLARITY, saponin can be used in the hydrogel monomer infusion process to facilitate diffusion of the hydrogel monomer and initiator into the tissue, particularly for samples in which cardiac perfusion is not feasible, such as post-mortem human tissues and zebrafish brains. Saponin shortens incubation time required in the hydrogel monomer infusion process. However, bubbles may form that could be linked to saponin use, so routine saponin is not suggested. (2) Distribute 40-ml aliquots into 50-ml conical tubes on ice. Each tissue sample will require the use of one 40-ml tube: 20 ml for perfusion and the remaining 20 ml for sample embedding. (3) Seal tubes tightly and keep in secondary containment (on ice) before removing them from the hood. Transfer aliquots from ice to −20 °C. Store these solutions at −20 °C until they are ready to be used. They can be stored at −20 °C indefinitely if all components were kept sufficiently cold during the preparation process.
Solution preparation: (1) Combine and mix 123.66 g boric acid, 400 g sodium dodecyl sulphate, and 9 l dH2O. To avoid skin contact or inhalation, prepare solution in a fume hood in proper PPE. Paying special attention to safety, combine water, boric acid and SDS while stirring. Add dH2O to 10 l and add NaOH until the pH has reached 8.5. This solution can be made, stored and used at room temperature (20 °C). Caution: SDS is a toxin and irritant to the skin and respiratory system.
Transcardial perfusion with hydrogel solution: (1) Before perfusing, thaw the frozen hydrogel monomer solution in the refrigerator or on ice. (2) When the solution is completely thawed and transparent (but still ice cold), gently invert to mix. Make sure that there is no precipitate and avoid introducing any bubbles into the solution. (3) Prepare perfusion materials within a fume hood. (4) Deeply anaesthetize adult mouse with Beuthanasia-D. (5) Perfuse the animal transcardially, first with 20 ml of ice-cold 1× PBS and then 20 ml of the ice-cold hydrogel solution. Maintain a slow rate of perfusion (about 2 min for the 20 ml of each solution). (6) Immediately place the tissue (for example, brain) in 20 ml cold hydrogel solution in a 50-ml conical tube. Keep the sample in solution on ice until it can be moved to a 4 °C refrigerator. (7) Cover sample in aluminium foil if it contains fluorophores and incubate at 4 °C for 2–3 days to allow for further diffusion of the hydrogel solution into the tissue.
Hydrogel tissue embedding: (1) De-gas the 50-ml conical tube containing the sample in the desiccation chamber (in a fume hood) to replace all of the gas in the tube with nitrogen (as oxygen impedes hydrogel formation), as follows: place the 50-ml conical tube on a rack in the desiccation chamber; twist the 50-ml conical tube open sufficiently to allow gas exchange; turn on the nitrogen tank and adjust the control valve such that the inlet to the desiccation chamber fills with nitrogen gas; switch the desiccation chamber valve from nitrogen gas flow to the vacuum. Turn on the vacuum pump; verify that the chamber is under full vacuum by testing the chamber lid. Keep vacuum on for 10 min; switch the vacuum off and slowly turn the valve to fill the chamber with nitrogen gas; carefully open the chamber just enough to reach the tubes while purging with nitrogen gas, taking great care to minimize exposure to air, and quickly and tightly close the sample tube. (2) Submerge the tube in 37 °C water bath in a 37 °C room or incubator on the rotator. Incubate for 3 h or until solution has polymerized. (3) In a fume hood, extract the embedded sample from the gel (carefully take the sample out and remove extra gel pieces with gloved fingers). Hydrogel waste disposal should be conducted in accordance with all institutional, state and country regulations for hydrogel monomers and crosslinkers (for example, acrylamide and PFA). (4) Wash the sample with 50 ml clearing solution for 24 h at room temperature to dialyse out extra PFA, initiator and monomer. Wash the sample twice more with 50 ml for 24 h, each at 37 °C to further reduce residual PFA, initiator and monomer. Take care to dispose of this waste solution carefully as a biohazard.
ETC: (1) Construct the ETC chamber as described (http://CLARITYresourcecenter.org): place the sample in the chamber, and circulate the clearing solution through the chamber using the temperature-controlled water circulator, with 10–60V applied across the tissue (for example, brain) at 37–50 °C for several days to clear the sample. The clearing process is faster at higher voltage and temperature, but requires more power, limiting the number of clearing set-ups simultaneously operable by one power supply (typical power output maximum, 300 W). For example, four set-ups can be run simultaneously at 37 °C and 30 V, whereas only two set-ups can be run at 50 °C and 60 V, so circulator temperature and voltage should be chosen to meet practical considerations. In addition, too-high voltage operation could cause bubble formation in the tissue and deposit of black particles on the surface of tissue. Therefore, low voltage (10–40 V) is recommended. Note that lipids and biomolecules lacking functional groups required for conjugation, such as native phosphatidylinositol 4,5-bisphosphate or exogenous dextrans used for labelling, may be lost during this process. (2) After several days, wash the sample twice with of PBST (0.1% Triton X-100 in 1× PBS) twice for 24 h each.
Preparing the sample for imaging: (1) Incubate the sample in FocusClear or 80% glycerol solution for 2 days before imaging; these have refractive indices matching that of clarified tissue. Ensure there is sufficient solution surrounding the sample, and that evaporative losses do not occur. Protect the sample from light. (2) Take a clean glass slide and gently place it on a dust-free surface. (3) Take a small piece of BluTack putty and prepare constant-diameter worm shapes using gloved hands. Make the thickness uniform and about 1.5× the thickness of the sample (for example, if the sample is 1 mm, make the putty tube diameter 1.5 mm). (4) Place two tubes of putty horizontally across the vertical slide, leaving space in between for the tissue sample. Cut excess putty that protrudes off the slide. (5) Using a spatula, carefully take the sample and place it between the putty tubes in the middle of the slide. Pipette ~20 µl of FocusClear medium on top of the sample. (6) Carefully place a Willco dish (with the lipped side facing up) on top of the putty tubes. Press down on the glass part of the dish (keeping fingers over the putty to avoid glass shattering) carefully and slowly until contact is made with the sample and FocusClear medium. Ensure that there are no bubbles between the sample, medium, slide and dish. (7) Using a P200 pipette, carefully introduce more FocusClear to either side of the sealed chamber (from the liquid that surrounded the sample for incubation as it has been optically matched). Take great care not to introduce any bubbles. (8) Now that the whole chamber is filled with FocusClear, use PDMS sealant (Kwik-Sil) across both vertical openings between the putty, dish and slide to fully seal the chamber and prevent evaporation. (9) Place aluminium foil on top of the chamber and place it on a level surface (shielded from light to minimize photodamage). Leave the sample for 10–15 min to allow the PDMS sealant to polymerize fully. (10) Preparation is now ready for imaging.
Supplementary Material
Acknowledgements
This work was funded by a National Institutes of Health (NIH) Director’s Transformative Research Award (TR01) to K.D. from NIMH, as well as by NSF, the Simons Foundation, and the President and Provost of Stanford University. K.D. is also funded by NIDA, the DARPA REPAIR program, and the Wiegers, Snyder, Reeves, Gatsby, and Yu Foundations. K.C. is supported by the Burroughs Wellcome Fund Career Award at the Scientific Interface. S.-Y.K. is supported by a Samsung Scholarship, A.S.A. by the Helen Hay Whitney Foundation, K.A.Z. and A.K.D. by an NSF Graduate Research Fellowship and J.M. by the NIH MSTP. We acknowledge H. Vogel, L. Luo, L. Schwarz, M. Monje, S. Hestrin and D. Castaneda for advice, and the Autism Tissue Program for providing human brain tissue, as well as J. J. Perrino, J. Mulholland and the Cell Sciences Imaging Facility at Stanford for electron microscopy imaging and advice. We would also like to thank the entire Deisseroth laboratory for discussions and support. CLARITY resources and protocols are freely supported online (http://CLARITYresourcecenter.org).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions K.C. and K.D. conceived and designed the experiments and wrote the paper. K.C. led development of the CLARITY technology and its implementation. K.C. and S.K. clarified samples. K.C. imaged samples. K.C., S.-Y.K., S.K., J.W., K.A.Z., S.P., J.J.M., J.M., V.G. and H.B. prepared animals. J.W. performed tracing. T.J.D. and A.S.A. performed image processing. K.C., J.W., J.J.M. and A.K.D. wrote the CLARITY protocol. A.K.D. created AutoCAD drawings. K.C. and L.G. created Supplementary Videos. A.S.A. contributed to in situ and zebrafish data. C.R., L.G. and V.G. contributed to set-up of the relevant laboratory infrastructure. K.D. supervised all aspects of the work.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
References
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 3.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 4.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 7.Li A, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–1408. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- 8.Botcherby EJ, et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl Acad. Sci. USA. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFelipe J. From the connectome to the synaptome: an epic love story. Science. 2010;330:1198–1201. doi: 10.1126/science.1193378. [DOI] [PubMed] [Google Scholar]
- 10.Petersen CCH. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Kasthuri N, Lichtman JW. The rise of the “projectome”. Nature Methods. 2007;4:307–308. doi: 10.1038/nmeth0407-307. [DOI] [PubMed] [Google Scholar]
- 12.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nature Rev. Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mombaerts P, et al. Visualizing an olfactory sensorymap. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 14.Insel TR, Young LJ. The neurobiology of attachment. Nature Rev. Neurosci. 2001;2:129–136. doi: 10.1038/35053579. [DOI] [PubMed] [Google Scholar]
- 15.Micheva KD, Busse B, Weiler NC, O’Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock DD, et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 18.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodt HU, Leischner U, Schierloh A. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nature Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 20.Ertürk A, Mauch C, Hellal F, Förstner F. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nature Med. 2012;18:166–171. doi: 10.1038/nm.2600. [DOI] [PubMed] [Google Scholar]
- 21.Hama H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 22.Cheong W, Prahl S, Welch A. A review of the optical properties of biological tissues. IEEE J. Quant. Electron. 1990;26:2166–2185. [Google Scholar]
- 23.Sykova E, Nicholson C. Diffusion in brain extracellular space. Physiol Rev. 2008;88:1277–1340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker K, Jährling N, Saghafi S, Weiler R, Dodt H-U. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS ONE. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porrero C, Rubio-Garrido P, Avendaño C, Clascá F. Mapping of fluorescent protein-expressing neurons and axon pathways in adult and developing Thy1-eYFP-H transgenic mice. Brain Res. 2010;1345:59–72. doi: 10.1016/j.brainres.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 26.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 28.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Two-photon absorption properties of fluorescent proteins. Nature Methods. 2011;8:393–399. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldenthal KL, Hedman K, Chen JW, August JT, Willingham MC. Postfixation detergent treatment for immunofluorescence suppresses localization of some integral membrane proteins. J. Histochem. Cytochem. 1985;33:813–820. doi: 10.1177/33.8.3894499. [DOI] [PubMed] [Google Scholar]
- 30.Tramu G. An efficient method of antibody elution for the successive or simultaneous localization of two antigens by immunocytochemistry. J. Histochem. Cytochem. 1978;26:322–324. doi: 10.1177/26.4.207771. [DOI] [PubMed] [Google Scholar]
- 31.Wählby C, Erlandsson F, Begntsson E, Zetterberg A. Sequential immunofluorescence staining and image analysis for detection of large numbers of antigens in individual cell nuclei. Cytometry. 2002;71:32–41. [PubMed] [Google Scholar]
- 32.Kolodziejczyk E, Baertschi AJ. Multiple immunolabeling in histology: a new method using thermo-inactivation of immunoglobulins. J. Histochem. Cytochem. 1986;34:1725–1729. doi: 10.1177/34.12.3023476. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Q, Homma KJ, Poo M. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ. The architecture of active zone material at the frog’s neuromuscular junction. Nature. 2001;409:479–484. doi: 10.1038/35054000. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, et al. Organization of the core structure of the postsynaptic density. Proc. Natl Acad. Sci. USA. 2008;105:4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macdonald R. Zebrafish immunohistochemistry. Methods Mol. Biol. 1999;127:77–88. doi: 10.1385/1-59259-678-9:77. [DOI] [PubMed] [Google Scholar]
- 37.Sporns O, Tononi G, Kötter R. Thehuman connectome: a structural description of the human brain. PLOS Comput. Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickersham IR, Finke S, Conzelmann K-K, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyamichi K, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condé F, Lund JS, Jacobowitz DM, Baimbridge KG, Lewis DA. Local circuit neurons immunoreactive for calretinin, calbindin D-28k or parvalbumin in monkey prefrontal cortex: distribution and morphology. J. Comp. Neurol. 1994;341:95–116. doi: 10.1002/cne.903410109. [DOI] [PubMed] [Google Scholar]
- 45.Cheng CM, et al. Biochemical and morphometric analyses show that myelination in the insulin-like growth factor 1 null brain is proportionate to its neuronal composition. J. Neurosci. 1998;18:5673–5681. doi: 10.1523/JNEUROSCI.18-15-05673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grossfeld RM, Shooter EM. A study of the changes in protein composition of mouse brain during ontogenetic development. J. Neurochem. 1971;18:2265–2277. doi: 10.1111/j.1471-4159.1971.tb00183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.