Abstract
Background. Live attenuated influenza vaccine (LAIV) and trivalent inactivated influenza vaccine (TIV) are effective for prevention of influenza virus infection in children, but the mechanisms associated with protection are not well defined.
Methods. We analyzed the differences in B-cell responses and transcriptional profiles in children aged 6 months to 14 years immunized with these 2 vaccines.
Results. LAIV elicited a significant increase in naive, memory, and transitional B cells on day 30 after vaccination, whereas TIV elicited an increased number of plasmablasts on day 7. Antibody titers against the 3 vaccine strains (H1N1, H3N2, and B) were significantly higher in the TIV group and correlated with number of antibody-secreting cells. Both vaccines induced overexpression of interferon (IFN)–signaling genes but with different kinetics. TIV induced expression of IFN genes on day 1 after vaccination in all age groups, and LAIV induced expression of IFN genes on day 7 after vaccination but only in children <5 years old. IFN-related genes overexpressed in both vaccinated groups correlated with H3N2 antibody titers.
Conclusions. These results suggest that LAIV and TIV induced significantly different B-cell responses in vaccinated children. Early induction of IFN appears to be important for development of antibody responses.
Keywords: Influenza vaccine, LAIV, TIV, children, interferon, HI antibodies, neutralizing antibodies
Influenza viruses cause annual epidemics and intermittent pandemics worldwide. Studies have demonstrated that in addition to children with underlying conditions, influenza also causes significant morbidity and mortality in healthy children [1–4]. In the United States, influenza vaccination is recommended for all individuals aged ≥6 months [5].
There are 2 major types of influenza vaccines approved for children in United States: trivalent inactivated influenza vaccine (TIV) and live attenuated influenza vaccine (LAIV). Both vaccines are efficacious in reducing influenza virus infection in children [6–9]. Some studies, however, suggest that LAIV has a higher efficacy than TIV in young children [10–12]. The immune mechanisms by which these vaccines induce immune protection are not well understood. We conducted a prospective study in children immunized with TIV or LAIV to characterize the differences in (1) B-cell populations by flow cytometry, (2) serum antibody titers by hemagglutination inhibition (HAI) assay and virus neutralization assay (VNA), and (3) whole-blood transcriptional profiles to determine whether early changes in expression of certain immune related genes correlated with antibody responses.
MATERIALS AND METHODS
Experimental Design
This was a prospective cohort of previously healthy children aged 6 months to 14 years enrolled from October 2011 to February 2012. Forty children were initially randomly assigned to receive 1 dose of LAIV (FluMist, MedImmune) or TIV (Fluzone, Sanofi Pasteur). Three children in the TIV cohort withdrew from the study, so the analyzed study cohort included 37 children (LAIV, n = 20; TIV, n = 17). Two exceptions were applied to the randomization: 4 children <2 years old and 4 children with controlled asthma received TIV, in accordance with Centers for Disease Control and Prevention recommendations. One child aged 6 months at enrollment received 2 vaccine doses with a 1-month interval, since it was his first influenza vaccination. In this child, all immunologic studies were performed only after the initial vaccine dose. All other children had received influenza vaccine during the previous season (ie, 2010–2011), and precise data on the previous season's vaccination were collected. Demographic data are presented in Table 1. Exclusion criteria included chronic conditions, except asthma, immunodeficiencies, and use of systemic steroids in the previous 2 weeks. Children with history of fever or respiratory symptoms within 4 weeks before enrollment were also excluded.
Table 1.
Demographic Data of Enrolled Subjects Who Received Live Attenuated Influenza Vaccine (LAIV) or Trivalent Inactivated Influenza Vaccine (TIV)
| Characteristic | LAIV Group (n = 20) | TIV Group (n = 17) | P |
|---|---|---|---|
| Age, y | |||
| Mean ± SD | 9.89 ± 4.6 | 8.47 ± 4.98 | .24a |
| Median (IQR) | 11.44 (4.2–14.0) | 10.94 (3.7–12.9) | |
| Group | .29b | ||
| 5–14 | 15 (75) | 10 (58.9) | |
| <5 | 5 (25) | 7 (41.2) | |
| Male sex | 10 (50) | 8 (47.1) | .85b |
| Race | .70b | ||
| White | 16 (80) | 13 (76.5) | |
| Asian | 1 (5) | 2 (11.8) | |
| African American | 1 (5) | 0 (0) | |
| White/African American | 2 (1) | 2 (11.8) | |
| Non-Hispanic ethnicity | 19 (95) | 17 (100) | .35b |
| Received influenza vaccine in 2010 | 19 (95) | 16 (94) | .9b |
Abbreviation: IQR, interquartile range.
a Mann–Whitney U test.
b χ2 test.
Blood samples were collected on day 0 before vaccination and 1 day, 7 days (range, 6–8 days), and 30 days (range, 27–33 days) after vaccination to measure B cells, antibody responses, and gene expression profiles. This study used a 2011–2012 vaccine formulation containing the following influenza virus strains: A/California/7/2009(H1N1)-like, A/Perth/16/2009(H3N2)-like, and B/Brisbane/60/2008-like. Data on adverse events were collected by the investigators at each study visit. The study was approved by the institutional review board at Nationwide Children's Hospital (IRB09-00262) and Baylor Institute for Immunology Research (IRB011-221). Written informed consent was obtained from the parents of all participants.
Flow Cytometry
Peripheral-whole-blood samples were analyzed with the following antibody panel to identify the B-cell populations: CD45 (Pacific Orange, Invitrogen), CD14 (Alexa Fluor 700), CD20 (Pe-Cy5), immunoglobulin D (FITC), CD24 (PE, BD Pharmingen), CD19 (ECD, Beckman-Coulter), CD27 (APC-Cy7, BioLegend), CD38 (Pe-Cy7), and CD138 (APC, BD BioScience). The staining protocol is described elsewhere [13]. Events were collected on a LSRII instrument (BD Biosciences, San Jose, CA), and analysis was performed using FlowJo software (Treestar, version 9.4.11). Total cell numbers were calculated using the routine white blood cell counts collected at each time point.
HAI Assay and VNA
Serum samples were collected at 2 time points, day 0 and day 30 after vaccination. The HAI assay was performed as previously described [14]. The hemagglutination inhibition (HI) titer was defined as the reciprocal of the highest dilution of serum that inhibits red blood cell hemagglutination. VNA was performed as previously described [15]. The viral neutralization (VN) titer was defined as the reciprocal of the highest dilution of serum that neutralizes 200 plaque-forming units of influenza virus. Seroconversion was based on the following criteria: a 4-fold increase in antibody titers between the prevaccination and the convalescent-phase serum samples or an increase of antibody titers from <10 to ≥40 for the prevaccination and convalescent-phase serum samples [16–18].
Microarray Data and Statistical Analysis
Blood samples were collected in Tempus tubes (Applied Biosystems, CA) and stored at −20°C. RNA was hybridized into Illumina Human WG-6 V4 beadchips (47 323 probes) and scanned on the Illumina Beadstation 500 [19, 20]. Data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE52005).
Analyses were performed using GeneSpring GX 7.3 software (Agilent Technologies) [21, 22]. Transcripts were first selected if present in >10% of all samples and were then filtered to select the most variable probes. We used several analytical tools. First, we used GeneSpring for supervised analysis with all samples (140 samples), using statistical filtering and class comparisons to identify transcripts differentially expressed between study groups. The nonparametric Mann–Whitney test for comparisons across groups was used, with a P < .05.
Second, we performed functional analyses of differentially expressed genes using modular analysis [23]. Gene expression levels were compared between each time point after vaccination and baseline (before vaccination) on a module-by-module basis. Modular transcript content and annotations are available at: http://www.biir.net/public_wikis/module_annotation/V2_Trial_8_Modules.
Third, to confirm the findings from the modular analyses, gene expression values were log2 transformed and analyzed using a linear mixed model for the LAIV and TIV cohorts separately. Specifically, time was included in the model as a categorical variable, with a spatial power covariance matrix to account for correlation because subjects had repeated measurements. Each time point was then compared to baseline. JMP Genomics 6.0 software (SAS Institute, Cary, NC) was used for this analysis. Genes with a P value of <.01 were considered differentially expressed [24].
Statistical analysis of demographic data was performed using Graph Pad Prism. Mann–Whitney tests were used for continuous variables, and χ2 and Fisher exact tests were used for categorical variables. Antibody titer correlations were performed using the Spearman nonparametric test on Sigma Plot. Flow cytometry data were analyzed using the linear mixed model approach described for microarray data. The data were log2 transformed, and a Bonferroni correction was used to adjust for multiple testing.
RESULTS
Immunization With LAIV and TIV Induce Distinct Changes in B Cell Populations
We immunized 20 children who received LAIV and 17 children who received TIV (Table 1). Sequential flow cytometry samples from all 4 time points were available for 16 in the LAIV group and 13 in the TIV group. Fourteen of 37 children (7 from each group) had adverse events after vaccination, all of which were upper respiratory tract symptoms.
In the LAIV group, we found a significant increase at 30 days after vaccination in the absolute numbers of naive, memory, and transitional B cells, compared with baseline (P < .05), but there were no significant differences in the numbers of plasmablasts and plasma cells. Children immunized with TIV showed a significant increase in absolute numbers of plasmablasts at day 7 after vaccination (P < .01) but no significant changes in the numbers of naive, memory, and transitional B cells (Figure 1A–J).
Figure 1.
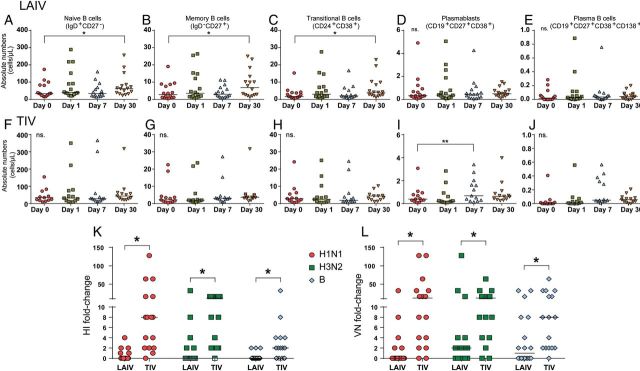
A–I, B-cell immune responses after immunization with live attenuated influenza vaccine (LAIV) or trivalent inactivated influenza vaccine (TIV). Whole-blood samples obtained from healthy children were stained for naive B cells (CD19+IgD+CD27−), memory B cells (CD19+IgD−CD27+), transitional B cells (CD19+CD24+CD38+), plasmablasts (CD19+CD27+CD38+), and plasma B cells (CD19+CD27+CD38+CD138) and analyzed by flow cytometry on day 0 (ie, baseline) and days 1, 7, and 30 after vaccination. Data are 16 LAIV vaccinees and 13 TIV vaccinees. The graphs consist of column scatter plots representing absolute numbers of B-cell populations over the different 4 time points. A, Naive B cells in LAIV vaccinees. B, Memory B cells in LAIV vaccinees. C, Transitional B cells in LAIV vaccinees. D, Plasmablasts in LAIV vaccinees. E, Plasma B cells in LAIV vaccinees. F, Naive B cells in TIV vaccinees. G, Memory B cells in TIV vaccinees. H, Transitional B cells in TIV vaccinees. I, Plasmablasts in TIV vaccinees. J, Plasma B cells in TIV vaccinees. Linear mixed model with Bonferroni correction (*P < .05 and **P < .01; NS, no statistical significance). Bars represent median values. K and L, Fold-increase of hemagglutination inhibition (HI) and viral neutralization (VN) titers before and after vaccination in LAIV and TIV recipients. Serum samples obtained on day 0 and 30 days after vaccination from each subject were assayed for HI and VN titers. Data are for 18 LAIV vaccinees and 16 TIV vaccinees. K, Column scatter plot representing fold-increase of HI titers among TIV and LAIV vaccinees for H1N1, H3N2, and B vaccine strains. L, Column scatter plot representing fold-increase of VN titers among TIV and LAIV vaccinees for H1N1, H3N2, and B vaccine strains. *P < .05, by the Mann–Whitney test. Bars represent median values.
TIV Induces a More Robust Serum Antibody Response Than LAIV
Antibody responses were measured in 34 of 37 subjects enrolled (LAIV group, n = 18; TIV group, n = 16). HI and VN titers for each of the 3 influenza virus strains contained in both vaccines were measured in serum samples obtained at days 0 and 30.
TIV induced a significantly greater increase in HI titers than LAIV for the 3 vaccine strains (P < .01; Figure 1K). TIV elicited a greater rate of seroconversion (70% for H1N1, 70% for H3N2, and 30% for B) than LAIV (5% for H1N1, 2% for H3N2, and 0% for B). Children <5 years old had the highest rate of seroconversion among TIV vaccinees. When we measured the maximum fold-change in HI titers against any of the 3 vaccine strains, the mean HI response to TIV was 8-fold higher than the response to LAIV (P < .001).
With respect to VN titers, TIV vaccinees showed a significantly greater increase in serum VN titers for the 3 vaccine strains, compared with subjects in the LAIV group (P < .05). In LAIV vaccines, there was a significant increase in titers against the B strain (P < .05) and an increasing trend for the H3N2 strain (P = .05), which was not observed on HAI analysis (Figure 1L). The rates of seroconversion for VNA were 75% for H1N1, 81% for H3N2, and 62% for B among TIV vaccinees and 16% for H1N1, 27% for H3N2, and 33% for B among LAIV vaccines (Table 2).
Table 2.
Hemagglutinin Inhibition and Viral Neutralization Antibody Titers at Baseline and 30 Days After Receipt of Live Attenuated Influenza Vaccine (LAIV) or Trivalent Inactivated Influenza Vaccine (TIV)
| Assay | Titer |
Fold-Increase |
||||||
|---|---|---|---|---|---|---|---|---|
| LAIV Group |
TIV Group |
LAIV Group |
TIV Group |
|||||
| Baseline | Day 30 | Baseline | Day 30 | Mean | Median | Mean | Median | |
| HAI assay | ||||||||
| H1N1 | 34.2 | 44.8 | 29.5 | 293.4a | 0.5 | 0.0 | 20.9 | 8.0 |
| H3N2 | 32.9 | 54.4 | 21.8 | 190.2a | 2.6 | 0.0 | 9.7 | 12.0 |
| B | 10.0 | 12.1 | 11.3 | 30.8a | 0.3 | 0.0 | 3.8 | 2.0 |
| VNA | ||||||||
| H1N1 | 96.9 | 100.7 | 64.4 | 640.0a | 2.6 | 0.0 | 28.7 | 12.0 |
| H3N2 | 68.5 | 172.8 | 91.1 | 794.7a | 10.7 | 2.0 | 15.1 | 12.0 |
| B | 63.4 | 166.2a | 83.5 | 586.8a | 5.8 | 1.0 | 14.3 | 8.0 |
Titers are geometric means.
Abbreviations: HAI, hemagglutinin inhibition; VNA, virus neutralization assay.
a P < .05.
We examined the impact of age on antibody responses and found that children younger than 5 years had similar antibody responses than older children. Likewise, children with asthma (n = 4) showed similar antibody responses than the rest of the children immunized with TIV.
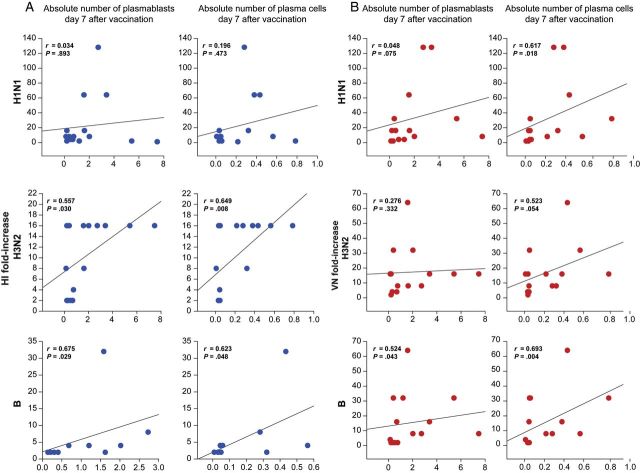
Antibody-Secreting Cells Correlate With Antibody Titers on TIV Recipients
Next, we examined the correlations between the number of plasmablasts and plasma cells with antibody titers. For correlations with HI titers, we only included TIV vaccinees, because of the low responses after LAIV. We found a significant correlation between the number of plasmablasts and plasma cells on day 7 after immunization and fold-increases of HI titers to H3N2 and B strains. Despite good rates of seroconversion for the H1N1 strain among TIV vaccinees, we did not find any significant correlation between the number of plasmablasts and plasma cells and H1N1 titers (Figure 2A). We also found significant correlations between the numbers of plasma cells on day 7 after TIV receipt and VN titers against the 3 vaccine strains, as well as the number of plasmablasts 7 days after vaccination and VN titers only against the B strain (Figure 2B). There was a significant correlation in the fold-increase of HI and VN titers among TIV vaccinees for the 3 vaccine strains (H1N1, r = 0.594 [P < .05]; H3N2, r = 0.815 [P < .001]; and B, r = 0.694 [P < .05]). Although there were no significant differences in the numbers of antibody-secreting cells over time in LAIV vaccinees, we observed a significant correlation between absolute numbers of plasmablasts on day 7 after LAIV receipt and H3N2 VN titers 30 days after vaccination (r = 0.668; P < .05).
Figure 2.
Correlations between hemagglutination inhibition (HI) and viral neutralization (VN) titers and plasmablast and plasma B cells among trivalent inactivated influenza vaccine (TIV) vaccinees. A, Each plot represents the correlation between absolute numbers of plasmablasts and plasma B cells 7 days after vaccination with TIV and fold-increases of H1N1, H3N2, and B HI titers from day 0 (ie, baseline) to day 30 after vaccination. B, Each plot represents the correlation between absolute numbers of plasmablasts and plasma B cells 7 days after vaccination with TIV and fold-increases of H1N1, H3N2, and B VN titers from day 0 to day 30 after vaccination.
TIV and LAIV Induce Distinct Immune Transcriptional Profiles
We analyzed whole blood gene expression profiles in 32 children (18 in the LAIV group, and 14 in the TIV group) at baseline (day 0) and at days 1, 7, and 30 after vaccination (Supplementary Figure 1A and 1B). Samples obtained at baseline were used as the reference for subsequent comparisons.
Statistical group comparisons identified a significantly larger number of differentially expressed transcripts among TIV vaccinees on days 1 and 7 after vaccination, compared with children who received LAIV (day 1, 777 genes in the TIV group vs 402 in the LAIV group; day 7, 812 genes in TIV group vs 453 in the LAIV group). On day 30, we observed the opposite pattern, as LAIV vaccinees had 790 differentially expressed transcripts, whereas TIV recipients had 462. Overall, for LAIV vaccinees, there was a predominance of underexpressed genes at all 3 time points, compared with day 0, whereas for TIV vaccines, we observed a higher proportion of overexpressed genes on days 1 and 7 but a predominant pattern of underexpressed genes on day 30 (Supplementary Figure 1C)
Next, we compared the significant top 10 overexpressed and underexpressed transcripts per time point per vaccine type. Among the top overexpressed transcripts identified on day 1 after TIV receipt, the majority were interferon (IFN)–related genes, whereas in the LAIV group there was overexpression of genes related to cell cycle activity. On day 7 after vaccination, we found overexpression of plasma cell–related genes and underexpression of inflammation genes in the TIV group, whereas LAIV vaccinees displayed overexpression of IFN and cell cycle–related transcripts. On day 30 after vaccination, cell cycle–related transcripts were overexpressed and inflammation transcripts were underexpressed in LAIV vaccinees, whereas in the TIV group we did not identify significant underexpression or overexpression of transcripts with well-defined function (Supplementary Table 1).
TIV and LAIV Induce Expression of IFN-Related Genes With Different Kinetics
To better understand the differences of immune-related genes induced by both vaccines, we performed modular analysis [23, 25]. Gene expression levels were compared between samples obtained at the different time points and at baseline (day 0) on a module-by-module basis. For each module, the percentage of genes significantly expressed (P < .05, by the Mann–Whitney test) are shown in the modular map, derived independently for LAIV and TIV vaccinees (Figure 3A–I) [23]. In the TIV group but not the LAIV group, the IFN-related modules (M1.2, M3.4, and M5.12) were overexpressed on day 1 after vaccination and included genes coding for IFN-inducible proteins, such as OASL, OAS3, IFIT1, IFIT3, IFI44L, RSAD2, ISG15, the guanylate-binding proteins GBP1 GBP4, and LOC400759 (Figure 3B). To assess whether age influenced the host response to LAIV and TIV, subjects were stratified in 2 groups: those <5 years old and those ≥5 years old (LAIV group, n = 4 and 14, respectively; TIV group, n = 4 and 10, respectively). TIV induced overexpression of the IFN-related modules in both age groups 1 day after vaccination, although the IFN overexpression was more pronounced in younger children (median expression, 2.11 [interquartile range {IQR}, 1.52–2.91] in younger children vs 1.35 [IQR, 1.14–1.59] in older children; P < .001; Figure 3C–D). On the other hand, only in TIV vaccinees >5 years of age was there overexpression of B-cell–related genes (M4.10; including genes related to B-cell differentiation, such as BLK, CD79A, and CD79B) 7 days after vaccination, which coincided with the peak antibody-secreting cell count measured by flow cytometry.
Figure 3.
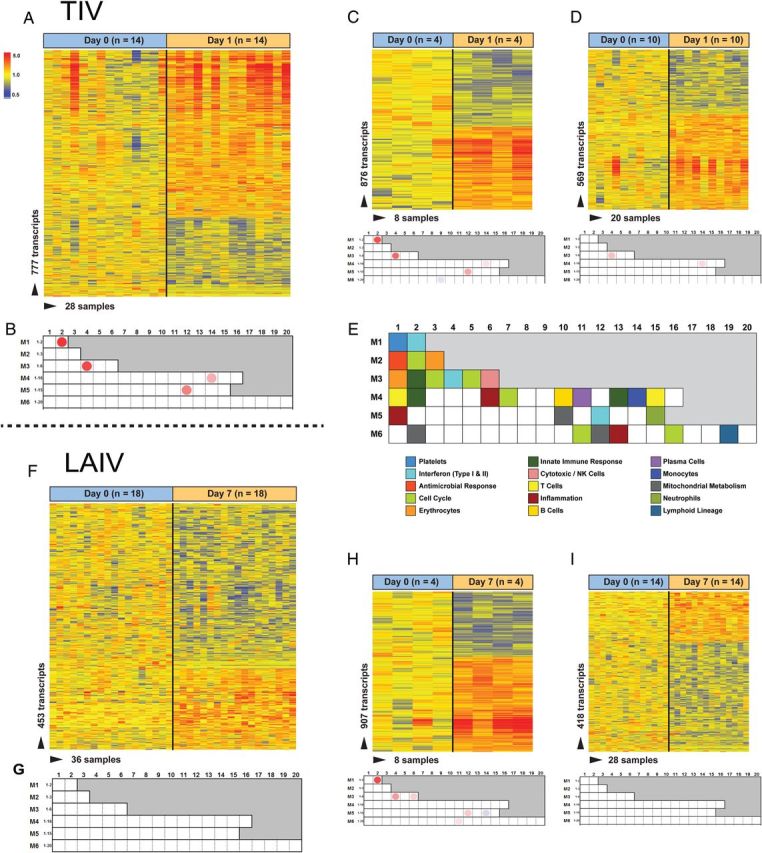
Transcriptional profiles of children immunized with trivalent inactivated influenza vaccine (TIV) and live attenuated influenza vaccine (LAIV) are characterized by activation of interferon (IFN). A, Statistical group comparison (P < .05, by the Mann Whitney test) between the TIV group (n = 14) on day 1 after vaccination versus baseline (ie, day 0) yielded 777 differentially expressed genes. Transcripts were organized by hierarchical clustering, where each row represents a single transcript and each column represents an individual subject. Normalized expression levels are indicated by red (overexpressed) or blue (underexpressed), compared with the median expression at baseline (yellow). B, Modular analysis 1 day after TIV receipt showed significant overexpression of IFN-related modules (M1.2, M3.4, and M5.12). The color intensity of the spots represents the percentage of significantly overexpressed genes (red), the percentage of significantly underexpressed genes (blue), or no differences (blank), compared with healthy controls (P < .05, by the Mann–Whitney test). C and D, Heat maps and modular maps from TIV vaccinees stratified by age. Both groups displayed overexpression of the IFN-related modules, with overexpression more pronounced in the younger group (n = 4). E, Modular map legend. F, Statistical group comparison (P < .05, by the Mann–Whitney test) between the LAIV group (n = 18) on day 7 after vaccination versus day 0 yielded 453 differentially expressed genes. G, Modular analysis 7 days after LAIV receipt. No modules were found to be significantly overexpressed or underexpressed. H and I, Heat maps and modular maps from LAIV vaccinees stratified by age. Only younger children (n = 4) displayed significant overexpression of the IFN-related modules (M1.2, M3.4, and M5.12). Abbreviation: NK, natural killer.
Transcriptional profiles in LAIV vaccinees were more attenuated. We observed overexpression of the IFN-related modules on day 7 after vaccination and only for younger children (median expression, 2.25 [IQR, 1.45–4.41] in younger children vs 1.00 [IQR, 0.96–1.08] in older children; P < .001), which also included genes coding for IFN-inducible antiviral proteins, such as OASL, OAS1, OAS2, IFIT1, IFIT3, IFITM3, RSAD2, RTP4, GBP1, GBP5, and tumor necrosis factor–induced protein TNFAIP6 (Figure 3F–I).
Expression of IFN-related modules on day 30 after vaccination was not significantly different than in baseline samples in children vaccinated with both LAIV and TIV. Kinetics of IFN-related modules expression stratified by age and vaccine type is summarized on Supplementary Figure 2.
To validate the IFN profile induced by both vaccines over time, we used linear mixed-model analyses. As observed with the modular analysis, IFN-related genes were overexpressed on day 1 after vaccination with TIV and on day 7 after vaccination with LAIV (Figure 4). Linear mixed models identified 25 and 6 IFN-related genes after TIV and LAIV receipt, respectively, that passed the statistical cutoff P value of <.01, confirming that TIV induces a more robust overexpression of IFN genes in blood. These significantly expressed genes were also identified in the modular analyses described above.
Figure 4.
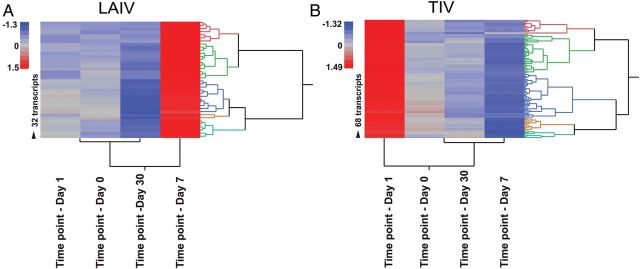
Interferon (IFN)–related gene expression after vaccination with trivalent inactivated influenza vaccine (TIV) and live attenuated influenza vaccine (LAIV). Significantly expressed genes derived from linear mixed-model analyses (P < .01) are displayed in a heat-map format. A, IFN-related genes were significantly overexpressed and different on day 7 after LAIV receipt, compared with other time points (day 0 [ie, baseline] and days 1 and 30 after vaccination). B, IFN genes were overexpressed and significantly different on day 1 after TIV vaccination, compared with all other time points.
Expression of IFN Genes Correlates With Antibody Production
Next, we examined the correlations between IFN-inducible gene expression and antibody production. For this analysis, we correlated the fold-increase of HI and VN antibody titers with IFN-inducible gene expression per individual modules (M1.2, 36 transcripts; M3.4, 62 transcripts; and M5.12, 63 transcripts), combinations of IFN modules (161 transcripts), and specific top 10 overexpressed IFN genes on day 1 after TIV vaccination and on day 7 after LAIV vaccination (Table 3). For LAIV vaccinees, we were able to perform correlations only with VN titers, since HI titers were too low. Among TIV vaccinees, H3N2 HI titers consistently correlated with all the IFN-related modules and 6 of 10 genes tested. Only expression of IFN-induced GBP1 significantly correlated with H3N2 and B VN titers. Among LAIV vaccinees, we also observed significant correlations only between H3N2 VN titers and all IFN-related modules, as well as 9 of the top 10 selected genes.
Table 3.
Correlations Between Fold-Increase of Hemagglutinin Inhibition and Virus Neutralization Titers and Interferon (IFN)–Related Modules and Genes, by Vaccine and Time After Vaccine Receipt
| Variable | Median Expression, Fold-Increase |
|||||
|---|---|---|---|---|---|---|
| H1N1 |
H3N2 |
B |
||||
| r | P | r | P | r | P | |
| HI titer 1 d after TIV receipt | ||||||
| IFN (161 genes) | −0.22 | .444 | 0.531 | .049 | −0.112 | .742 |
| M1.2 | −0.134 | .637 | 0.633 | .013 | −0.0745 | .844 |
| M3.4 | −0.244 | .39 | 0.531 | .049 | −0.0373 | .913 |
| M5.12 | −0.179 | .532 | 0.592 | .024 | 0.0745 | .844 |
| OASL | −0.141 | .615 | 0.695 | <.01 | −0.186 | .612 |
| OAS3 | −0.271 | .34 | 0.493 | .069 | −0.0373 | .913 |
| IFIT1 | −0.0941 | .738 | 0.571 | .032 | −0.112 | .742 |
| IFIT3 | −0.114 | .693 | 0.585 | .026 | 0 | .983 |
| IFI44L | −0.184 | .521 | 0.578 | .029 | −0.0373 | .913 |
| RSAD2 | −0.224 | .435 | 0.507 | .061 | −0.0373 | .913 |
| ISG15 | 0.0291 | .916 | 0.659 | .010 | −0.149 | .676 |
| GBP1 | 0.125 | .659 | 0.566 | .033 | 0.224 | .55 |
| GBP4 | −0.152 | .594 | 0.469 | .087 | 0.0373 | .913 |
| LOC400759 | −0.213 | .453 | 0.393 | .157 | 0.0373 | .913 |
| VN titer 1 d after TIV receipt | ||||||
| IFN (161 genes) | 0.075 | .792 | 0.372 | .199 | 0.346 | .219 |
| M1.2 | 0.0417 | .878 | 0.468 | .102 | 0.487 | .074 |
| M3.4 | 0.0861 | .764 | 0.372 | .199 | 0.373 | .184 |
| M5.12 | 0.136 | .643 | 0.476 | .093 | 0.459 | .094 |
| OASL | 0.158 | .591 | 0.378 | .192 | 0.466 | .087 |
| OAS3 | 0.114 | .696 | 0.358 | .221 | 0.403 | .148 |
| IFIT1 | 0.161 | .591 | 0.4 | .166 | 0.444 | .109 |
| IFIT3 | 0.0611 | .835 | 0.471 | .097 | 0.351 | .212 |
| IFI44L | 0.103 | .723 | 0.42 | .148 | 0.496 | .069 |
| RSAD2 | 0.0667 | .821 | 0.4 | .166 | 0.493 | .069 |
| ISG15 | 0.211 | .481 | 0.48 | .093 | 0.505 | .0636 |
| GBP1 | 0.0139 | .949 | 0.664 | .012 | 0.568 | .032 |
| GBP4 | −0.0889 | .764 | 0.414 | .154 | 0.38 | .173 |
| LOC400759 | −0.383 | .186 | 0.324 | .269 | 0.199 | .482 |
| VN titer 7 d after LAIV receipt | ||||||
| IFN (161 genes) | −0.616 | .233 | 0.934 | <.001 | 0.581 | .087 |
| M1.2 | −0.41 | .45 | 0.889 | <.001 | 0.641 | .058 |
| M3.4 | −0.462 | .45 | 0.921 | <.001 | 0.359 | .308 |
| M5.12 | −0.616 | .233 | 0.83 | <.001 | 0.419 | .243 |
| OASL | −0.462 | .45 | 0.937 | <.001 | 0.641 | .058 |
| OAS1 | −0.359 | .517 | 0.74 | <.01 | 0.197 | .58 |
| OAS2 | 0.205 | .683 | 0.762 | <.01 | 0.65 | .050 |
| IFIT1 | −0.103 | .783 | 0.762 | <.01 | 0.522 | .138 |
| IFIT3 | −0.41 | .45 | 0.921 | <.001 | 0.41 | .243 |
| IFITM3 | −0.616 | .233 | 0.791 | <.01 | 0.624 | .067 |
| RSAD2 | −0.41 | .45 | 0.889 | <.001 | 0.641 | .058 |
| RTP4 | −0.718 | .133 | 0.525 | .107 | 0.539 | .124 |
| GBP1 | −0.872 | .083 | 0.869 | <.001 | 0.641 | .058 |
| GBP5 | −0.205 | .683 | 0.701 | .021 | −0.00855 | .948 |
IFN, 161 genes (M1.2 + M3.4 + M5.12). M1.2: IFN module. M3.4: IFN module. M5.12: IFN module.
Abbreviations: LAIV, live attenuated influenza vaccine; TIV, trivalent inactivated influenza vaccine.
DISCUSSION
We still lack a full understanding of the signaling pathways responsible for the protective immune responses induced by these 2 influenza vaccines. The goal of this study was to compare the B-cell populations, antibody responses, and transcriptional immune profiles elicited after immunization with LAIV and TIV in children. Our results show that both vaccines evoked different responses in all 3 parameters analyzed. While TIV elicited an early increase in antibody-secreting cells and robust antibody titers, LAIV elicited an increase in naive and memory B cells, no increase in antibody-secreting cells, and less robust antibody responses. Both vaccines induced expression of IFN-related genes, but with different kinetics. TIV induced overexpression of IFN genes 1 day after vaccination, whereas the effect of LAIV on IFN-inducible genes was detected 7 days after vaccination.
Previous studies showed that TIV induced greater increase in plasmablasts and antibody responses than LAIV [26, 27]. Likewise, we found higher antibody titers and a significant increase in numbers of plasmablasts in children immunized with TIV, but we also found a significant increase in numbers of naive, memory, and transitional B cells at day 30 after LAIV receipt, which, to our knowledge, is a novel observation. LAIV is a live vaccine, and therefore its efficacy depends on viral replication in the nasal mucosa. Whether and when antibody-secreting cells appear in the blood after mucosal immunization is not known, and we may have not detected these cells because of our sampling times. An alternative hypothesis would be that LAIV-induced responses are predominantly local, in the upper respiratory tract, so only part of these cells would actually recirculate in blood.
In TIV vaccinees, the increase in antibody titers significantly correlated with increased numbers of antibody-secreting cells 7 days after vaccination, with the exception of the H1N1 strain. It is intriguing that, despite the significant increase in H1N1 titers measured with 2 different assays (an HAI assay and a VNA), these titers did not correlate with the number of plasmablasts. Fewer previous exposures to this virus, as this strain corresponds to the 2009 pandemic strain, or unique characteristics of this particular antigen may account for these findings. Although LAIV did not elicit good rates of seroconversion when measured by the HAI assay, we observed substantially better rates when we measured VN titers, supporting the concept that HAI may underestimate the protection provided by LAIV. Some authors suggest that neutralizing assays are more sensitive than HAI, especially for measuring H1N1 antibody responses to LAIV in adults and children [28–30], although the titers induced by LAIV cannot be compared directly with titers induced by TIV as indicative of protection. A good way to assess LAIV antibody production would be to measure HI titers in the nasal wash, but these assays are not yet fully developed or standardized. Some authors have shown that IgA titers in the nasal mucosa correlate with protection after LAIV immunization [31–34], supporting the idea that the relevant response to LAIV takes place locally.
Applying systems immunology to the characterization of vaccine responses is becoming more common, although few studies have addressed influenza vaccine responses in children. Previous studies have showed conflicting results with respect to the ability of these 2 vaccines to induce IFN responses. Nakaya et al showed altered expression of IFN-related genes in adults 3 days after LAIV receipt but not after TIV receipt [27]. Zhu et al showed that children immunized with LAIV had overexpression of IFN-inducible genes 7–10 days after vaccination, but these findings were not observed in TIV recipients [35]. Others showed that TIV in adults induced an IFN signature 24 hours after vaccination [25, 36]. Our study is the first conducted in children that evaluated both vaccines on days 1, 7, and 30 after immunization and correlated IFN expression with B-cell responses and antibody titers. Our results demonstrate that both vaccines induce expression of IFN genes but with different kinetics. TIV induced expression of IFN genes on day 1 after vaccination and LAIV induced expression on day 7, and this was more evident in younger children. It is important to note that children <5 years of age displayed higher expression of IFN-related genes in response to both vaccines. This might be related to their fewer previous exposures to influenza virus infection and/or immunization, compared with older children. Our results also suggest that the correlation between early expression of IFN genes and antibody titers, which was significant for H3N2 and other vaccine strains as previously shown in adults [25, 27], might be different for the 2009 H1N1 strain. This is a potentially important observation that will need confirmation by studies involving a larger number of subjects of different ages.
Our study has some limitations. First, there was a relatively small number of subjects enrolled, mainly in the younger group, because of the challenge of obtaining sequential samples in infants. Nevertheless, the robustness and consistency of the biological changes and immune responses measured before and after vaccination indicate that LAIV and TIV elicit different blood transcriptional, cellular, and serological responses. We were not able to define whether all of the vaccine strains or a predominant strain was driving the IFN responses, but the correlations among fold-changes in antibody titers and IFN-related genes were found mainly with the H3N2 strain, suggesting that this strain is a more robust vaccine antigen or that the vaccinees have been exposed more often to this antigen. Last, we were unable to measure responses in nasal wash samples, owing to the challenge of consistently obtaining a sufficient number of cells for flow cytometry analyses and to the lack of standardized assays to characterize mucosal antibody responses.
In summary, we have uncovered profound differences in B-cell populations elicited by 2 influenza vaccines over time and have established their relatedness to antibody responses to the unique components of these vaccines. In addition, we have found significant kinetic differences in IFN responses between these vaccines and showed the influence of age in these responses.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the study participants, for their enrollment into this study; and S. Farley and P. Davies, for enrolling the study participants.
R. C. performed and analyzed flow cytometry experiments. G. O., E. F., and S. L. developed panels for the flow cytometry assay. R. C. and N. S. performed microarray analysis. R. C., S. L., and S. M. organized and managed the vaccine study and the collection of samples. R. A. and A. G.-S. analyzed antibody titers and reviewed the manuscript. D. B. and G. O. contributed to the statistical analysis. R. C. and O. R. wrote the manuscript. G. O., E. F., A. M., K. P., V. P., and O. R. designed the study and reviewed the manuscript.
Financial support. This work was supported by the National Institutes of Health (grant U19 AI089987), Nationwide Children's Hospital (intramural funds), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant 201277/2011-5 to R. C., from the Brazilian government).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Izurieta HS, Thompson WW, Kramarz P, et al. Influenza and the rates of hospitalization for respiratory disease among infants and young children. N Engl J Med. 2000;342:232–9. doi: 10.1056/NEJM200001273420402. [DOI] [PubMed] [Google Scholar]
- 2.Neuzil KM, Zhu Y, Griffin MR, et al. Burden of interpandemic influenza in children younger than 5 years: a 25-year prospective study. J Infect Dis. 2002;185:147–52. doi: 10.1086/338363. [DOI] [PubMed] [Google Scholar]
- 3.Heikkinen T, Silvennoinen H, Peltola V, et al. Burden of influenza in children in the community. J Infect Dis. 2004;190:1369–73. doi: 10.1086/424527. [DOI] [PubMed] [Google Scholar]
- 4.Poehling KA, Edwards KM, Weinberg GA, et al. The underrecognized burden of influenza in young children. N Engl J Med. 2006;355:31–40. doi: 10.1056/NEJMoa054869. [DOI] [PubMed] [Google Scholar]
- 5.Fiore AE, Uyeki TM, Broder K, et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep. 2010;59:1–62. [PubMed] [Google Scholar]
- 6.Negri E, Colombo C, Giordano L, Groth N, Apolone G, La Vecchia C. Influenza vaccine in healthy children: a meta-analysis. Vaccine. 2005;23:2851–61. doi: 10.1016/j.vaccine.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 7.Rhorer J, Ambrose CS, Dickinson S, et al. Efficacy of live attenuated influenza vaccine in children: A meta-analysis of nine randomized clinical trials. Vaccine. 2009;27:1101–10. doi: 10.1016/j.vaccine.2008.11.093. [DOI] [PubMed] [Google Scholar]
- 8.Zangwill KM, Belshe RB. Safety and efficacy of trivalent inactivated influenza vaccine in young children: a summary for the new era of routine vaccination. Pediatr Infect Dis J. 2004;23:189–97. doi: 10.1097/01.inf.0000116292.46143.d6. [DOI] [PubMed] [Google Scholar]
- 9.Jefferson T, Rivetti A, Harnden A, Di Pietrantonj C, Demicheli V. Vaccines for preventing influenza in healthy children. Cochrane Database Syst Rev. 2008 doi: 10.1002/14651858.CD004879.pub3. CD004879. [DOI] [PubMed] [Google Scholar]
- 10.Ashkenazi S, Vertruyen A, Aristegui J, et al. Superior relative efficacy of live attenuated influenza vaccine compared with inactivated influenza vaccine in young children with recurrent respiratory tract infections. Pediatr Infect Dis J. 2006;25:870–9. doi: 10.1097/01.inf.0000237829.66310.85. [DOI] [PubMed] [Google Scholar]
- 11.Fleming DM, Crovari P, Wahn U, et al. Comparison of the efficacy and safety of live attenuated cold-adapted influenza vaccine, trivalent, with trivalent inactivated influenza virus vaccine in children and adolescents with asthma. Pediatr Infect Dis J. 2006;25:860–9. doi: 10.1097/01.inf.0000237797.14283.cf. [DOI] [PubMed] [Google Scholar]
- 12.Belshe RB, Edwards KM, Vesikari T, et al. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356:685–96. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 13.Wrammert J, Smith K, Miller J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Taaffe J, Parker C, et al. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J Virol. 2006;80:11628–37. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steel J, Lowen AC, Pena L, et al. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J Virol. 2009;83:1742–53. doi: 10.1128/JVI.01920-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady RC, Treanor JJ, Atmar RL, et al. Safety and immunogenicity of a subvirion inactivated influenza A/H5N1 vaccine with or without aluminum hydroxide among healthy elderly adults. Vaccine. 2009;27:5091–5. doi: 10.1016/j.vaccine.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keitel WA, Campbell JD, Treanor JJ, et al. Safety and immunogenicity of an inactivated influenza A/H5N1 vaccine given with or without aluminum hydroxide to healthy adults: results of a phase I-II randomized clinical trial. J Infect Dis. 2008;198:1309–16. doi: 10.1086/592172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Center for Biologics Evaluation and Research, Food and Drug Administration, Department of Health and Human Services. Guidance for industry: clinical data needed to support the licensure of seasonal inactivated influenza vaccines. US Department of Health and Human Services, FDA, Rockville, MD. 2007. pp. 1–16.
- 19.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–7. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banchereau R, Jordan-Villegas A, Ardura M, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7:e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allantaz F, Chaussabel D, Stichweh D, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–44. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Chen X, Wolfinger RD, Franklin JL, Coffey RJ, Zhang B. A unified mixed effects model for gene set analysis of time course microarray experiments. Stat Appl Genet Mol Biol. 2009;8 doi: 10.2202/1544-6115.1484. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obermoser G, Presnell S, Domico K, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–44. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki S, Jaimes MC, Holmes TH, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81:215–28. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakaya HI, Wrammert J, Lee EK, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MS, Mahmood K, Adhikary L, et al. Measuring antibody responses to a live attenuated influenza vaccine in children. Pediatr Infect Dis J. 2004;23:852–6. doi: 10.1097/01.inf.0000137566.87691.3b. [DOI] [PubMed] [Google Scholar]
- 29.Veguilla V, Hancock K, Schiffer J, et al. Sensitivity and specificity of serologic assays for detection of human infection with 2009 pandemic H1N1 virus in US populations. J Clin Microbiol. 2011;49:2210–5. doi: 10.1128/JCM.00229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenburg J, Baz M, Hamelin ME, et al. Evaluation of serological diagnostic methods for the 2009 pandemic influenza A (H1N1) virus. Clin Vaccine Immunol. 2011;18:520–2. doi: 10.1128/CVI.00449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belshe RB, Gruber WC, Mendelman PM, et al. Correlates of immune protection induced by live, attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J Infect Dis. 2000;181:1133–7. doi: 10.1086/315323. [DOI] [PubMed] [Google Scholar]
- 32.Clements ML, Betts RF, Tierney EL, Murphy BR. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol. 1986;24:157–60. doi: 10.1128/jcm.24.1.157-160.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrose CS, Wu X, Jones T, Mallory RM. The role of nasal IgA in children vaccinated with live attenuated influenza vaccine. Vaccine. 2012;30:6794–801. doi: 10.1016/j.vaccine.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Barria MI, Garrido JL, Stein C, et al. Localized mucosal response to intranasal live attenuated influenza vaccine in adults. J Infect Dis. 2013;207:115–24. doi: 10.1093/infdis/jis641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W, Higgs BW, Morehouse C, et al. A whole genome transcriptional analysis of the early immune response induced by live attenuated and inactivated influenza vaccines in young children. Vaccine. 2010;28:2865–76. doi: 10.1016/j.vaccine.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 36.Bucasas KL, Franco LM, Shaw CA, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:921–9. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.