Abstract
Background
In recent years advances have been made in the investigative methods of molecular background of canine heart disease. Studies have been conducted to identify specific genes which, when pathologically expressed, could lead to the dysfunction of the canine heart or are correlated with heart failure. For this purpose genome wide microarray experiments on tissues from failing hearts have been performed. In the presented study a whole genome microarray analysis was used for the first time to describe the transcription profile of peripheral blood nuclear cells in dogs with heart failure. Dogs with recognized heart disease were classified according the ISACHC (International Small Animal Cardiac Health Council) classification scheme as class 1 (asymptomatic) - 13 dogs, class 2 (mild to moderate heart failure) - 13 dogs and class 3 (severe heart failure) - 12 dogs. The control group consisted of 14 healthy dogs. The clinical picture of the animals included: animal history, clinical examination, echocardiographic examination and where applicable electrocardiographic and radiographic examinations.
Results
In the present study we identified four sets of differentially expressed genes, namely heart-failure-specific genes and ISACHC1-specific genes, ISACHC2-sepcific genes and ISACHC-3 specific genes. The most important set consisted of genes differentially expressed in all dogs with heart failure, despite the ISACHC stage. We identified 71 heart-failure-specific genes which were involved in two statistically significant receptor signalling pathways, namely angiotensinR - > CREB/ELK-SRF/TP53 signalling and ephrinR - > actin signalling. The number of ISACHC1-specific genes was 83; ISACHC2-specific genes - 1247 and ISACHC3-specific - 200.
Conclusions
The transcriptomic profile of peripheral blood nuclear cells in dogs with heart failure seems to reflect the presence of clinical signs of the disease in patients based on the observation that the largest number of differentially expressed genes was identified in ISACHC 2 group of patients. This group consists of dogs just starting to show clinical signs of heart failure. A set of genes was also found to have changed expression in all dogs with heart failure, despite the stage of the disease.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-509) contains supplementary material, which is available to authorized users.
Keywords: Dogs, Heart failure, ISACHC, Transcriptomic profile, Microarrays
Background
In recent years some advances have been made in the methods of investigation of the molecular background of canine heart disease. The etiopathogenesis of one of the most frequent heart diseases, myxomatous valve disease, in dogs may be different compared to humans although some similarities have been noted [1]. Some research has been conducted to identify specific genes which, when pathologically expressed, could lead to the dysfunction of the canine heart or are correlated with heart failure. For this purpose genome wide microarray experiments on tissues from failing hearts have been performed. Their results revealed that expression of some genes differ between tissue samples from failing hearts and healthy hearts, including: the matrix metalloproteinases and their tissue inhibitors (mainly MMP9, MMP1, MMP2, TIMP1, TIMP3) as well as genes involved in Ca2+ handling in cardiomyocytes, e.g. genes encoding the components of the cardiac ryanodyne receptor (RyR2) [2–6]. Other examples of differentially expressed genes are TGF-β (TGF-βR2, TGF-βR3) and serotonin receptors (5HT-R2B) [2]. The genome pattern of heart tissues from dogs suffering from heart failure were also investigated with the Real-time PCR method and the level of proteins encoded by differentially expressed genes was examined by immunohistochemical techniques. These findings gave similar results and pointed out different expression of extracellular matrix (ECM) genes, TGF-β receptors and thyroid hormone receptors (TRβ1 and TRβ2) in failing hearts compared to healthy control tissues [7–12].
The most important obstacle of the mentioned types of studies is access to diagnostic material. That is why researchers are attempting to investigate gene expression of peripheral blood nuclear cells taken from patients with cardiac disease. Blood nuclear cells are not a perfect source of diagnostic material in patients with heart failure but easy access makes them a very interesting source of potential disease markers. It has turned out that identification of such markers is possible since Wang et al. described the prognostic value of B-natriuretic peptide (BNP) in humans [13]. The results of a few studies suggest that expression of Na + −Ca2+ exchanger (NCX-1) gene in peripheral blood nuclear cells could have prognostic or diagnostic value [14, 15]. The other genes proposed as genomic markers in dogs are phospholamban (PLN) and haematopoietic lineage cell-specific protein 1 associated protein X-1 (HAX-1). Fonfara et al. also pointed out that the blood cells of dogs with heart failure could be appropriate material for searching for genomic markers. Their results showed increased levels of proinflammatory cytokines (Il-1, −2, −8, INF), MMP-1, MMP-3 and TIMP-3 and decreased levels of TNF-10, Il-10, TGF-b1, TIMP-1 and TIMP-2 in dogs with congestive heart failure [16].
In the present paper a whole genome microarray analysis was used for the first time to describe the transcription profile of peripheral blood nuclear cells in dogs with heart failure. The potential identification of any prognostic marker of canine cardiac disease would be of great importance since it may not only give new insight into the pathogenesis or staging of the disease but also help in the effective planning of treatment.
Results
The number of differentially expressed genes
Comparison of gene expression between all study groups (healthy dogs, ISACHC 1, ISACHC 2, ISACHC 3) revealed 4579 differentially expressed transcripts. Using the Tukey's HSD Post-hoc test it was possible to identify differentially expressed transcripts between individual study groups (Table 1).
Table 1.
The number of differentially expressed genes in dogs with different ISACHC stages of heart disease
| Name of group | ISACHC 2 | ISACHC 1 | Healthy dogs |
|---|---|---|---|
| ISACHC 3 | 2140 | 696 | 1170 |
| ISACHC 2 | - | 3499 | 2964 |
| ISACHC 1 | - | - | 426 |
Heart failure specific and ISACHC stage specific genes
The next step of our analysis was to identify heart-failure-specific genes. This means genes which were differentially expressed in all dogs with heart disease, despite the stage of the disease. The comparison between healthy and ISACHC 1 group revealed 426 differentially expressed transcripts; between healthy and ISACHC 2 dogs revealed 2964 differentially expressed transcripts and between healthy and ISACHC 3 dogs revealed 1170 differentially expressed transcripts. Among these three sets of differentially expressed genes there were 117 common transcripts. However, since some genes are represented on the array by more than one spot, we verified the consistency of differential expression in all the spots representing the same genes to eliminate false positive results. Moreover, the ontologies of all the transcripts are not known. Taking into consideration these two facts, a list of 71 heart - failure - specific genes of known ontologies was obtained (Table 2). The direction of changes in expression (up or down) of all 71 genes was the same in each disease stage. If a gene was down-regulated, this means that it was down regulated in all three comparisons (healthy vs ISACHC 1; healthy vs ISACHC 2; healthy vs ISACHC 3).
Table 2.
The list of the differentially regulated heart-failure-specific genes in dogs with ISACHC heart failure
| GeneSymbol | Adjusted p-values | Regulation | Description |
|---|---|---|---|
| ACTA1 | 0.014573121 | Up | Actin, alpha 1, skeletal muscle |
| ACTA2 | 0.039442863 | Up | Actin, alpha 2, smooth muscle, aorta |
| ACTC1 | 0.024042822 | Up | Actin, alpha, cardiac muscle 1 |
| ACTL7A | 0.024659734 | Up | Actin-like 7A |
| ADA | 0.01982955 | Up | Adenosine deaminase F |
| ATF4 | 0.044055864 | Up | Activating transcription factor 4 |
| CAMP | 0.018274972 | Up | Cathelicidin antimicrobial peptide |
| CAPG | 0.046687745 | Up | Capping protein (actin filament), gelsolin-like |
| CCL14 | 0.024452273 | Up | Chemokine (C-C motif) ligand 14 |
| CD151 | 0.012394691 | Up | CD151 molecule (Raph blood group) |
| CD177 | 0.010730711 | Up | CD177 molecule |
| CD300E | 0.0064891023 | Up | CD300e molecule |
| CORO1B | 0.03894925 | Up | Coronin, actin binding protein, 1B |
| CRYL1 | 0.0078813145 | Up | Crystallin, lambda 1 |
| CYBA | 0.031601883 | Up | Cytochrome b-245, alpha polypeptide |
| CYP27A1 | 0.034636706 | Up | Cytochrome P450, family 27, subfamily A, polypeptide 1 |
| DPEP2 | 0.008431375 | Up | Dipeptidase 2 |
| DSTN | 0.008163466 | Up | Destrin (actin depolymerizing factor) |
| EAF1 | 0.03957692 | Up | ELL associated factor 1 |
| EPHX2 | 0.018156866 | Up | Epoxide hydrolase 2, cytoplasmic |
| FAM107A | 0.020942843 | Up | Family with sequence similarity 107, member A |
| FANCG | 0.0043643974 | Down | Fanconi anemia, complementation group G |
| FLOT2 | 0.017366651 | Up | Flotillin 2 |
| GP9 | 0.0075741783 | Up | Glycoprotein IX (platelet) |
| GSN | 0.006870092 | Up | Gelsolin |
| HK3 | 0.0073725334 | Up | Hexokinase 3 (white cell) |
| HP | 0.016630936 | Up | Haptoglobin |
| HPCAL1 | 0.030948874 | Up | Hippocalcin-like 1 |
| IL10RA | 0.046400532 | Up | Interleukin 10 receptor, alpha |
| KDELR1 | 0.0037026592 | Up | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| KLHDC8B | 0.015807072 | Up | Kelch domain containing 8B |
| LOC475113 | 0.008475645 | Up | Lymphocyte antigen 6B-like |
| LOC491090 | 0.027546933 | Up | Similar to tubulin, alpha 1 |
| LSM10 | 0.0052581653 | Up | LSM10, U7 small nuclear RNA associated |
| MAPK1 | 0.03080859 | Up | Mitogen-activated protein kinase 1 |
| ME1 | 0.016186723 | Up | Malic enzyme 1, NADP(+)-dependent, cytosolic |
| MMP8 | 0.008103578 | Up | Matrix metallopeptidase 8 (neutrophil collagenase) |
| MMRN1 | 0.005678207 | Up | Multimerin 1 |
| MOV10L1 | 0.012869541 | Up | Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse) |
| MRPL37 | 0.026399544 | Up | Mitochondrial ribosomal protein L37 |
| MVP | 0.008610116 | Up | Major vault protein |
| MYL6 | 0.032878894 | Up | Myosin, light chain 6, alkali, smooth muscle and non-muscle |
| MYL6B | 0.008475645 | Up | Myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| N4BP2 | 0.01824313 | Down | NEDD4 binding protein 2 |
| NAGPA | 0.038744777 | Up | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| NEIL1 | 0.030278882 | Down | Nei endonuclease VIII-like 1 (E. coli) |
| NQO2 | 0.037860617 | Up | NAD(P)H dehydrogenase, quinone 2 |
| NRGN | 0.014320144 | Up | Neurogranin (protein kinase C substrate, RC3) |
| NSG1 | 0.009202879 | Up | Neuron specific gene family member 1 |
| PARVB | 0.025306692 | Up | Parvin, beta |
| PCGF2 | 0.015807072 | Up | Polycomb group ring finger 2 |
| PGLYRP1 | 0.0151561815 | Up | Peptidoglycan recognition protein 1 |
| PPP1R3D | 0.0077612605 | Up | Protein phosphatase 1, regulatory (inhibitor) subunit 3D |
| PTGER3 | 0.0045230556 | Up | Prostaglandin E receptor 3 (subtype EP3) |
| PTPN9 | 0.008163466 | Up | Protein tyrosine phosphatase, non-receptor type 9 |
| RSU1 | 0.046061113 | Up | Ras suppressor protein 1 |
| SEPT5 | 0.0062189046 | Up | Septin 5 |
| SHC2 | 0.02669537 | Up | SHC (Src homology 2 domain containing) transforming protein 2 |
| SLC11A1 | 0.0045230556 | Up | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| SLC27A2 | 0.006745509 | Up | Solute carrier family 27 (fatty acid transporter), member 2 |
| SLC2A6 | 0.0045230556 | Up | Solute carrier family 2 (facilitated glucose transporter), member 6 |
| TBC1D30 | 0.030404283 | Up | TBC1 domain family, member 30 |
| TBXAS1 | 0.0049617672 | Up | Thromboxane A synthase 1 (platelet) |
| TCIRG1 | 0.023556981 | Up | T-cell, immune regulator 1, ATPase, H + transporting, lysosomal V0 subunit A3 |
| TIMP1 | 0.0043643974 | Up | TIMP metallopeptidase inhibitor 1 |
| TOP3B | 0.02519754 | Up | Topoisomerase (DNA) III beta |
| TRPV2 | 0.0049617672 | Up | Transient receptor potential cation channel, subfamily V, member 2 |
| TSPO | 0.004600876 | Up | Translocator protein (18 kDa) |
| TTC19 | 0.0075741783 | Down | Tetratricopeptide repeat domain 19 |
| VNN1 | 0.031601883 | Down | Vanin 1 |
| ZNF248 | 0.004831977 | Down | Zinc finger protein 248 |
After identifying heart - failure - specific genes we applied the same approach to a selection of each stage - specific genes. The 83 genes specific for ISACHC class 1 were identified (data not shown). The list of ISACHC class 2 - specific genes was the longest and consisted on 1247 genes (data not shown). In cases of most advanced disease, ISACHC 3, the list of specific genes was 200 (data not shown). These four lists of genes were subjected to further analysis.
Pathways connected with differentially expressed genes
Next we examined the differentially regulated genes for enrichment in any known canonical pathway. For this purpose the Pathway Studio software was used. The most interesting results were obtained from analysis of the Ariadne Receptor Signaling Pathways. The results of this analysis are presented in Tables 3, 4, 5, 6.
Table 3.
Ariadne pathways based on heart-failure-specific genes differentially regulated in dogs with ISACHC heart failure
| Ariadne receptor signaling pathways | The genes engaged in listed pathways | p-value |
|---|---|---|
| EphrinR - > actin signaling | ACTA2, GSN, ACTA1, ACTC1, DSTN | 0.0179871 |
| AngiotensinR - > CREB/ELK-SRF/TP53 signaling | MAPK1, CYBA, NQO2 | 0.0359593 |
Table 4.
Ariadne pathways based on ISACHC1-specific genes differentially regulated in dogs with ISACHC1 heart failure
| Ariadne receptor signaling pathways | The genes engaged in listed pathways | p-value |
|---|---|---|
| PTPRC - > BCL6 signaling | 2 | 0.0221563 |
| BradykininR - > STAT3 signaling | 1 | 0.0349728 |
| TNFR - > AP-1/ATF/TP53 signaling | 2 | 0.0462323 |
Table 5.
Ariadne pathways based on ISACHC2-specific genes differentially regulated in dogs with ISACHC2 heart failure
| Ariadne receptor signaling pathways | The number of genes | p-value |
|---|---|---|
| UrokinaseR - > ELK-SRF signaling | 9 | 0.0010517 |
| IL4R - > ELK-SRF/HMGY signaling | 9 | 0.00118083 |
| PDGFR - > FOXO3A signaling | 5 | 0.00176524 |
| IL2R - > ELK-SRF/MYC signaling | 8 | 0.00234544 |
| AGER - > NF-kB signaling | 6 | 0.00241572 |
| IL15R - > NF-kB/NFATC signaling | 6 | 0.0028208 |
| IL7R - > FOXO/NF-kB signaling | 6 | 0.00327501 |
| InsulinR - > CTNNB/FOXA/FOXO signaling | 5 | 0.0038699 |
| FibronectinR - > CTNNB signaling | 6 | 0.00434523 |
| TNFRSF5/13B - > NFATC1 signaling | 5 | 0.00459712 |
| VEGFR - > CTNNB signaling | 5 | 0.00459712 |
| PTPRC - > STAT6 signaling | 2 | 0.00520887 |
| FGFR - > AP-1/CREB/CREBBP/ELK-SRF/MYC signaling | 10 | 0.00699557 |
| HGFR - > FOXO3A signaling | 4 | 0.00709744 |
| AdenosineR - > NF-kB signaling | 7 | 0.0071087 |
| ErythropoietinR - > FOXO3A signaling | 4 | 0.00859443 |
| TLR3 - > NF-kB signaling | 3 | 0.00948063 |
| ICAM2 - > CTNNB/FOXO/STAT3 signaling | 5 | 0.00974526 |
| CD19 - > NF-kB signaling | 5 | 0.00974526 |
| InsulinR - > ELK-SRF/SREBF signaling | 7 | 0.0104641 |
| FibronectinR - > AP-1/ELK-SRF/SREBF signaling | 10 | 0.0106044 |
| Notch - > NF-kB signaling | 5 | 0.0111183 |
| GHR - > NF-kB signaling | 5 | 0.0111183 |
| IL12R - > NF-kB/NFATC signaling | 5 | 0.0126187 |
| DopamineR2 - > NF-kB signaling | 5 | 0.0126187 |
| ThrombinR - > NF-kB signaling | 5 | 0.0126187 |
| ErythropoietinR - > NF-kB signaling | 6 | 0.0138912 |
| AGER - > CREB/SP1 signaling | 5 | 0.0142517 |
| AngiopoietinR - > FOXO signaling | 4 | 0.0142826 |
| VEGFR - > FOXO3A signaling | 4 | 0.0166053 |
| CholinergicRm - > CREB/ELK-SRF signaling | 10 | 0.0174704 |
| B-cell receptor - > NFATC signaling | 5 | 0.0179347 |
| ErythropoietinR - > AP-1/CREB/MYC signaling | 8 | 0.0182859 |
| EDG2 - > ELK-SRF signaling | 7 | 0.0189356 |
| SerotoninR1 - > FOS signaling | 10 | 0.019617 |
| HGFR - > AP-1/CREB/MYC signaling | 8 | 0.0209776 |
| IGF1R - > CEBPA/FOXO1A signaling | 4 | 0.0219319 |
| FcIgER - > ELK-SRF signaling | 7 | 0.0220582 |
| TLR1/2/6 - > NF-kB signaling | 5 | 0.0222033 |
| DopamineR2 - > AP-1/CREB/ELK-SRF signaling | 9 | 0.024631 |
| GDNF - > HSF1 signaling | 7 | 0.025526 |
| ThrombopoietinR - > AP-1/CREB/ELK-SRF/MYC signaling | 8 | 0.0255337 |
| VEGFR - > AP-1/CREB/MYC signaling | 8 | 0.0289362 |
| MacrophageR - > CEBPB/NF-kB signaling | 5 | 0.0326186 |
| TLR3 - > IRF signaling | 2 | 0.0326546 |
| FibronectinR - > NF-kB signaling | 6 | 0.032985 |
| GHR - > ELK-SRF/MYC signaling | 6 | 0.0355346 |
| B-cell receptor - > NF-kB signaling | 6 | 0.0382096 |
| CholinergicRn - > CREB signaling | 5 | 0.0388136 |
| NTRK - > FOXO/MYCN signaling | 4 | 0.0394373 |
| EGFR - > AP-1/CREB/ELK-SRF/MYC signaling | 8 | 0.0410289 |
| IL6R - > STAT signaling | 2 | 0.0423644 |
| PDGFR - > AP-1/MYC signaling | 7 | 0.043159 |
| CD38 - > NF-kB signaling | 7 | 0.043159 |
| KIT - > MITF signaling | 6 | 0.043943 |
| NTRK - > AP-1/CREB/ELK-SRF/MYC/SMAD3/TP53 signaling | 8 | 0.0457237 |
| PTAFR - > AP-1/ATF1/CREB/ERK-SRF signaling | 9 | 0.0466592 |
| IGF1R - > MEF/MYOD/MYOG signaling | 6 | 0.0470043 |
| CD19 - > AP-1/ELK-SRF signaling | 6 | 0.0470043 |
| PTAFR - > NF-kB signaling | 6 | 0.0470043 |
| ThrombopoietinR - > SPI1 signaling | 4 | 0.0481701 |
Table 6.
Ariadne pathways based on ISACHC3-specific genes differentially regulated in dogs with ISACHC3 heart failure
| Ariadne receptor signaling pathways | The number of genes | p-value |
|---|---|---|
| HGFR - > FOXO3A signaling | 3 | 0.00563574 |
| ERBB2/3 - > EP300/ETS/ETV/SP1 signaling | 5 | 0.01015 |
| EGFR/ERBB2 - > HIF1A signaling | 5 | 0.0128385 |
| TNFRSF5/13B - > NFATC1 signaling | 3 | 0.0152928 |
| HGFR - > AP-1/CREB/MYC signaling | 5 | 0.0216208 |
| AdenosineR - > NF-kB signaling | 4 | 0.0217062 |
| ICAM2 - > CTNNB/FOXO/STAT3 signaling | 3 | 0.0242659 |
| NeurotensinR - > ELK-SRF/AP-1/EGR signaling | 5 | 0.0248821 |
| IL2R - > ELK-SRF/MYC signaling | 4 | 0.0259176 |
| InsulinR - > ELK-SRF/SREBF signaling | 4 | 0.0274252 |
| GHR - > ELK-SRF/MYC signaling | 4 | 0.0274252 |
| KIT - > MITF signaling | 4 | 0.032265 |
| IGF1R - > MEF/MYOD/MYOG signaling | 4 | 0.033985 |
| UrokinaseR - > ELK-SRF signaling | 4 | 0.0357589 |
| IL4R - > ELK-SRF/HMGY signaling | 4 | 0.0375869 |
| FibronectinR - > CTNNB signaling | 3 | 0.0381165 |
| EDG2 - > ELK-SRF signaling | 4 | 0.0394692 |
| EGFR/ERBB3 - > MEF/MYOD/NFATC/MYOG signaling | 6 | 0.0414187 |
| FcIgER - > ELK-SRF signaling | 4 | 0.0433973 |
| EGFR - > NCOR2 signaling | 4 | 0.0454432 |
| GDNF - > HSF1 signaling | 4 | 0.047544 |
| ErythropoietinR - > ELK-SRF/FOS signaling | 5 | 0.0477064 |
Real-time RT-PCR
To validate microarray data we selected three genes for Real-time PCR analysis (ACTA2, TIMP1, MMP8). The changes in the expression of these three genes measured using Real-time qPCR were similar to gene expression changes observed in the microarray studies (Figures 1, 2, 3).
Figure 1.
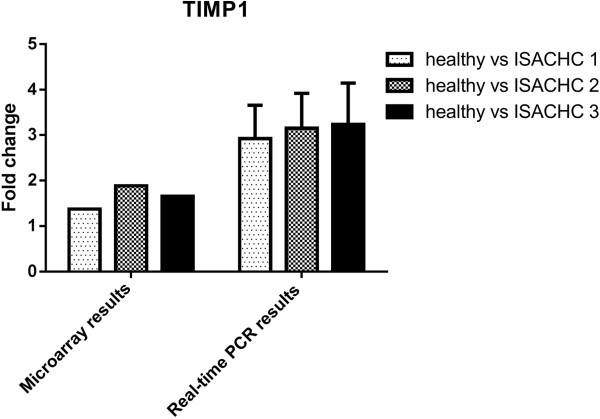
The expression TIMP1 gene measured with microarray and Real-time PCR in dogs with different ISACHC stages of heart disease.
Figure 2.
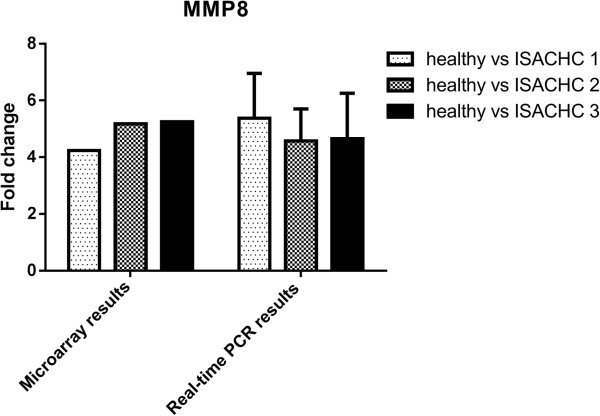
The expression MMP8 gene measured with microarray and Real-time PCR in dogs with different ISACHC stages of heart disease.
Figure 3.
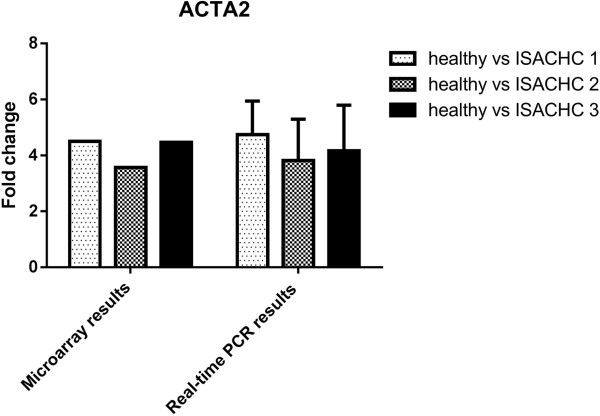
The expression ACTA2 gene measured with microarray and Real-time PCR in dogs with different ISACHC stages of heart disease.
Discussion
Heart failure in dogs does not occur as frequently as it does in humans and is mainly caused by two diseases, namely endocardiosis (also referred to as myxomatous valve disease or chronic degenerative valvular disease) and dilated cardiomyopathy. Both these diseases can lead to congestive heart failure and the associated clinical signs. Since the molecular background of these diseases, especially endocardiosis, is still far from fully elucidated, the search for any specific markers (prognostic, therapeutic) of this disease would be of great value.
In the present study we evaluated the transcriptomic profile of peripheral blood nuclear cells of dogs with different stages of heart disease. Since the whole analysis resulted in the huge number of differentially expressed transcripts (4579) we decided to identify two types of genes - 1) genes differentially expressed in all affected dogs, despite the ISACHC stage (so called heart-failure-specific genes) and 2) genes differentially expressed in each of ISACHC stages (so called ISACHC stage-specific genes). This approach seems reasonable because if the gene is regulated in all patients with heart failure, despite the stage of the disease, it could be a potential marker of heart disease in dogs. On the other hand the identification of each ISACHC stage-specific gene give the opportunity to identify markers of heart disease progression (i.e. form stage I to stage II or from stage II to stage III).
The list of heart-failure specific genes included 117 transcripts for which we were able to identify 71 specific genes. Among these genes we were able to identify three α-SMA genes which are considered as a markers of muscle cells. This phenomenon could suggest the presence of myofibroblasts (circulating fibroblasts) in the peripheral blood nuclear cells of dogs with heart failure. Similar observation has been recently published in humans with atrial fibrillation what could indicate that heart disease could be related with the presence of α-SMA containing cells in the blood [17]. However, α-SMA containing cells could also be of hematopoietic progenitor cells origin so their source in the blood of heart disease patients is still unclear [18]. Moreover, our results of flow cytometer analysis showed that the percentage of α-SMA positive cells in peripheral blood mononuclear cells of three dogs with ISACHC class 2 heart failure was between 0.2-0.5% (Additional files 1, 2). Whether these cells could be a marker of heart disease remains a pure speculation. Other genes present among 71 heart failure specific genes have been previously related to heart failure or involved in heart disease development. Examples include matrix metallopeptidase 8 (MMP8) and TIMP metallopeptidase inhibitor 1 (TIMP1). It is known that remodelling of the extracellular matrix (ECM) occurs during heart failure and the matrix metalloproteinases and their tissue inhibitors are involved in this process [19]. Initially MMP-8 was considered to be only expressed in peripheral inflammatory cells, but its myocardial sources could be endogenous cell types such as mast cells [20]. The level of MMP8 is increased during left ventricular remodelling following myocardial infarction in rats [21]. Our study showed that the expression of MMP8 is increased in all study groups in comparison to healthy control dogs (over a 4 fold change). Moreover, these results were confirmed with Real-time PCR (Figure 2). The study conducted by Oyama and Chittur revealed that the expression of TIMP1 was over 4 times higher in dogs with endocardiosis than in healthy dogs [4]. Our results (both microarray and Real-time PCR) showed a similar tendency. Another gene identified as heart - failure - specific one was haptoglobin (HP). The expression of HP was higher (over 2.4 fold change) in diseased dogs than in control dogs. In humans the functional allelic polymorphism in the HP gene plays a key role in determining survival and presence of congestive heart failure after myocardial infarction [22]. However, it seems quite impossible that single gene expression could influence the complex processes of heart disease development. Rather relationships and interactions between differentially expressed genes could play a key role. In order to evaluate these possibilities we used the Pathway Studio software. This database gives the opportunity to find genes involved in different kinds of signalling pathways. Pathway Studio software can also group differentially expressed genes on the basis of their molecular function and biological processes in which they are involved. The genes differentially expressed with statistical significance identified as heart-failure-specific were involved in regulation of two canonical pathways, namely angiotensinR - > CREB/ELK-SRF/TP53 signalling and ephrinR - > actin signalling. The first signalling pathway shows the relation between the expression of angiotensin receptors and CREB/ELK-SRF/TP53 transcription factors and indicates that the combined activation of CREB/ELK-SRF/TP53 leads to angiotensin receptor activation during the disease process. Serum response factor (SRF) is a transcription factor which regulates many immediate-early genes and is required for the appearance of beating sarcomeres in the heart [23]. SRF activates genes involved in smooth muscle differentiation and proliferation by recruiting muscle-restricted cofactors, such as ternary complex factors (TCFs) of the ETS-domain family, for example ELK-1 [24]. Changed expression of ELK-4 in peripheral blood nuclear cells in patients with refractory ischemic end-stage heart failure were also seen in our previous study [25] and Kuner et al. recognized ELK-4 as potential interaction partner necessary for creating network of regulatory genes in ischemic cardiomyopathy [26]. Since CREB/ELK-SRF/TP53 transcription factors target angiotensin receptors in dogs with heart failure, it clearly proves that the function of the failing heart is directly correlated with renal function and that in cases of heart disease we could observe changes in the function of renin-angiotensin-aldosterone axis. Such a conclusion was also drawn from one of our previous studies in which we were able to identify that transcriptomic profile of peripheral blood nuclear cells in humans after acute heart failure is correlated with the creatinine (marker of kidney function) level in the blood [27].
Since the lists of genes specific for each heart failure stage were much longer than the list of heart-failure-specific genes we also ran the Pathway Studio analysis to obtain information on significantly regulated signalling pathways in each ISACHC stage (Tables 4, 5, 6). While analysing these results one has to remember that ISACHC 1 heart failure stage encompasses patients without clinical signs of heart disease, ISACHC 2 stage includes dogs with compensated clinical signs and ISACHC 3 stage includes dogs with uncompensated heart failure. Having this in mind one could conclude the very long list of differentially regulated genes as well as significantly regulated signalling pathways in ISACHC 2 patients is not only the results of heart disease but also the result of many different compensatory mechanisms which were triggered in order to restore homeostasis. On the other hand in ISACHC 3 patients, the majority of these mechanisms are no longer in place so the list of regulated genes and pathways is much shorter. Based on our results it could be concluded that for the ISACHC 1 group, the specific changes in the transcriptome include the regulation of protein tyrosine phosphatase kinase receptors, bradykinin receptor and tumor necrosis factor receptor. These changes seem to be rather unspecific for heart disease and probably reflect only the reaction of the body to progressed disease, not the progression of the disease itself. In case of ISACHC 2 specific genes and pathways the interpretation of the obtained results is complex but in our opinion it is worth to point out that in addition to the statistically significant regulation of many inflammatory pathways and growth factors, we also see changes in the regulation of transcription factors targeted at platelet-mediated mediators and PTAFR (platelet-activating factor receptor). Anti-platelet therapy is one of the standard approaches in the treatment of patients with heart failure in humans and the regulation of this receptor was also identified in our previous study on patients with end-stage ischemic heart failure [25]. Platelet-activating factor may play a critical role in the development of severe heart failure coupled with resulting multiple organ failure, as seen in the Munakata et al. study performed on dogs with left ventricular assistance post coronary ligation [28]. This study showed that it may also be beneficial for dogs with clinical signs of severe heart disease to receive some form of anti-platelet therapy.
In the pathways significantly regulated in ISACHC 3 patients, we were able to identify for the first time the presence of transcription factors targeting EGFR/ERBB2. The ErbB2(HER2)-dependent signalling pathway is involved in proper myocardium function, structure and contractility [29–31]. The ErbB2 (HER2) receptor is responsible for the activation of several transcription factors, including AP-1 (also in heart muscle hypertrophy) and nuclear factor kappa B (involved in the response to oxidative stress) [32]. It could play a key role in dilated cardiomyopathy prevention [33]. In mice with ErbB2 gene deletion a gradual development of systolic myocardial dysfunction has been observed, occurring with myocardium thinning and heart ventricles distension [34]. In mice the binding of neuregulin-1 (NRG-1) to the HER2 receptor is cardioprotective [35]. Since there is no data in the literature in this field obtained from dogs, we could only speculate that the regulation of ErbB2 signalling pathway in dogs with ISACHC 3 heart failure could be the feature of end-stage heart failure also in this species. But this observation requires further investigation.
Conclusions
The transcriptomic profile of peripheral blood nuclear cells in dogs with heart failure seems to reflect the presence of clinical signs of the disease in patients. Such a conclusion could be drawn from the observation that the longest list of differentially expressed genes as well as signalling pathways were identified in ISACHC 2 group of patients, those first to show clinical signs of heart failure. The development of heart failure in dogs results in changed expression of genes involved in the regulation of the renin-angiotensin-aldosterone axis in peripheral blood nuclear cells whereas end-stage heart failure could be reflected by activation of transcription factors targeting at the ErbB2 receptor.
Methods
Animals
This study complies with national and institutional guidelines on the use of animals in clinical research according to the Polish legal act from January 21st, 2005 (Ustawa o doświadczeniach na zwierzętach z dnia 21 stycznia 2005 r. (Dz. U. z 2005 r. Nr 33, poz. 289 z późn.zm.)), concerning experiments performed on client owned animals. Before enrolling a dog into the study an informed consent from its owner was obtained and a high standard of care was adhered to throughout each examination. A prospective analysis and studies were carried out on dogs submitted to the Cardiology Service of the Small Animal Clinic, Faculty of Veterinary Medicine, School of Agriculture in Warsaw. The clinical picture of the animals included: animal history, clinical examination, echocardiographic examination and where applicable electrocardiographic and radiographic examinations. Dogs with recognized heart disease were classified according the ISACHC (International Small Animal Cardiac Health Council) classification scheme as class 1 (asymptomatic) - 13 dogs, class 2 (mild to moderate heart failure) - 13 dogs and class 3 (severe heart failure) - 12 dogs. The control group consisted of 14 healthy dogs. Study population characteristics are shown in Table 7. Statistical analyses of clinical parameters were performed using GraphPad Prism version 5.00 (GraphPad Software, Inc., USA).
Table 7.
Study population characteristics (data are shown as mean ± SD)
| ISACHC | Healthy | ISACHC 1 | ISACHC 2 | ISACHC 3 | |
|---|---|---|---|---|---|
| n = 52 | 14 | 13 | 13 | 12 | |
| Breeds | Dachshund (5), German Shepherds (6), Labrador retriever (1), German Pointer (1), Rottweiler (1) | Dachshund (10), Labrador retriever (1), Miniature poodle (1), Standard schnauzer (1) | Dachshund (9), Cocker spaniel (2), Airedale terrier (1), Standard schnauzer (1) | Dachshund (12) | |
| Age range | 1.4 - 10.5 years | 5.75 - 14.0 years | 9.8 - 15.2 years | 7.25 - 15.2 years | |
| Sex | Males | 8 | 7 | 7 | 10 |
| Females | 6 | 6 | 6 | 2 | |
| EF | 66.08 ± 7.99 | 72.93 ± 9.33 | 77.0 ± 8,90** | 83.08 ± 4.96*** | |
| HR | 107.3 ± 18.90 | 113.7 ± 26.08 | 134.0 ± 21.29* | 140.7 ± 19.43** | |
*Statistically significant difference vs healthy dogs with p<0.05; **statistically significant difference vs healthy dogs with p<0.01; ***statistically significant difference vs healthy dogs with p<0.001.
Clinical and laboratory examinations
The clinical and laboratory examinations included: complete physical examination, including thoracic auscultation, assessment of dyspnoea, cough, heart rate, presence of heart murmurs, mucosal membrane colour, pulse, palpation of thorax and abdomen as well as additional cardiologic examinations: echocardiography, electrocardiography (I, II,III, aVL, aVF and aVR leads) in dogs with arrhythmias, radiography in dogs with suspicion of congestive heart failure, and blood pressure measurement (Doppler method). All of above mentioned examinations were carried out by one clinician from the Cardiology Service of Small Animal Clinic at Faculty of Veterinary Medicine. The blood morphology and biochemistry analyses were conducted in the Laboratory of Small Animal Clinic.
All examinations were performed at rest without pharmacological restraint. A transthoracic echocardiographic (TTE) examination was performed in all dogs with an Aloka 4000 ultrasound machine equipped with a cardiology programs and 2.5 - 7-megahertz (mHz) sector transducers. Electrocardiographic examination (leads I, II, III, aVL, aVF and aVR) were performed with a BTL-08 MD machine. Radiological examination of the chest were performed with the G&E Prestige II X-ray machine. The blood samples for the transcription profile analysis were collected from client-owned dogs during routine veterinary examinations.
RNA isolation, validation, labelling, and hybridization
Blood samples were drawn from the cephalic or jugular veins and collected in Rneasy Protect Animal Blood Tubes (Qiagen, USA). Total RNA from peripheral blood nuclear cells was isolated using a Rneasy Protect Animal Blood Kit (Qiagen, USA). Isolated RNA samples were dissolved in 30 μl of REB Buffer from the test kit. Next the RNA quantity was measured spectrophotometrically using a NanoDrop (NanoDrop Technologies, USA). The analysis of final RNA quality and integrity was performed with a BioAnalyzer (Agilent, USA). To ensure optimal data quality, only RNA samples with RIN number ≥7.5 were included in the analysis.
The analysis of gene-expression profile was performed using Canine (V2) Gene Expression Microarray, 4x44K (Agilent Technologies, USA). Each slide contained 4 microarrays representing about 45000 canine predicted mRNAs. The Quick Amp Labeling Kit (Agilent, USA) was used to amplify and label target RNA to generate complementary RNA (cRNA) for oligo microarrays used in gene expression profiling. Experiment was performed using a common reference design, where the common reference was a pool of equal amounts of RNA from 10 healthy dogs. On each two-color microarrays, we hybridized 825 ng of cRNA from the pool (labelled Cy3) and 825 ng of cRNA (labelled Cy5). In total we ran 52 microarrays - one for each patient. Microarray hybridization was performed with the Gene Expression Hybridization Kit (Agilent Technologies, USA), according to the manufacturer's protocols. RNA Spike In Kit (Agilent Technologies, USA) was used as an internal control. Acquisition and analysis of hybridization intensities were performed using the Agilent DNA microarray scanner.
Signal detection and statistical analysis
Data were extracted and background subtracted using the standard procedures contained in the Agilent Feature Extraction (FE) Software version 10.7.3.1. FE performs a Lowess normalization. The statistical analysis was performed using Gene Spring 12 software (Agilent, USA). The samples underwent quality control and the results showed that each sample had a similar QC metric profile. The next step was filtering probesets by flags to remove poor quality probes (absent flags). The statistical significance of the differences was evaluated using a one-way ANOVA and Tukey's HSD Post-hoc test (p < 0.05). A multiple testing correction was performed using Benjamini and Hochberg False Discovery Rate (FDR) < 5%. Microarray data were deposited at the Gene Expression Omnibus data repository under the number GSE48319 and followed MIAME requirements.
To identify signalling pathways and gene function the microarray data was analyzed using Pathway Studio 9.0 (Ariadne Genomics). This is a database consisting of millions of individually modelled relationships between proteins, genes, complexes, cells, tissues, drugs and diseases [36].
Real-time RT-PCR
To verify microarray results on the expressions of ACTA2 (the gene for the actin, alpha 2, smooth muscle, aorta), MMP8 (the gene for the matrix metallopeptidase 8 (neutrophil collagenase)), TIMP1 (the gene for the TIMP metallopeptidase inhibitor 1) were checked by real-time RT-PCR. The sequences of these genes were obtained from the Ensembl database. Primers were designed using Primer-Blast software (NCBI database) and then checked for secondary structures using Oligo Calculator (free on-line access). The secondary structures of the amplicon were examined using the mfold Web Server (free on-line access). GAPDH was used as a house keeping gene [37]. The sequences of the primers are listed in Table 8.
Table 8.
The sequences of the primers used in the study
| Primer | Forward | Reverse | Annealing conditions |
|---|---|---|---|
| ACTA2 | GTGGGGATGGGACAAAAGG | GAAAGCACCGCCTGAATAG | 60°C for 9 s |
| MMP8 | CGATGCAGAAGAAACATGGA | TTGCTTGAAGGACCGTAGAT | 60°C for 10 s |
| TIMP1 | CTTAAACCGGCGTTATGAGA | GGGGATGGATGAACAGGTAA | 60°C for 10 s |
| GAPDH | CTGGGGCTCACTTGAAAGG | CAAACATGGGGGCATCAG | 59°C for 4 s |
cDNA was synthesized using the Enhanced Avian HS RT-PCR Kit (Sigma-Aldrich, St. Louis, Missouri). All analyses were performed on individual samples of total RNA using a LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics GmBH, Germany) as follows: Mg2+ was added to a final concentration of 3 mM; pre-incubation at 95°C for 10 minutes; amplification (40 cycles) including denaturation at 95°C for 10 seconds, annealing – temperature and time specified in Table 8; melting curve including denaturation at 95°C for 0 seconds, annealing at 65°C for 15 seconds, continuous melting at 95°C for 0 seconds (slope = 0.1°C/s); cooling at 40°C for 30 seconds. Results were calculated using the 2-ΔΔCT method [38].
Electronic supplementary material
Additional file 1: Representative results of flow cytometric analysis showing negative control of immunofluorescence staining of peripheral blood mononuclear cells (PBMC) from dog number 1 with ISACHC 2 class heart failure. PBMC were fixed, permeabilized and stained with secondary antibodies: chicken anti-mouse IgG conjugated with Alexa Fluor 488 (Molecular Probes, Life Technologies). The percentage of α-SMA positive cells is 0.0%; a) morphological flow cytometric parameters of analyzed cells gated on the basis of their size (forward scatter: FSC) and granularity (side scatter: SSC); b) cytogram of Alexa Fluor 488 positive cells (gate P2); c) histogram of Alexa Fluor 488 positive cells (gate P4); d) table showing population hierarchy of cells in cytograms a, b, and histogram c. (PDF 72 KB)
Additional file 2: Representative results of flow cytometric analysis of peripheral blood mononuclear cells (PBMC) from dog number 1 with ISACHC 2 class heart failure, which were stained with antibodies against alpha smooth muscle actin (α-SMA). PBMC were fixed, permeabilized and stained with monoclonal mouse anti- α-SMA antibodies (clone 1A4, Dako, Denmark), followed by incubation with secondary antibodies: chicken anti-mouse IgG conjugated with Alexa Fluor 488 (Molecular Probes, Life Technologies). The percentage of α-SMA positive cells is around 0.3%; a) morphological flow cytometric parameters of analyzed cells gated on the basis of their size (forward scatter: FSC) and granularity (side scatter: SSC); b) cytogram showing α-SMA –positive cells gated in gate P2; c) histogram showing α-SMA –positive cells gated in gate P4; d) table showing population hierarchy of cells in cytograms a, b, and histogram c. (PDF 85 KB)
Acknowledgements
The preparation of this paper was supported by grant no. N N308 607538 received from the Polish Ministry of Science.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MH was responsible for all transcriptomic and statistical analyses and preparing of manuscript. MG was responsible for clinical part of the study – patients examination, disease staging and final draft of the text. MP-J assisted with clinical examination of patients and took blood samples. MJ designed the study protocol, coordinated the whole study, assisted in statistical analyses and was involved in final draft of the text. All authors read and approved the final manuscript.
Contributor Information
Magdalena Hulanicka, Email: magda.loj@gmail.com.
Magdalena Garncarz, Email: magdagarncarz@yahoo.com.
Marta Parzeniecka-Jaworska, Email: mparzeniecka@gmail.com.
Michał Jank, Email: majank@hotmail.com.
References
- 1.Disatian S, Lacerda C, Orton EC. Tryptophan hydroxylase 1 expression is increased in phenotype-altered canine and human degenerative myxomatous mitral valves. J Heart Valve Dis. 2010;19:71–78. [PubMed] [Google Scholar]
- 2.Zheng J, Chen Y, Pat B, Dell'italia LA, Tillson M, Dillon AR, Powell PC, Shi K, Shah N, Denney T, Husain A, Dell'Italia LJ. Microarray identifies extensive downregulation of noncollagen extracellular matrix and profibrotic growth factor genes in chronic isolated mitral regurgitation in the dog. Circulation. 2009;119:2086–2095. doi: 10.1161/CIRCULATIONAHA.108.826230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiczak L, Tomaszek A, Bania J, Paslawska U, Zacharski M, Noszczyk-Nowak A, Janiszewski A, Skrzypczak P, Ardehali H, Jankowska EA, Ponikowski P. Expression and complex formation of MMP9, MMP2, NGAL, and TIMP1 in porcine myocardium but not in skeletal muscles in male pigs with tachycardia-induced systolic heart failure. Biomed Res Int. 2013;2013:283856. doi: 10.1155/2013/283856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oyama MA, Chittur SV. Genomic expression patterns of mitral valve tissues from dogs with degenerative mitral valve disease. Am J Vet Res. 2006;67:1307–1318. doi: 10.2460/ajvr.67.8.1307. [DOI] [PubMed] [Google Scholar]
- 5.Oyama M, Trafny D, Meurs K, Chittur S: Comparative blood-based transcriptional activity in healthy Doberman Pinschers and occult cardiomyopathy or congestive heart failure.Proceedings of the 20th Annual ECVIM-CA Congress (ESVC, European Society of Veterinary Cardiology): 9–11 September 2010; Toulouse34. http://www.worldcat.org/title/congress-proceedings-the-european-college-of-veterinary-internal-medicine-companion-animals-2010–20th-ecvim-ca-congress-9th-11th-september/oclc/690901860&referer=brief_results
- 6.Oyama MA, Chittur SV, Reynolds CA. Decreased triadin and increased calstabin2 expression in great Danes with dilated cardiomyopathy. J Vet Intern Med. 2009;23:1014–1019. doi: 10.1111/j.1939-1676.2009.0360.x. [DOI] [PubMed] [Google Scholar]
- 7.Aupperle H, Thielebein J, Kiefer B, März I, Dinges G, Schoon HA, Schubert A. Expression of genes encoding matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMPs) in normal and diseased canine mitral valves. J Comp Pathol. 2009;140:271–277. doi: 10.1016/j.jcpa.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Aupperle H, Thielebein J, Kiefer B, März I, Dinges G, Schoon HA. An immunohistochemical study of the role of matrix metalloproteinases and their tissue inhibitors in chronic mitral valvular disease (valvular endocardiosis) in dogs. Vet J. 2009;180:88–94. doi: 10.1016/j.tvjl.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Disatian S, Ehrhart EJ, Zimmerman S, Orton EC. Interstitial cells from dogs with naturally occurring myxomatous mitral valve disease undergo phenotype transformation. J Heart Valve Dis. 2008;17:402–411. [PubMed] [Google Scholar]
- 10.Aupperle H, März I, Thielebein J, Schoon HA. Expression of transforming growth factor-beta1, −beta2 and -beta3 in normal and diseased canine mitral valves. J Comp Pathol. 2008;139:97–107. doi: 10.1016/j.jcpa.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Obayashi K, Miyagawa-Tomita S, Matsumoto H, Koyama H, Nakanishi T, Hirose H. Effects of transforming growth factor-β3 and matrix metalloproteinase-3 on the pathogenesis of chronic mitral valvular disease in dogs. Am J Vet Res. 2011;72:194–202. doi: 10.2460/ajvr.72.2.194. [DOI] [PubMed] [Google Scholar]
- 12.Shahrara S, Tidholm A, Drvota V, Häggstöm J, Sylvén C. Upregulation of thyroid hormone receptor beta 1 and beta 2 messenger RNA in the myocardium of dogs with dilated cardiomyopathy or chronic valvular disease. Am J Vet Res. 1999;60:848–852. [PubMed] [Google Scholar]
- 13.Wang TJ, Larson MG, Levy D, Benjamin EJ, Leip EP, Omland T, Wolf PA, Vasan RS. Plasma natriuretic peptide levels and the risk of cardiovascular events and death. N Engl J Med. 2004;350:655–663. doi: 10.1056/NEJMoa031994. [DOI] [PubMed] [Google Scholar]
- 14.Moon HS, Choi E, Hyun C. The cardiac sodium-calcium exchanger gene (NCX-1) is a potential canine cardiac biomarker of chronic mitral valvular insufficiency. J Vet Intern Med. 2008;22:1360–1365. doi: 10.1111/j.1939-1676.2008.0209.x. [DOI] [PubMed] [Google Scholar]
- 15.Nam SJ, Han SH, Kim HW, Hyun C. The cardiac biomarker sodium-calcium exchanger (NCX-1) can differentiate between heart failure and renal failure: a comparative study of NCX-1 expression in dogs with chronic mitral valvular insufficiency and azotemia. J Vet Intern Med. 2010;24:1383–1387. doi: 10.1111/j.1939-1676.2010.0628.x. [DOI] [PubMed] [Google Scholar]
- 16.Fonfara S, Tew S, Clegg P, Dukes-McEwan J. Cytokine and matrix metalloproteinase expression in blood samples of dogs with congestive heart failure [abstract] J Vet Intern Med. 2010;24:671. doi: 10.1111/j.1939-1676.2009.0458.x. [DOI] [Google Scholar]
- 17.Xie X, Liu Y, Gao S, Wu B, Hu X, Chen J. Possible involvement of fibrocytes in atrial fibrosis in patients with chronic atrial fibrillation. Circ J. 2014;78:338–344. doi: 10.1253/circj.CJ-13-0776. [DOI] [PubMed] [Google Scholar]
- 18.Goette A, Jentsch-Ullrich K, Lendeckel U, Röcken C, Agbaria M, Auricchio A, Mohren M, Franke A, Klein HU. Effect of atrial fibrillation on hematopoietic progenitor cells: a novel pathophysiological role of the atrial natriuretic peptide? Circulation. 2003;108:2446–2449. doi: 10.1161/01.CIR.0000102968.19341.FC. [DOI] [PubMed] [Google Scholar]
- 19.Moore L, Fan D, Basu R, Kandalam V, Kassiri Z. Tissue inhibitor of metalloproteinases (TIMPs) in heart failure. Heart Fail Rev. 2012;17:693–706. doi: 10.1007/s10741-011-9266-y. [DOI] [PubMed] [Google Scholar]
- 20.Spinale FG. Myocardial matrix remodeling and the matrix metalloproteinases: influence on cardiac form and function. Physiol Rev. 2007;87:1285–1342. doi: 10.1152/physrev.00012.2007. [DOI] [PubMed] [Google Scholar]
- 21.Peterson JT, Li H, Dillon L, Bryant JW. Evolution of matrix metalloprotease and tissue inhibitor expression during heart failure progression in the infarcted rat. Cardiovasc Res. 2000;46:307–315. doi: 10.1016/S0008-6363(00)00029-8. [DOI] [PubMed] [Google Scholar]
- 22.Asaf R, Blum S, Roguin A, Kalet-Litman S, Kheir J, Frisch A, Miller-Lotan R, Levy AP. Haptoglobin genotype is a determinant of survival and cardiac remodeling after myocardial infarction in diabetic mice. Cardiovasc Diabetol. 2009;8:29. doi: 10.1186/1475-2840-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 25.Szmit S, Jank M, Maciejewski H, Grabowski M, Glowczynska R, Majewska A, Filipiak KJ, Motyl T, Opolski G. Gene expression profiling in peripheral blood nuclear cells in patients with refractory ischaemic end-stage heart failure. J Appl Genet. 2010;51:353–368. doi: 10.1007/BF03208866. [DOI] [PubMed] [Google Scholar]
- 26.Kuner R, Barth AS, Ruschhaupt M, Buness A, Zwermann L, Kreuzer E, Steinbeck G, Poustka A, Sültmann H, Nabauer M. Genomic analysis reveals poor separation of human cardiomyopathies of ischemic and nonischemic etiologies. Physiol Genomics. 2008;34:88–94. doi: 10.1152/physiolgenomics.00299.2007. [DOI] [PubMed] [Google Scholar]
- 27.Szmit S, Jank M, Maciejewski H, Balsam P, Majewska A, Loj M, Grabowski M, Filipiak KJ, Motyl T, Opolski G. White blood cell transcriptome correlates with renal function in acute heart failure. Int Heart J. 2012;53:117–124. doi: 10.1536/ihj.53.117. [DOI] [PubMed] [Google Scholar]
- 28.Munakata M, Ono Y, Koyama M, Fukui K, Satoh K, Suzuki S. Nafamostat mesilate modulates the release of platelet-activating factor during left ventricular assistance with hemofiltration in canine heart failure. Jpn J Thorac Cardiovasc Surg. 2000;48:106–111. doi: 10.1007/BF03218100. [DOI] [PubMed] [Google Scholar]
- 29.Negro A, Brar BK, Lee KF. Essential roles of Her2/erbB2 in cardiac development and function. Recent Prog Horm Res. 2004;59:1–12. doi: 10.1210/rp.59.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Sawyer DB, Zuppinger C, Miller TA, Eppenberger HM, Suter TM. Modulation of anthracycline-induced myofibrillar disarray in rat ventricular myocytes by neuregulin-1beta and anti-erbB2: potential mechanism for Trastuzumab-induced cardiotoxicity. Circulation. 2002;105:1551–1554. doi: 10.1161/01.CIR.0000013839.41224.1C. [DOI] [PubMed] [Google Scholar]
- 31.Chien KR. Herceptin and the heart—a molecular modifier of cardiac failure. N Engl J Med. 2006;354:789–790. doi: 10.1056/NEJMp058315. [DOI] [PubMed] [Google Scholar]
- 32.Speyer J. Cardiac dysfunction in the trastuzumab clinical experience. J Clin Oncol. 2002;20:1156–1157. doi: 10.1200/JCO.2002.20.5.1156. [DOI] [PubMed] [Google Scholar]
- 33.Crone SA, Zhao YY, Fan L, Gu Y, Minamisawa S, Liu Y, Peterson KL, Chen J, Kahn R, Condorelli G, Ross J, Jr, Chien KR, Lee KF. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med. 2002;8:459–465. doi: 10.1038/nm0502-459. [DOI] [PubMed] [Google Scholar]
- 34.Ozcelik C, Erdmann B, Pilz B, Wettschureck N, Britsch S, Hübner N, Chien KR, Birchmeier C, Garratt AN. Conditional mutation of the ErbB2 (HER2) receptor in cardiomyocytes leads to dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2002;99:8880–8885. doi: 10.1073/pnas.122249299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu FF, Stone JR, Schuldt AJ, Okoshi K, Okoshi MP, Nakayama M, Ho KK, Manning WJ, Marchionni MA, Lorell BH, Morgan JP, Yan X. Heterozygous knockout of neuregulin-1 gene in mice exacerbates doxorubicin-induced heart failure. Am J Physiol Heart Circ Physiol. 2005;289:H660–H666. doi: 10.1152/ajpheart.00268.2005. [DOI] [PubMed] [Google Scholar]
- 36.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio–the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 37.Fonfara S, Hetzel U, Tew SR, Dukes-McEwan J, Cripps P, Clegg PD. Leptin expression in dogs with cardiac disease and congestive heart failure. J Vet Intern Med. 2011;25:1017–1024. doi: 10.1111/j.1939-1676.2011.00782.x. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Representative results of flow cytometric analysis showing negative control of immunofluorescence staining of peripheral blood mononuclear cells (PBMC) from dog number 1 with ISACHC 2 class heart failure. PBMC were fixed, permeabilized and stained with secondary antibodies: chicken anti-mouse IgG conjugated with Alexa Fluor 488 (Molecular Probes, Life Technologies). The percentage of α-SMA positive cells is 0.0%; a) morphological flow cytometric parameters of analyzed cells gated on the basis of their size (forward scatter: FSC) and granularity (side scatter: SSC); b) cytogram of Alexa Fluor 488 positive cells (gate P2); c) histogram of Alexa Fluor 488 positive cells (gate P4); d) table showing population hierarchy of cells in cytograms a, b, and histogram c. (PDF 72 KB)
Additional file 2: Representative results of flow cytometric analysis of peripheral blood mononuclear cells (PBMC) from dog number 1 with ISACHC 2 class heart failure, which were stained with antibodies against alpha smooth muscle actin (α-SMA). PBMC were fixed, permeabilized and stained with monoclonal mouse anti- α-SMA antibodies (clone 1A4, Dako, Denmark), followed by incubation with secondary antibodies: chicken anti-mouse IgG conjugated with Alexa Fluor 488 (Molecular Probes, Life Technologies). The percentage of α-SMA positive cells is around 0.3%; a) morphological flow cytometric parameters of analyzed cells gated on the basis of their size (forward scatter: FSC) and granularity (side scatter: SSC); b) cytogram showing α-SMA –positive cells gated in gate P2; c) histogram showing α-SMA –positive cells gated in gate P4; d) table showing population hierarchy of cells in cytograms a, b, and histogram c. (PDF 85 KB)


