Abstract
Gene vectors derived from DNA transposable elements have become powerful molecular tools in biomedical research and are slowly moving into the clinic as carriers of therapeutic genes. Conventional uses of DNA transposon-based gene vehicles rely on the intracellular production of the transposase protein from transfected nucleic acids. The transposase mediates mobilization of the DNA transposon, which is typically provided in the context of plasmid DNA. In recent work, we established lentiviral protein transduction from Gag precursors as a new strategy for direct delivery of the transposase protein. Inspired by the natural properties of infecting viruses to carry their own enzymes, we loaded lentivirus-derived particles not only with vector genomes carrying the DNA transposon vector but also with hundreds of transposase subunits. Such particles were found to drive efficient transposition of the piggyBac transposable element in a range of different cell types, including primary cells, and offer a new transposase delivery approach that guarantees short-term activity and limits potential cytotoxicity. DNA transposon vectors, originally developed and launched as a non-viral alternative to viral integrating vectors, have truly become viral. Here, we briefly review our findings and speculate on the perspectives and potential advantages of transposase delivery by lentiviral protein transduction.
Keywords: DNA transposition, protein transduction, piggyBac, Sleeping Beauty, lentiviral vector, IDLV
It is fascinating how ancient and extremely primitive DNA transposable elements are making an impact in a world of modern and advanced genetics that is constantly climbing to new technological heights. In spite of their genetic simplicity, parasitic mobile DNA elements have infiltrated and colonized the genomes of all living creatures, exploiting the capacity to relocate between genomic loci and multiply in numbers. Fossil remnants of these once actively jumping elements have lost the mobility due to accumulating mutations, and hide in genomes across the animal kingdom as inactive marks of a past genetic history. The piggyBac DNA transposon is one of the exceptions to the rule. This element was first identified when it actively jumped from its insect host, the cabbage looper moth Trichoplusia ni, into the genome of a baculovirus.1 The active mobility of the piggyBac transposon led to its use in genetic analyses of insects,2 but its capacity to insert genes in mammalian cells was not unveiled until recently.3,4 Screening of a library of mutant transposases led to the identification of a hyperactive piggyBac transposase, hyPBase, with even higher activity in human systems. Another DNA transposon, the Sleeping Beauty element was the first transposable element shown to efficiently transpose in mammalian cells.5 This element, a member of the Tc1/mariner transposon family, was reconstructed from salmonid transposable elements by consecutive steps of mutagenesis, facilitating its subsequent use in genetic engineering,6,7 animal transgenesis,8-10 forward genetic screens,11,12 and therapeutic gene transfer.13-15 Engineering of the hyperactive SB100X transposase variant, generated by a high-throughput PCR-based DNA shuffling approach,16 and vectors with improved inverted repeats17,18 have added further efficacy to the system.
In their most typical composition, DNA transposons contain a single gene encoding the transposase protein. The gene is flanked by two inverted terminal repeats that serve as binding sites for transposase protein during cut-and-paste transposition. Interaction of transposase subunits binding to each of the terminal regions of the transposon facilitates the close association of the transposon ends allowing first excision from a donor and insertion into an acceptor site. Conventional DNA transposon-based vector systems are based on the co-delivery of (i) donor plasmids carrying the DNA transposon vector including the gene of interest and (ii) plasmids carrying the transposase expression cassette. Alternatively, in vitro-transcribed RNA molecules may serve as a source of the transposase.19 Also, viral vectors, including lentiviral, adenoviral, and adeno-associated viral vectors, have been adapted as carriers of the transposase expression cassette.20-23
Common for all current DNA transposon systems is the need for intracellular production of the transposase. Ideally, DNA transposition is achieved within a short time-frame defined by a short-term boost of protein production and activity. It is challenging, however, to control the level and longevity of expression after plasmid transfection or viral vector transduction, and even transient expression strategies may cause sustained expression of the transposase, at least in slowly proliferating tissues and cell types. Also, in many cell types DNA and RNA transfection or nucleofection procedures tend to create massive nuclear accumulation of the transposase, which may on one hand be desired for optimal efficacy but on the other increase the risk of inserting numerable copies of the transposon or harm the cells otherwise. Adding to this, delivery of transposase-encoding plasmid DNA or viral vectors comes with the inherent risk of stably integrating the transposase expression cassette (often driven by a strong promoter) in the genome of the treated cells. It is known that transposases may interfere with normal cell cycle progression24 and cause premitotic cell cycle arrest and even apoptosis through mechanisms involving activation of p53 and c-Jun.25 By processes that are potentially linked to such toxicity, too high transient transposase expression levels lead to reduced efficacy of DNA transposition.14,26 We have previously discussed these aspects in detail.27 This type of regulation, normally referred to as overproduction inhibition (OPI), may possibly reflect natural regulatory mechanisms or, alternatively, that artificial overproduction of the transposase may harm the cells and even cause cell death.
In light of the uncertainties and potential safety precautions associated with intracellular production of effector proteins, like transposases and endonucleases, it is important to scrutinize other means of delivering such proteins to cells. Direct delivery of protein itself is an obvious alternative that, if successful, guarantees short-term activity and limits potential cytotoxicity. Fused to proteins of interest, cell-penetrating peptides (CPPs), also referred to as protein transduction domains (PTDs), can facilitate cellular protein uptake28 and hold the potential to carry drugs into the interior of cells. A recent study unveiled intrinsic cell-penetrating capacities of zinc-finger nucleases (ZFNs), allowing direct delivery of recombinant ZFN protein to mammalian cells in vitro.29 However, recombinant production of functional Sleeping Beauty and piggyBac transposases has turned out to be extremely challenging, and the few published attempts did not successfully demonstrate efficacy of DNA transposition catalyzed by recombinant transposases.30,31 Low levels of activity may be explained by problems related to production and purification of active transposase as well as to reduced cellular uptake potentially caused by entrapment of protein in the endosomes.32
To establish effective DNA transposition without the need of transferring transposase-encoding nucleic acids, we recently set out to investigate an alternative route for delivering transposase proteins to cells.33 Inspired by the capacity of viruses to carry their own enzymatic proteins, we sought to examine the ability of lentivirus-like particles to incorporate heterologous proteins like the piggyBac and Sleeping Beauty transposases. Here, we briefly review our findings, published earlier this year in Nucleic Acids Research,33 and speculate on the perspectives and further applications of transposase delivery by lentiviral protein transduction.
Early studies demonstrated that the structure of gamma-retroviruses is sufficiently flexible to allow incorporation of foreign proteins fused to the Gag polypeptide.34,35 More recently, this approach was successfully employed to deliver the Flp recombinase ferried within Gag precursors in murine leukemia virus (MLV) particles.36 In accordance with these findings, HIV-1-derived lentiviral particles were found to tolerate the inclusion of heterologous proteins fused to Gag. Work by the Komano group demonstrated release of proteins like β-lactamase, GFP, and caspase 3 upon virion maturation37,38 and established a basis for exploiting lentiviral Gag precursors as carriers of proteins and drugs. Using a related approach, Schenkwein and coworkers fused heterologous proteins to the integrase protein within the Pol region of the GagPol polypeptide.39 Despite the fact that lentiviral particles contain considerably fewer GagPol than Gag polypeptides, the strategy effectively supported transfer of the mCherry reporter protein and p53, the latter of which was found to trigger apoptosis in virus-treated cells. Hitch-hiking of proteins in Gag and GagPol polypeptides offers attractive alternatives to previous approaches based on the viral incorporation of proteins fused to the HIV-1 accessory protein Vpr.40,41 Vpr-based fusions have been successfully used to deliver reporter proteins, Cre recombinase, and I-SceI meganuclease to virus-treated cells,40,42,43 but the relatively few copies of Vpr in the particles44 and the potential toxicity of Vpr45 may restrict the applicability of this approach. Table 1 provides an overview of the strategies that have been successfully utilized to incorporate and transfer foreign proteins of interest (POIs) in lentiviral particles.
Table 1. Overview of strategies used to deliver proteins of interest (POIs) by lentiviral protein transduction.
| POI incorporation strategy | Transferred POI |
|---|---|
| Gag-POI (POI fused to p6) | GFP,46 YFP,47,48 CFP,47,48 mCherry,49 pHluorin49 |
| POI-Gag-Pol (POI fused to MA) | GFP,37 β-lactamase,37 Caspase-338 |
| MA-POI-CA (POI inserted between MA and CA) | GFP50 |
| Gag-Pol-POI (POI fused to IN) | mCherry,39 p53,39 λ repressor,51 LexA,52 I-PpoI,53 Zif268,54 E2C55,56 |
| Vpr-POI (POI fused to Vpr) | GFP,57,58 SN,41 CAT,41 IN,59,60 RT,60 PR,61 Cre,43 I-SceI,42 Luc,62 A3G,63 HSV-TK,64 linamarase64 |
| POI-WXXF (POI fused to Vpr-binding WXXF-motif) | CAT,65 IN,66 scAb67 |
| Nef-POI (POI fused to Nef) | GFP,75,76 HSV-TK76,77 |
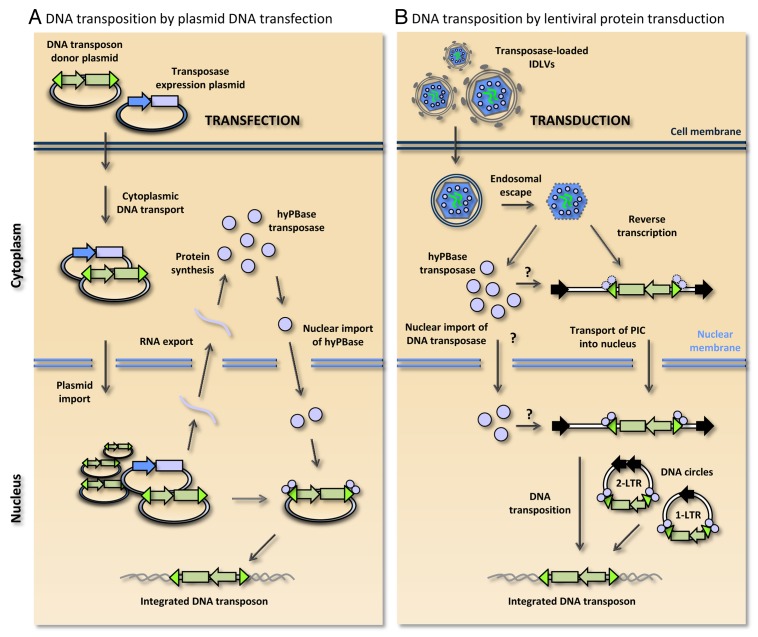
To convert lentiviral particles into transposase protein delivery vehicles, we introduced the transposase in the N-terminus of the Gag polypeptide at a position between the matrix protein and an artificially introduced myristoylation signal derived from the Lyn kinase.37 Also, in the modified packaging construct, we introduced the D64V mutation in the integrase gene,68 certifying that the normal lentiviral integration machinery of such particles was not active. Assisted by HIV-1 protease cleavage sites flanking the transposase protein, the transposase was effectively liberated from Gag and GagPol polypeptides upon virion maturation. Western analyses demonstrated potent incorporation of both SB100X and hyPBase transposasases in virions. Our first indications of the efficacy of virally delivered transposases came from observations of high levels of piggyBac DNA transposition. In HeLa cells, treatment with hyPBase-loaded virus particles triggered higher levels of transposition than we normally observe with a standardized plasmid co-transfection protocol. A schematic overview of DNA transposition driven by a conventional plasmid-based system and by lentiviral protein transduction is provided in Figure 1.
Figure 1. Schematic comparison of piggyBac DNA transposition by plasmid DNA transfection and lentiviral protein transduction. (A) DNA transposition by co-transfection of the DNA transposon donor plasmid and transposase-encoding plasmid. Transport through the cytoplasm and nuclear uptake lead to production of hyPBase transposase, which is subsequently imported into the nucleus. Within the nucleus the transposon-based gene vector (indicated in green) is excised from the donor plasmid and inserted into a genomic locus. (B) DNA transposition by lentiviral protein transduction in integrase-defective lentiviral vectors (IDLVs). Engineered lentiviral particles carry both the hyPBase protein (indicated by small light-purple circles) and the diploid RNA vector genome (indicated by green lines). Cell entry mediated by the VSV-G surface protein occurs through endocytosis and subsequent endosomal escape. Reverse-transcribed double-stranded DNA intermediates serve as transposon donors. Along with linear DNA substrates, 1-LTR and 2-LTR circles generated by homologous recombination and non-homologous end joining, respectively, may serve as transposon donors. Question marks indicate that it is not currently known whether transposase subunits are associated with the transposon in the cytoplasm or are imported into the nucleus prior to association with the transposon terminal repeats. It is currently unclear whether the transposase remains part of the pre-integration complex (PIC) during nuclear entry or is released from the PIC during cytoplasmic transport.
We have previously shown that DNA transposons are effectively mobilized from lentiviral DNA intermediates generated by reverse transcription of integrase-defective lentiviral vectors (IDLVs).22,23 This led to the idea that lentiviral particles could potentially accommodate both the transposase and the transposon, essentially mimicking conventional lentiviral vectors carrying both integrase protein and the recognition sites for active gene insertion. During the analysis of transposase-loaded IDLVs, we encountered the obstacle that transfer of the vector was markedly restricted by the load of Gag-fused transposase molecules. However, this problem was solved by generating particles composed of both wildtype and transposase-containing Gag and GagPol (still carrying the D64V mutation). Such chimeric particles supported quite significant levels of piggyBac DNA transposition in a panel of cell types including human primary keratinocytes and normal human dermal fibroblasts. Notably, such transposition was not evident when the particles were loaded with a mutated, inactive variant of the hyPBase and when the particles were not able to get access to the cells due to lack of VSV-G pseudotyping.
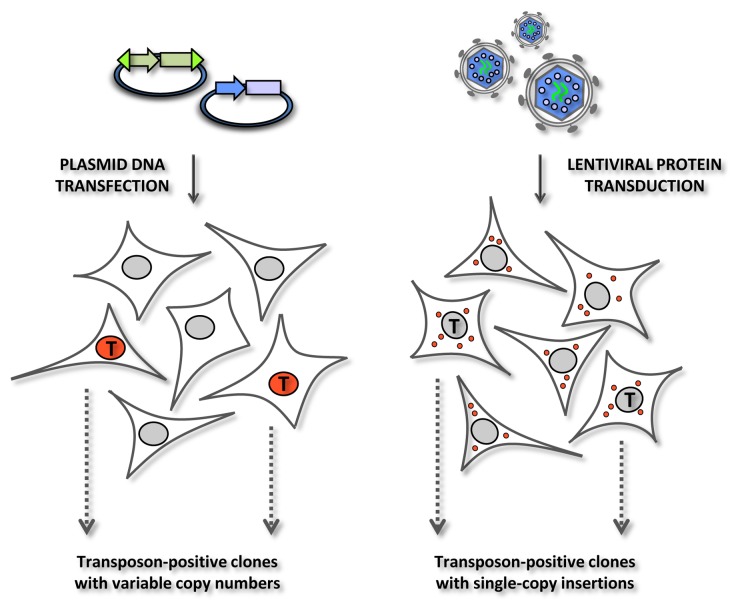
With experimental evidence for this new gene delivery concept, we went on to characterize the transduced cells in more detail using confocal microscopy. This analysis showed, perhaps surprisingly, quite low cellular levels of the transposase, which was visible only in few concentrated foci within transduced cells. In comparison, cells transfected with hyPBase-encoding plasmid displayed massive nuclear accumulation of the transposase. Still, the level of transposition, as measured by colony formation after mobilization of a transposon containing the puromycin resistance gene, was higher using the viral approach. We believe that this observation reflects that more or less all the cells were transduced by the viral vector, whereas a lower percentage of cells were transfected with plasmid DNA. Following this reasoning, a high transduction rate may compensate for the potential limitations of DNA transposition from virally delivered substrates. In line with this notion, we made an interesting observation when genomic vector copy numbers were determined after DNA transposition. Notably, all analyzed puromycin-resistant clones generated after virus-mediated transposition contained a single copy of the transposon, whereas transposition after plasmid transfer resulted in clones with variable copy numbers, half of them with 5 or more (and up to 12) copies of the transposon. The background for such key differences between plasmid- and virus-based transposase delivery is schematically depicted in Figure 2.
Figure 2. Comparative models of DNA transposition observed after plasmid DNA transfection and lentiviral protein transduction. Red marking indicates schematically the patterns of immunostaining that were observed by confocal microscopy of cells stained with an antibody specific for HA-tagged hyPBase transposase.33 Plasmid DNA transfection leads to dramatic overexpression of the transposase in successfully transfected cells, whereas lentiviral protein transduction results in much lower overall levels of the transposase in virus-treated cells. Small red dots in transduced cells indicate that the transposase is present in all cells, but only observed in concentrated foci primarily within the cytoplasm. ‘T’ indicates cells in which successful transposition is achieved. Effective DNA transposition is a likely result of robust nuclear levels of transposase, potentially leading to several transposon insertions in a single cell/clone, whereas DNA transposition supposedly is less efficient and does not occur in all transposase-positive cells after protein transduction. As a result, however, all resulting clones contain only a single integrated copy of the transposon. See text for further details.
Our data confirmed that DNA transposition after plasmid transfection may result in multiple insertions. This returning observation in the transposon field is the expected result of both strong and prolonged expression of the transposase as well as concomitant high levels of transposon donor plasmid in cells that are fairly easy to transfect. In clones developing from a transfected cell, DNA transposition is likely to keep going for several cell divisions until the plasmids are lost. This means that DNA transposons that are initially inserted in chromosomal DNA can be re-excised from the genome or, likely more frequent, that additional DNA transposons in a developing clone may be transferred from plasmid DNA to locations within the chromosomes. As a result, such ‘clones’ may be composed of subclones containing a variable number of insertions and, hence, may not be truly clonal. This will lead to clonal heterogeneity in terms of transgene expression. Much like diluted preps of conventional lentiviral vectors, transposase-loaded IDLVs balance the level of gene insertion in each transduced cell leading only to a single insertion. Thus, this technique can support applications where only one insertion of the transposon is desired. As a direct consequence, clones resulting from the short-term action of the transposase during protein transduction are more likely to display homogenous expression of the transgene.
Our data show that hyPBase delivered in lentiviral particles can get access to DNA transposons localized either in co-transfected plasmid DNA or in co-transduced IDLVs. This argues that transposase subunits at a certain stage during transduction are liberated from the invading pre-integration complex (PIC), allowing formation of transposition complexes on substrates delivered in trans. Still, the efficiency of the approach seems to be further improved by incorporating transposases in transposon-carrying IDLVs. In analogy with conventional lentiviral vectors carrying both the integrase and the substrate for the integration process, we believe that the proximity between transposases and the reverse-transcribed substrate within the PIC is supporting the overall efficiency of the process. In this way, the PIC is potentially offering an intracellular environment with high, local concentration of the transposase and, hence, compensating for the overall low levels of transposase in virus-treated cells.
As part of our endeavor to establish DNA transposition by protein delivery in human cells, we repeatedly tried to incorporate the hyperactive SB100X transposase16 in lentiviral particles. We have seen that Gag-fused SB100X is indeed effectively packaged into virions and also liberated from Gag during virion maturation, but so far we have not been able to demonstrate efficacy of such virally delivered SB100X proteins. The lack of function is at least partially explained by the negative effect of the C-terminal 4-amino acid tag originating from the protease cleavage site between SB100X and Gag. The cleavage site is required to ensure proper release of the SB100X during virion maturation, but the N-terminal part of the cleavage site remains an integrated part of the protein after cleavage. In separate analyses of different plasmid-encoded SB100X variants with such small peptide tags fused to the C-terminus of the protein, we have seen that the function is completely abolished. This confirms the general notion that the C-terminus of SB transposases needs to be left untouched for full activity of the protein69 and that piggyBac transposases are more flexible and less vulnerable to such changes.70 To facilitate lentiviral SB100X protein transduction, it would be an alternative option to fuse SB100X to the C-terminal end of GagPol, allowing release of a protein with an additional tag only in the N-terminus.
DNA transposons are by definition non-viral transposable elements, and derived gene vectors were originally developed and launched as a non-viral alternative to viral integrating vectors. As such, they are already powerful tools in biomedicine and now moving into the clinic.71 Why then bother adapting virus particles as carriers of transposases? By showing proof-of-concept that genomic engineering tools can be directly delivered by lentiviral particles, we are addressing one of the major potential challenges related to current systems of genomic engineering and editing. Is it appropriate and sufficiently safe to overexpress enzymes like transposases and site-targeted nucleases within treated cells? Lentiviral protein transduction is the first approach that allows efficacious DNA transposition in human cells after direct protein delivery. Previous attempts to produce and deliver recombinant transposases have had limited success,30,31 and lentiviral particles furthermore offer the opportunity of delivering transposases and transposon substrates simultaneously in a single vehicle. As an additional bonus, such hybrid particles generally seem to generate only a single genomic insertion of the DNA transposon, although this phenomenon needs to be further scrutinized in additional cell types. Also, the option of targeting DNA transposition to certain cell types by pseudotyping of the lentiviral particles deserves more attention in the future.
Early studies showed the applicability of plasmid-based DNA transposition system in vivo with initial focus on the mouse liver.14,26 Many other tissues are significantly less accessible to plasmid DNA transfection, and low transfection rates are challenging in terms of wider in vivo use of both Sleeping Beauty- and piggyBac-based systems. It is tempting to speculate that transposase-loaded IDLVs could be paving the way for several new in vivo applications of piggyBac DNA transposition. One such application could be gene insertion by DNA transposition in skin. We have previously demonstrated efficient gene delivery and prolonged transgene expression in human skin intradermally injected with lentiviral vectors.72,73 Future studies will show whether injection of IDLVs carrying the PB transposase can facilitate in vivo DNA transposition in skin progenitor cells. As we explore new ways of exploiting this transposase delivery approach, we are hard at work stretching the boundaries of lentiviral particles as transporters of foreign cargo. Most recently, we demonstrated targeted genome editing by lentiviral protein transduction of programmable nucleases co-delivered in virus particles with substrates for homology-directed recombination.74
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Work utilizing and exploring DNA transposon-based technologies in the laboratory of JGM is made possible through support of the Lundbeck Foundation, the Novo Nordisk Foundation, EU FP6 (INTHER), Grosserer L. F. Foghts Fond, the Hørslev Foundation, Aase og Ejnar Danielsens Fond, Agnes og Poul Friis Fond, Aage Bangs Fond, Grosserer A. V. Lykfeldt og Hustrus Legat, Else og Mogens Wedell-Wedellsborgs Fond, Fonden af 17-12-1981, Civilingeniør Frode V. Nyegaard og hustrus Fond, Helga og Peter Kornings Fond, Oda og Hans Svenningsens Fond, Snedkermester Sophus Jacobsen & Hustru Astrid Jacobsens Fond, Familien Hede Nielsens Fond, and. JGM is head of Gene Therapy Initiative Aarhus (GTI-Aarhus) funded by the Lundbeck Foundation and a member of the Aarhus Research Center for Innate Immunology (ARCII) established through funding by the AU-Ideas program at Aarhus University.
References
- 1.Fraser MJ, Smith GE, Summers MD. Acquisition of Host Cell DNA Sequences by Baculoviruses: Relationship Between Host DNA Insertions and FP Mutants of Autographa californica and Galleria mellonella Nuclear Polyhedrosis Viruses. J Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handler AM, McCombs SD, Fraser MJ, Saul SH. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc Natl Acad Sci U S A. 1998;95:7520–5. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Wilson MH, Coates CJ, George AL., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–45. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 5.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 6.Sharma N, Cai Y, Bak RO, Jakobsen MR, Schrøder LD, Mikkelsen JG. Efficient sleeping beauty DNA transposition from DNA minicircles. Mol Ther Nucleic Acids. 2013;2:e74. doi: 10.1038/mtna.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staunstrup NH, Sharma N, Bak RO, Svensson L, Petersen TK, Aarenstrup L, Kristiansen K, Bolund L, Mikkelsen JG. A Sleeping Beauty DNA transposon-based genetic sensor for functional screening of vitamin D3 analogues. BMC Biotechnol. 2011;11:33. doi: 10.1186/1472-6750-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, Schmidt M, Winther KD, Schyth BD, Holm IE, et al. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res. 2011;20:533–45. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- 9.Staunstrup NH, Madsen J, Primo MN, Li J, Liu Y, Kragh PM, Li R, Schmidt M, Purup S, Dagnæs-Hansen F, et al. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human β1 and α2 integrin. PLoS One. 2012;7:e36658. doi: 10.1371/journal.pone.0036658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Mashhadi RH, Sørensen CB, Kragh PM, Christoffersen C, Mortensen MB, Tolbod LP, Thim T, Du Y, Li J, Liu Y, et al. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant. Sci Transl Med. 2013;5:ra1. doi: 10.1126/scitranslmed.3004853. [DOI] [PubMed] [Google Scholar]
- 11.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 12.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 13.Aronovich EL, Bell JB, Khan SA, Belur LR, Gunther R, Koniar B, Schachern PA, Parker JB, Carlson CS, Whitley CB, et al. Systemic correction of storage disease in MPS I NOD/SCID mice using the sleeping beauty transposon system. Mol Ther. 2009;17:1136–44. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 15.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, Hackett PB, Kohn DB, Shpall EJ, Champlin RE, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–71. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–61. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 17.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–35. doi: 10.1016/S0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 18.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–47. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilber A, Wangensteen KJ, Chen Y, Zhuo L, Frandsen JL, Bell JB, Chen ZJ, Ekker SC, McIvor RS, Wang X. Messenger RNA as a source of transposase for sleeping beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol Ther. 2007;15:1280–7. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
- 20.Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W, Solanki M, Müther N, Ebel M, Wang J, Sun C, Izsvak Z, Ehrhardt A. Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells. PLoS One. 2013;8:e76771. doi: 10.1371/journal.pone.0076771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N, Mátés L, Izsvák Z, Chen W, Ivics Z, et al. Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol Ther. 2011;19:1499–510. doi: 10.1038/mt.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, Izsvák Z, Mikkelsen JG. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther. 2009;17:1205–14. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walisko O, Izsvák Z, Szabó K, Kaufman CD, Herold S, Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc Natl Acad Sci U S A. 2006;103:4062–7. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galla M, Schambach A, Falk CS, Maetzig T, Kuehle J, Lange K, Zychlinski D, Heinz N, Brugman MH, Göhring G, et al. Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res. 2011;39:7147–60. doi: 10.1093/nar/gkr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA. Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther. 2003;8:654–65. doi: 10.1016/S1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 27.Skipper KA, Andersen PR, Sharma N, Mikkelsen JG. DNA transposon-based gene vehicles - scenes from an evolutionary drive. J Biomed Sci. 2013;20:92. doi: 10.1186/1423-0127-20-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bechara C, Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013;587:1693–702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9:805–7. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CY, Li JF, Liou JS, Charng YC, Huang YW, Lee HJ. A gene delivery system for human cells mediated by both a cell-penetrating peptide and a piggyBac transposase. Biomaterials. 2011;32:6264–76. doi: 10.1016/j.biomaterials.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Järver P, Fernaeus S, El-Andaloussi S, Tjörnhammer M-L, Langel Ü. Co-transduction of sleeping beauty transposase and donor plasmid via a cell-penetrating peptide: a simple one step method. Int J Pept Res Ther. 2008;14:58–63. doi: 10.1007/s10989-007-9114-z. [DOI] [Google Scholar]
- 32.Mellert K, Lamla M, Scheffzek K, Wittig R, Kaufmann D. Enhancing endosomal escape of transduced proteins by photochemical internalisation. PLoS One. 2012;7:e52473. doi: 10.1371/journal.pone.0052473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, Bak RO, Krogh LB, Staunstrup NH, Moldt B, Corydon TJ, Schrøder LD, Mikkelsen JG. DNA transposition by protein transduction of the piggyBac transposase from lentiviral Gag precursors. Nucleic Acids Res. 2014;42:e28. doi: 10.1093/nar/gkt1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones TA, Blaug G, Hansen M, Barklis E. Assembly of gag-beta-galactosidase proteins into retrovirus particles. J Virol. 1990;64:2265–79. doi: 10.1128/jvi.64.5.2265-2279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weldon RA, Jr., Erdie CR, Oliver MG, Wills JW. Incorporation of chimeric gag protein into retroviral particles. J Virol. 1990;64:4169–79. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelkel C, Galla M, Maetzig T, Warlich E, Kuehle J, Zychlinski D, Bode J, Cantz T, Schambach A, Baum C. Protein transduction from retroviral Gag precursors. Proc Natl Acad Sci U S A. 2010;107:7805–10. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aoki T, Miyauchi K, Urano E, Ichikawa R, Komano J. Protein transduction by pseudotyped lentivirus-like nanoparticles. Gene Ther. 2011;18:936–41. doi: 10.1038/gt.2011.38. [DOI] [PubMed] [Google Scholar]
- 38.Miyauchi K, Urano E, Takizawa M, Ichikawa R, Komano J. Therapeutic potential of HIV protease-activable CASP3. Sci Rep. 2012;2:359. doi: 10.1038/srep00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenkwein D, Turkki V, Kärkkäinen HR, Airenne K, Ylä-Herttuala S. Production of HIV-1 integrase fusion protein-carrying lentiviral vectors for gene therapy and protein transduction. Hum Gene Ther. 2010;21:589–602. doi: 10.1089/hum.2009.051. [DOI] [PubMed] [Google Scholar]
- 40.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–52. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Liu H, Xiao H, Kim J, Seshaiah P, Natsoulis G, Boeke JD, Hahn BH, Kappes JC. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J Virol. 1995;69:3389–98. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izmiryan A, Basmaciogullari S, Henry A, Paques F, Danos O. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic Acids Res. 2011;39:7610–9. doi: 10.1093/nar/gkr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel G, Yu Y, Chang T, Yee JK. Site-specific gene insertion mediated by a Cre-loxP-carrying lentiviral vector. Mol Ther. 2010;18:1814–21. doi: 10.1038/mt.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson CM, Malim MH. SnapShot: HIV-1 proteins. Cell. 2008;133:742–, e1. doi: 10.1016/j.cell.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Tachiwana H, Shimura M, Nakai-Murakami C, Tokunaga K, Takizawa Y, Sata T, Kurumizaka H, Ishizaka Y. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res. 2006;66:627–31. doi: 10.1158/0008-5472.CAN-05-3144. [DOI] [PubMed] [Google Scholar]
- 46.Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci U S A. 2005;102:15453–8. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson DR, Ma YM, Vogt VM, Webb WW. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J Cell Biol. 2003;162:1233–44. doi: 10.1083/jcb.200303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Dou J, Ding L, Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J Virol. 2007;81:12899–910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–40. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H, Kräusslich HG. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol. 2004;78:10803–13. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci U S A. 1994;91:9233–7. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goulaouic H, Chow SA. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schenkwein D, Turkki V, Ahlroth MK, Timonen O, Airenne KJ, Ylä-Herttuala S. rDNA-directed integration by an HIV-1 integrase--I-PpoI fusion protein. Nucleic Acids Res. 2013;41:e61. doi: 10.1093/nar/gks1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bushman FD, Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–64. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–48. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan W, Zhu K, Segal DJ, Barbas CF, 3rd, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J Virol. 2004;78:1301–13. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muthumani K, Montaner LJ, Ayyavoo V, Weiner DB. Vpr-GFP virion particle identifies HIV-infected targets and preserves HIV-1Vpr function in macrophages and T-cells. DNA Cell Biol. 2000;19:179–88. doi: 10.1089/104454900314564. [DOI] [PubMed] [Google Scholar]
- 58.Waldhuber MG, Bateson M, Tan J, Greenway AL, McPhee DA. Studies with GFP-Vpr fusion proteins: induction of apoptosis but ablation of cell-cycle arrest despite nuclear membrane or nuclear localization. Virology. 2003;313:91–104. doi: 10.1016/S0042-6822(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 59.Fletcher TM, 3rd, Soares MA, McPhearson S, Hui H, Wiskerchen M, Muesing MA, Shaw GM, Leavitt AD, Boeke JD, Hahn BH. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16:5123–38. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, Liu H, Xiao H, Conway JA, Hunter E, Kappes JC. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–22. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X, Liu H, Xiao H, Kappes JC. Proteolytic activity of human immunodeficiency virus Vpr- and Vpx-protease fusion proteins. Virology. 1996;219:307–13. doi: 10.1006/viro.1996.0253. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez G, DaFonseca S, Errazuriz E, Coric P, Souquet F, Turcaud S, Boulanger P, Bouaziz S, Hong SS. Characterization of a novel type of HIV-1 particle assembly inhibitor using a quantitative luciferase-Vpr packaging-based assay. PLoS One. 2011;6:e27234. doi: 10.1371/journal.pone.0027234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ao Z, Yu Z, Wang L, Zheng Y, Yao X. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS One. 2008;3:e1995. doi: 10.1371/journal.pone.0001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Link N, Aubel C, Kelm JM, Marty RR, Greber D, Djonov V, Bourhis J, Weber W, Fussenegger M. Therapeutic protein transduction of mammalian cells and mice by nucleic acid-free lentiviral nanoparticles. Nucleic Acids Res. 2006;34:e16. doi: 10.1093/nar/gnj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.BouHamdan M, Xue Y, Baudat Y, Hu B, Sire J, Pomerantz RJ, Duan LX. Diversity of HIV-1 Vpr interactions involves usage of the WXXF motif of host cell proteins. J Biol Chem. 1998;273:8009–16. doi: 10.1074/jbc.273.14.8009. [DOI] [PubMed] [Google Scholar]
- 66.Kulkosky J, BouHamdan M, Geist A, Pomerantz RJ. A novel Vpr peptide interactor fused to integrase (IN) restores integration activity to IN-defective HIV-1 virions. Virology. 1999;255:77–85. doi: 10.1006/viro.1998.9544. [DOI] [PubMed] [Google Scholar]
- 67.Okui N, Sakuma R, Kobayashi N, Yoshikura H, Kitamura T, Chiba J, Kitamura Y. Packageable antiviral therapeutics against human immunodeficiency virus type 1: virion-targeted virus inactivation by incorporation of a single-chain antibody against viral integrase into progeny virions. Hum Gene Ther. 2000;11:537–46. doi: 10.1089/10430340050015725. [DOI] [PubMed] [Google Scholar]
- 68.Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, Buch P, MacLaren RE, Anderson PN, Barker SE, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–53. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 69.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kettlun C, Galvan DL, George AL, Jr., Kaja A, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol Ther. 2011;19:1636–44. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maiti SN, Huls H, Singh H, Dawson M, Figliola M, Olivares S, Rao P, Zhao YJ, Multani A, Yang G, et al. Sleeping beauty system to redirect T-cell specificity for human applications. J Immunother. 2013;36:112–23. doi: 10.1097/CJI.0b013e3182811ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bak RO, Stenderup K, Rosada C, Petersen LB, Moldt B, Dagnæs-Hansen F, Jakobsen M, Kamp S, Jensen TG, Dam TN, et al. Targeting of human interleukin-12B by small hairpin RNAs in xenografted psoriatic skin. BMC Dermatol. 2011;11:5. doi: 10.1186/1471-5945-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jakobsen M, Stenderup K, Rosada C, Moldt B, Kamp S, Dam TN, Jensen TG, Mikkelsen JG. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol Ther. 2009;17:1743–53. doi: 10.1038/mt.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai Y, Bak RO, Mikkelsen JG. Targeted genome editing by lentiviral protein transduction of zinc-finger and TAL-effector nucleases. Elife. 2014;3:e01911. doi: 10.7554/eLife.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muratori C, D’Aloja P, Superti F, Tinari A, Sol-Foulon N, Sparacio S, Bosch V, Schwartz O, Federico M. Generation and characterization of a stable cell population releasing fluorescent HIV-1-based Virus Like Particles in an inducible way. BMC Biotechnol. 2006;6:52. doi: 10.1186/1472-6750-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peretti S, Schiavoni I, Pugliese K, Federico M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol Ther. 2005;12:1185–96. doi: 10.1016/j.ymthe.2005.06.474. [DOI] [PubMed] [Google Scholar]
- 77.Peretti S, Schiavoni I, Pugliese K, Federico M. Selective elimination of HIV-1-infected cells by Env-directed, HIV-1-based virus-like particles. Virology. 2006;345:115–26. doi: 10.1016/j.virol.2005.09.054. [DOI] [PubMed] [Google Scholar]