Abstract
Real-time quantitative PCR (qRT-PCR) is one of the important methods for investigating the changes in mRNA expression levels in cells and tissues. Selection of the proper reference genes is very important when calibrating the results of real-time quantitative PCR. Studies on the selection of reference genes in goat tissues are limited, despite the economic importance of their meat and dairy products. We used real-time quantitative PCR to detect the expression levels of eight reference gene candidates (18S, TBP, HMBS, YWHAZ, ACTB, HPRT1, GAPDH and EEF1A2) in ten tissues types sourced from Boer goats. The optimal reference gene combination was selected according to the results determined by geNorm, NormFinder and Bestkeeper software packages. The analyses showed that tissue is an important variability factor in genes expression stability. When all tissues were considered, 18S, TBP and HMBS is the optimal reference combination for calibrating quantitative PCR analysis of gene expression from goat tissues. Dividing data set by tissues, ACTB was the most stable in stomach, small intestine and ovary, 18S in heart and spleen, HMBS in uterus and lung, TBP in liver, HPRT1 in kidney and GAPDH in muscle. Overall, this study provided valuable information about the goat reference genes that can be used in order to perform a proper normalisation when relative quantification by qRT-PCR studies is undertaken.
Keywords: Reference Gene, Expression Stability, Tissue, Goat
INTRODUCTION
Goat is an economically important animal in many communities worldwide, providing meat and milk products with abundant nutritional value (Dubeuf et al., 2004). Previous studies have shown that breeding, growth and other major economical traits of goats are highly regulated by functional genes. Studies involving temporal and spatial expression specificity of these genes in different goat tissues are helpful when screening the functional genes regulating important economic traits of goats. They are also vital in marker-assisted selection (MAS) studies and in breeding program improvements (Moioli et al., 2007; Domby et al., 2010; Fu et al., 2011; Zi et al., 2012).
Real-time quantitative PCR is a quantitative analytical method used to determine mRNA expression at the transcriptional level. It is rapid, highly reproducible and extremely sensitive, and is therefore widely used in the quantitative studies on gene expression (Dabek et al., 2010; Novinscak et al., 2011; Veys et al., 2012). However, the reliability of the results from qRT-PCR is affected by experimental conditions, yield and quality of the sample RNA, as well as the efficiency of reverse transcription (Fleige and Pfaffl, 2006). Therefore, specific reference genes are normally selected for calibration and normalization of target genes in the studies in order to reduce the deviation of the results (Mehta et al., 2010; Wang and Xu, 2010; Pierzchala et al., 2011). Studies on the screening of reference genes used in qRT-PCR have been reported in tissue sourced from humans, mice, pigs, cattle and other mammals (Garcia-Crespo et al., 2005; Spalenza et al., 2011; Chechi et al., 2012; Zhang et al., 2012), but studies in goat tissues are limited. The expression levels of frequently used reference genes may differ significantly among different species, cell types, tissues, different stages of cell proliferations and organ development, in vitro culture and other kinds of experimental conditions (Chechi et al., 2012; Chi et al., 2012). Therefore, it is of great importance to select suitable reference genes to carry out calibration and normalization for the expression of target genes in the specific experimental conditions.
We selected eight candidates of reference genes (18S, TBP, HMBS, YWHAZ, ACTB, HPRT1, GAPDH, and EEF1A2), and the differences in their expression levels in ten different tissues types (heart, liver, spleen, lung, kidney, stomach, uterus, ovaries, small intestines and muscles) of Boer goat were compared. geNorm, NormFinder and Bestkeeper analytical software packages were used to determine the gene combination showing the most stable expression. Identifying the best combination of reference genes will provide information to select the most suitable reference genes for calibrating qRT-PCR analysis of mRNAs from goats.
MATERIALS AND METHODS
Tissues
Ten different goat tissues (heart, liver, spleen, lung, kidney, stomach, small intestine, uterus, ovary and skeletal muscle) were collected from three 5-month-old female Boer goats. All the samples were immediately snap-frozen in liquid nitrogen and stored at −80°C and would be used for evaluating the stability of each candidate genes.
Total RNA isolation and cDNA synthesis
Total RNA was extracted using Trizol (Invitrogen, Gaithersburg, MD) according to the manufacturer’s instructions. RNA concentration was determined by spectrophotometry at 260 nm/280 nm absorbance ratio with expected values between 1.8–2.00 using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). RNA integrity was confirmed by gel electrophoresis using 1% agarose with ethidium bromide. RNA samples were run on a 1% agarose gel at 100 V for 30 min (Supplementary Figure 1). The isolated RNA was immediately frozen at −80°C. cDNA was synthesized from 1 μg of total RNA by using a PrimerScript RT reagent kit (TaKaRa, Osaka, Japan) with an oligo-dT primer. The mixture was incubated at 37°C for 15 min and 85°C for five seconds to terminate reaction in an ABI 9700 DNA Thermal Cycler (Applied Biosystems, Foster City, CA, USA).
Reference gene selection and primer designing
Eight genes were selected to be evaluated as reference genes according to the literature: 18S, TBP, HMBS, YWHAZ, ACTB, HPRT1, GAPDH, and EEF1A2 (Garcia-Crespo et al., 2005; Spalenza et al., 2011; Chechi et al., 2012; Zhang et al., 2012). Selection was done considering their stability values from published studies. The candidate genes and primers are listed in Table 1. The primers were designed using Primer Express 2.0 software (Yoon et al., 2002) and synthesized by Invitrogen Shanghai (Shanghai, China).
Table 1.
Selected candidate reference genes used in the qRT-PCR assay indicating name, GenBank accession number, annealing temperature (Ta), PCR efficiency, slope, regression coefficient and primers used for the expression study
| Gene | Full gene name | GenBank accession number | Ta (°C) | PCR efficiency (%) | Slope | Regression coefficient (r2) | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|---|---|---|---|
| ACTB | Beta-actin | DQ845171 | 62 | 99.66 | −3.33 | 0.998 | TGCCCTGAGGCTCTCTTCCA | TGCGGATGTCGACGTCACA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | AF017079 | 62 | 96.84 | −3.40 | 0.998 | GCAAGTTCCACGGCACAGTC | CCCACTTGATGTTGGCAGGA |
| TBP | TATA box-binding protein | DQ845178 | 62 | 94.92 | −3.45 | 0.993 | AAAGACCATTGCACTTCGTGC | ATATCAGTGCGGTGGTGCG |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | DQ845175 | 60 | 102.21 | −3.27 | 0.998 | ATTATGGACAGGACCGAACGG | CCAACAGGTCGGCAAAGAACT |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-Monooxygenase activation protein zeta polypeptide | DQ845179 | 60 | 98.84 | −3.35 | 0.999 | CCAACGCTTCACAAGCAGAGA | TGCTTGCTGTGACTGGTCCA |
| HMBS | Hydroxymethylbilane synthase mRNA | DQ845174 | 60 | 97.63 | −3.39 | 0.998 | AGCATGCCTTGGAGAGGAATG | CGCTTGCAGACAGCTCCAAT |
| 18S | 18 S ribosomal RNA | DQ222453.1 | 60 | 95.68 | −3.43 | 0.993 | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT |
| EEF1A2 | Eukaryotic translation elongation factor 1 alpha 2 | BC108110.1 | 60 | 101.78 | −3.29 | 0.997 | CGACTGGCCACCTCATCTACA | CCAGGCATACTTGAAGGAGCC |
Real-time quantitative PCR
Real-time quantitative PCR was performed using three biological replicates and technical triplicates of each cDNA sample, according to the manufacturer’s protocol. cDNA samples were made into seven dilution series (1:10), and each dilution was amplified in triplicate PCR amplifications and plotted as mean values to generate a standard curve. PCR was performed using the same volume of cDNA sample for each gene to record the cycle threshold (Ct) value. PCR experiments were performed with an ABI Prism 7900 HT sequence-detection system (Applied Biosystems), using SYBR Green PCR Master Mix according to the manufacturer’s protocol. A 50 μL reaction sample included 5 μL SYBR Green Real-time PCR Master Mix, 0.2 μL (10 μM) primers, 1 μL sample, 0.2 μL ROX reference dye, and 3.4 μL nuclease-free water. The following cycling conditions were used: 95°C for 30 s and 40 cycles of 95°C for five seconds, and 60°C for 30 s. Fluorescence signal was obtained at a temperature of 72°C extension.
Data analysis
All data are representative of experiments performed at least three times in duplicate. Data are mean±standard deviation (SD). The Ct value of each gene after qRT-PCR was input into an Excel datasheet (Microsoft Excel 2003) before importing the data into geNorm (Vandesompele et al., 2002), NormFinder (Pfaffl et al., 2004) and Bestkeeper (Andersen et al., 2004) software packages.
geNorm was used to evaluate the stability of reference genes and for screening the optimal combination by using two parameters M (average expression stability) and V (pairwise variation). geNorm generates an M value for each gene that is arbitrarily suggested to be lower than 1.5 (with a lower value indicating increased gene stability across samples), and a pairwise stability measure (V number) to determine the benefit of adding extra reference genes for the normalization process (again with a lower value indicating greater stability of the normalization factor). Lower M values for the references indicated higher stability. Paired variation analysis was carried out using geNorm after the optimal combination of two reference genes was obtained. If the paired variation coefficient was lower than 0.15, the combination of these two genes was sufficient to be used as the criteria for calibrating the results from quantitative analysis, and the next gene was not required. By contrast, if the paired variation coefficient was higher than 0.15, the next genes should be added to the analysis (Mehta et al., 2010).
Similarly, NormFinder generates a stability measure of which a lower value indicates increased stability in gene expression and group samples. This allows a direct estimation of expression variation, ranking genes according to the similarity of their expression profiles by using a model-based approach. Bestkeeper generates a pairwise correlation coefficient between each gene and the Bestkeeper index (the geometric mean of the threshold cycle values of all the reference genes grouped together) (Perez et al., 2008).
RESULTS
Amplification efficiency and specificity of qRT-PCR
As shown in Table 1, the regression coefficients of the standard curves for the eight candidates reference genes were all higher than 0.99. The amplification efficiencies were all higher than 94% and showed good linear correlation, indicating that the specificity of the primers was relatively good and that the results of the quantitative analysis were accurate and reliable. Mean Ct values and standard deviation for the genes in each tissue are shown in Table 2. The obtained Ct and amplification efficiency values were used in the geNorm, NormFinder and Bestkeeper algorithms to calculate expression stability of the investigated reference genes for each tissue.
Table 2.
Mean cycle threshold (Ct) values and standard deviation for candidate reference genes in examined tissues
| Tissues | ACTB | GAPDH | TBP | HMBS | HPRT1 | EEF1A2 | YWHAZ | 18S |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| ------------------------------------------------------ Mean±SD ------------------------------------------------------ | ||||||||
| Heart | 20.02±0.15 | 18.71±0.41 | 28.55±0.13 | 25.54±1.15 | 24.62±0.27 | 17.86±0.50 | 20.87±1.23 | 10.38±0.36 |
| Liver | 19.72±0.06 | 20.48±0.43 | 28.65±0.90 | 25.85±0.82 | 21.24±2.13 | 22.31±0.21 | 21.90±0.89 | 11.36±1.50 |
| Spleen | 17.40±0.17 | 21.38±0.43 | 28.29±0.39 | 27.31±0.69 | 23.79±0.24 | 21.84±0.41 | 20.66±0.95 | 10.74±0.40 |
| Lung | 17.91±0.48 | 22.11±0.06 | 28.07±0.38 | 26.80±0.39 | 23.64±0.45 | 21.67±0.25 | 20.48±0.63 | 10.93±0.91 |
| Kidney | 18.86±0.45 | 19.77±0.37 | 28.20±0.25 | 24.92±0.16 | 24.63±0.07 | 21.98±0.12 | 21.00±0.31 | 9.77±0.39 |
| Stomach | 17.94±0.17 | 21.19±0.53 | 28.13±0.29 | 25.43±0.73 | 22.39±0.91 | 21.45±0.31 | 18.86±0.66 | 10.26±0.31 |
| Muscle | 20.56±0.68 | 17.19±0.69 | 29.51±0.14 | 26.58±1.04 | 24.72±0.69 | 18.47±0.73 | 22.26±1.22 | 10.67±0.38 |
| Small intestine | 18.03±1.01 | 21.00±0.91 | 28.79±0.82 | 25.69±1.40 | 24.72±0.62 | 21.67±1.06 | 20.82±1.44 | 9.99±0.18 |
| Uterus | 18.34±0.04 | 20.96±0.18 | 27.77±0.17 | 26.21±0.16 | 24.06±0.22 | 21.20±0.11 | 20.48±0.75 | 10.39±0.16 |
| Ovary | 18.11±0.13 | 19.94±0.31 | 27.23±0.32 | 26.19±0.50 | 23.55±0.67 | 20.63±0.20 | 19.95±1.02 | 9.81±1.05 |
Stability of reference gene expression
Firstly, the stability was evaluated taking into account the entire data (all tissues and goats) with geNorm, NormFinder and Bestkeeper algorithms. Table 3 lists the stability of reference gene expression obtained from the geNorm analysis (from high to low): 18S, TBP, YWHAZ, HMBS, ACTB, HPRT1, GAPDH, and EEF1A2 (Figure 1). The stability of the reference gene expression obtained by NormFinder analysis is as follows (from high to low): 18S, TBP, HMBS, YWHAZ, ACTB, HPRT1, GAPDH, and EEF1A2. The stability of the reference gene expression obtained using Bestkeeper analysis is (from high to low): 18S, TBP, HMBS, YWHAZ, HPRT1, ACTB, GAPDH, and EEF1A2.
Table 3.
Expression stability values of the candidate reference genes calculated by geNorm, NormFinder and Bestkeeper algorithms (ranking in parentheses)
| Gene | Stability value geNorm | Stability value NormFinder | Stability value Bestkeeper |
|---|---|---|---|
| 18S | 0.634 (1) | 0.156 (1) | 0.396 (1) |
| TBP | 0.634 (2) | 0.467 (2) | 0.445 (2) |
| YWHAZ | 0.722 (3) | 0.813 (4) | 0.642 (4) |
| HMBS | 0.799 (4) | 0.584 (3) | 0.566 (3) |
| ACTB | 0.887 (5) | 1.203 (5) | 0.881 (6) |
| HPRT1 | 1.048 (6) | 1.297 (6) | 0.825 (5) |
| GAPDH | 1.297 (7) | 1.550 (7) | 1.096 (7) |
| EEF1A2 | 1.425 (8) | 1.585 (8) | 1.153 (8) |
Figure 1.
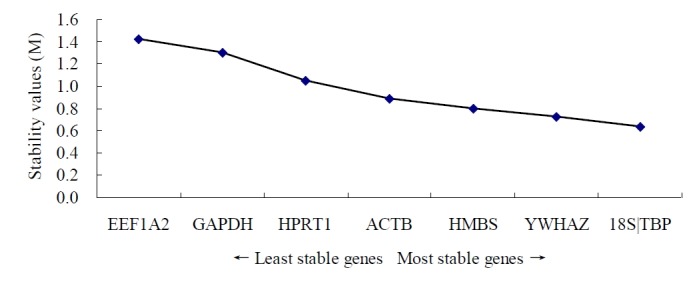
pGene expression stability of candidate reference genes. Gene expression stability of candidate reference genes in goat tissues analyzed by the geNorm program which proceeds to the stepwise exclusion of the genes whose relative expression levels are more variable among tissue samples. Threshold for eliminating a gene as unstable was M ≥ 1.5. Lower values of M correspond to the most stable genes, hence the most suitable for normalization.
From the analytical results of the three software packages, it can be deduced that 18S and TBP displayed the best gene expression stability, while GAPDH and EEF1A2 are the least stable. The gene expression stability of HMBS, YWHAZ, ACTB, and HPRT1 is moderate.
The paired variation coefficient of geNorm was higher than 0.15 (V2/3 = 0.234) when 18S and TBP are used in combination as the reference genes, indicating that the reference gene combination with 18S and TBP was not sufficient to be used as the calibration criteria for the quantitative analysis for gene expression, and the next candidate for reference gene was required (Figure 2). When YWHAZ, HMBS, and ACTB (ranked in third, fourth and fifth places in expression stability) were successively added, the variation coefficient showed 0.197 (V3/4), 0.183 (V4/5) and 0.210 (V5/6). As 0.183 (V4/5) was the smallest variation coefficient, so HMBS should be added and 18S, TBP, and HMBS should be the optimal combination of reference genes.
Figure 2.
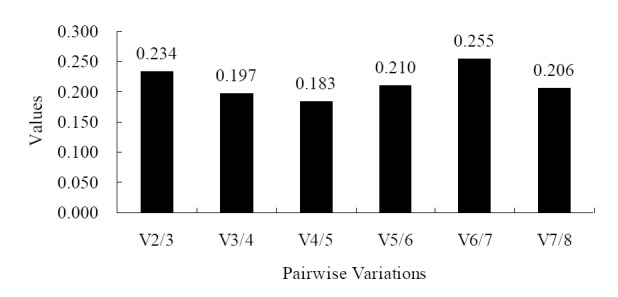
Evaluation of the optimum number of reference genes according to the geNorm software. The magnitude of the change in the normalization factor after the inclusion of an additional reference gene reflects the improvement obtained. Vi/i+1 represent the models being compared: those with i and i+1 reference genes.
The stability of the genes was also evaluated for each of the ten tissues analysed. As expected, the stability of the genes varies among tissues (Table 4). ACTB was the most stable gene in stomach, small intestine and ovary, 18S in heart and spleen, HMBS in uterus and lung, TBP in liver, HPRT1 in kidney and GAPDH in muscle, evidencing the specificity of each tissue developing characteristic biological functions and specific metabolic pathways. These results showed that tissue is an important variability factor that affects the stability genes expression. Thus, it is evidenced the necessity of using reference genes according the tissue analysed.
Table 4.
Stability values (M-values) for each reference gene calculated by using geNorm algorithm
| Tissues | ACTB | GAPDH | TBP | HMBS | HPRT1 | EEF1A2 | YWHAZ | 18S |
|---|---|---|---|---|---|---|---|---|
| Heart | 0.390(5) | 0.348(4) | 0.260(3) | 0.853(8) | 0.433(6) | 0.173(2) | 0.671(7) | 0.171(1) |
| Liver | 0.532(5) | 0.439(3) | 0.357(1) | 0.359(2) | 1.299(8) | 0.477(4) | 0.722(6) | 1.024(7) |
| Spleen | 0.227(3) | 0.294(4) | 0.206(2) | 0.394(5) | 0.471(6) | 0.536(7) | 0.610(8) | 0.203(1) |
| Lung | 0.413(6) | 0.354(4) | 0.311(3) | 0.242(1) | 0.245(2) | 0.387(5) | 0.623(8) | 0.518(7) |
| Kidney | 0.261(7) | 0.239(6) | 0.176(4) | 0.148(3) | 0.051(1) | 0.055(2) | 0.324(8) | 0.221(5) |
| Stomach | 0.148(1) | 0.407(5) | 0.301(3) | 0.512(6) | 0.640(8) | 0.152(2) | 0.577(7) | 0.373(4) |
| Muscle | 0.714(8) | 0.147(1) | 0.620(6) | 0.381(3) | 0.556(5) | 0.149(2) | 0.430(4) | 0.664(7) |
| Small intestine | 0.107(1) | 0.110(2) | 0.231(3) | 0.554(7) | 0.335(4) | 0.411(5) | 0.486(6) | 0.633(8) |
| Uterus | 0.123(3) | 0.188(5) | 0.218(7) | 0.058(1) | 0.202(6) | 0.062(2) | 0.359(8) | 0.181(4) |
| Ovary | 0.205(1) | 0.209(2) | 0.497(6) | 0.373(4) | 0.435(5) | 0.317(3) | 0.831(8) | 0.680(7) |
Numbers into brackets show stability values sorted out for each tissue from 1 (most stable) to 8 (less stable). The stability values of the most stable gene for each group are marked in bold.
DISCUSSION
The techniques for quantitative analysis of gene expression at mRNA level include RNA blotting, gene microarray, and qRT-PCR. The final results from the expression of target genes should be calibrated using reference genes (Kidd et al., 2007; Kortner et al., 2011; Nesvadbova and Knoll, 2011; Lardizabal et al., 2012; Rivera-Vega et al., 2012). Ideal reference genes should be stably expressed in the tissues, cells and under the experimental conditions used in the experimental system. However, the so-called constant expression of any reference gene only refers to the specific conditions under certain environments, and the expression levels in different cell types, tissues and different experimental systems can be significant (Radonic et al., 2004; Bustin et al., 2009; Lanoix et al., 2012). Therefore, it is particularly important to use reference genes suitable for the specific experimental conditions and species under investigation.
Studies on reference genes in mammals have previously been reported. Nygard et al. (2007) screened for reference genes in different tissues by using SYBR Green qRT-PCR technique. The results showed that the expression of ACTB, RPL4, TPB, and HPRT1 was the most stable; ACTB and RPL4 can be used as the reference genes for analyzing gene expression of high abundance genes, while TPB and HPRT1 can be used as the reference genes for analyzing gene expression of low abundance genes (Nygard et al., 2007). Lisowski et al. (2008) screened reference genes in different tissues sourced from cattle. The results showed that the stability of the expression of reference genes was not the same in different tissues. The expression of ACTB and TBP was the most stable in liver, the expression of GAPDH and YWHAZ was the most stable in kidney tissue, the expression of GAPDH and SDHA was the most stable in pituitary tissue, and the expression of TBP and HPRTI was the most stable in thyroid (Lisowski et al., 2008). Crespo et al. (2005) found that different combinations from six reference genes (ACTB, YWHAZ, RPL19, GAPDH, G6PDH, and SDHA) were applicable for the calibration of the results from qRT-PCR analysis in different sheep tissues. For example, GAPDH, SDHA, and ACTB were suitable for cerebrum, G6PDH, ACTB, and YWHAZ were suitable for cerebellum, while SDHA, RPL19, and GAPDH were suitable for mesenteric lymph node (Garcia-Crespo et al., 2005). As the optimal reference genes for different species and different tissues are not necessarily the same, it is essential to determine the optimal reference genes for specific species used in the experimental system.
The present study was carried out to examine the expression levels of eight candidates reference genes (18S, TBP, HMBS, YWHAZ, ACTB, HPRT1, GAPDH, and EEF1A2) in heart, liver, spleen, lung, kidney, stomach, uterus, ovaries, small intestines, and muscles tissues sourced from Boer goat. Real-time quantitative PCR technique and three software packages (geNorm, NormFinder and Bestkeeper) were used to analyze the results. Though the algorithms utilized by the three software were slightly different, they all sorted the candidate reference genes by their level of expression stability.
Even though the stability calculations of the eight reference genes obtained from geNorm, NormFinder and Bestkeeper were slightly different, the two groups of genes showing the best (18S, TBP) and the poorest (GAPDH, EEF1A2) stability were the same (Table 3). This implies that GAPDH and EEF1A2 are not suitable to be used as the reference for quantitative analysis of gene expression in goat tissues. In addition, the expression of the most frequently used reference gene ACTB was not sufficiently stable in the present study. The optimal combination of reference genes obtained by using geNorm was 18S and TBP, but the paired variation coefficient of them was higher than 0.15 (V2/3 = 0.234), indicating that they were not sufficient to be used as the criteria for calibrating the quantitative analysis for gene expression in goat tissues, and other candidates of reference genes were required (Figure 2). The paired variation analysis indicated that the variation coefficient showed 0.183 (V4/5, the smallest among all variation coefficients) for HMBS when its stability values were added. Using more than three reference gene candidates in combination may increase the level of difficulty and introduce errors into the experiment, and may also increase the cost of the experiment (Vandesompele et al., 2002; Schmid et al., 2003; Lardizabal et al., 2012). We therefore recommend that in the present study, the combination of three reference genes (18S, TBP, and HMBS) should be used for the calibration of the results from quantitative analysis of gene expression in goat tissues.
The present study was carried out to identify suitable reference genes (combination) in ten different tissue types sourced from Boer goats for the first time by using qRT-PCR technique. The results showed that the frequently used reference genes in other mammals (pigs, cattle, sheep and others) may not be suitable for goats. The expression of 18S, TBP, and HMBS was the most stable in the ten goat tissue types, and their combination was the most suitable to be used as the reference for quantitative analysis on gene expression in goat tissues. Our future studies will focus on consolidating the best optimal reference genes identified in the present study. We will use the results to calibrate the quantitative analysis of major functional genes in goats in order to obtain accurate, reliable and significant data. The data will provide a solid theoretical basis for illustrating molecular regulatory mechanisms of economically important traits in goats and accelerating variety/breed improvements in goats.
CONCLUSION
In conclusion, this work evaluated the stability of eight genes in different goat tissues showing that they could be used as reference genes. Stability values reflect that tissue is an important variability factor and it must be taken into consideration in the experimental design. It is recommended the use of three reference genes: 18S, TBP, and HMBS in studies which include multiple tissues. For studies in a specific tissue, the most stable reference gene varies between the tissues studied. In stomach, small intestine and ovary it is recommended ACTB. In heart and spleen the best option is 18S, while in uterus and lung we recommend using HMBS. If the study is focused in liver, we encourage using TBP, but if working with kidney and muscle, the most stable gene to be used as reference gene is HPRT1 and GAPDH, respectively.
Result of the gel electrophoresis of total RNA isolated from goat tissue samples. 1: heart, 2: liver, 3: spleen, 4: lung, 5: kidney, 6: stomach, 7: small intestine, 8: uterus, 9; ovary, 10: muscle. RNA samples were run on a 1% agarose gel at 100 V for 30 min. All the samples show high-quality RNA because there is a clear appearance of the 28S, 18S and 5S rRNA bands. The results indicated that total RNA was isolated successfully and with no degradation and could be used for further research.
Acknowledgments
This work was supported by grants from the National High Science and Technology Foundation of China “863” (No. 2011AA100307-4), the National Natural Science Foundation of China (No. 31171200), the Technology Innovation Project Special Program and the Science and Technology Program of Anhui Province (No. 11Z0101095 and No. 11010302108).
Footnotes
AUTHOR’S CONTRIBUTIONS
These studies were designed by Yu Zhang, Xiao-Dong Zhang and Yun-Hai Zhang. Yu Zhang and Xiao-Dong Zhang carried out all the experimental analyses and prepared all figures and tables. Yu Zhang, Xiao-Dong Zhang and Yun-Hai Zhang analyzed the data and drafted the manuscript. Xing Liu and Yun-Sheng Li contributed to revisions of the manuscript. Jian-Ping Ding and Xiao-Rong Zhang assisted in explaining the results and revised the final version of the manuscript. All authors have read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors report no conflicts of interest with this study.
REFERENCES
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chechi K, Gelinas Y, Mathieu P, Deshaies Y, Richard D. Validation of reference genes for the relative quantification of gene expression in human epicardial adipose tissue. PloS one. 2012;7:e32265. doi: 10.1371/journal.pone.0032265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Hu R, Yang Q, Zhang X, Pan L, Chen N, Chen M, Yang Z, Wang T, He Y, Yu S. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics. 2012;287:167–176. doi: 10.1007/s00438-011-0665-5. [DOI] [PubMed] [Google Scholar]
- Dabek J, Ligus J, Szota J. Oligonucleotide microarray and QRT-PCR study of adhesion protein gene expression in acute coronary syndrome patients. Inflammation. 2010;33:398–407. doi: 10.1007/s10753-010-9198-z. [DOI] [PubMed] [Google Scholar]
- Domby E, Engle T, Han H. The relationship of tissue copper concentrations and genes involved in copper homeostasis in the cow, pig, and goat. J Dairy Sci. 2010;93:403–404. [Google Scholar]
- Dubeuf JP, Morand-Fehr P, Rubino R. Situation, changes and future of goat industry around the world. Small Rumin Res. 2004;51:165–173. [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fu Y, Shi Z, Wu M, Zhang J, Jia L, Chen X. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder(Paralichthys olivaceus) Plos One. 2011;6:e22957. doi: 10.1371/journal.pone.0022957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Crespo D, Juste RA, Hurtado A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res. 2005;1:3. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M, Nadler B, Mane S, Eick G, Malfertheiner M, Champaneria M, Pfragner R, Modlin I. GeneChip, geNorm, and gastrointestinal tumors: novel reference genes for real-time PCR. Physiol Genomics. 2007;30:363–370. doi: 10.1152/physiolgenomics.00251.2006. [DOI] [PubMed] [Google Scholar]
- Kortner TM, Valen EC, Kortner H, Marjara IS, Krogdahl A, Bakke AM. Candidate reference genes for quantitative real-time PCR (qPCR) assays during development of a diet-related enteropathy in Atlantic salmon (Salmo salar L.) and the potential pitfalls of uncritical use of normalization software tools. Aquaculture. 2011;318:355–363. [Google Scholar]
- Lanoix D, Lacasse AA, St-Pierre J, Taylor SC, Ethier-Chiasson M, Lafond J, Vaillancourt C. Quantitative PCR pitfalls: The case of the human placenta. Mol Biotechnol. 2012;52:234–243. doi: 10.1007/s12033-012-9539-2. [DOI] [PubMed] [Google Scholar]
- Lardizabal MN, Nocito AL, Daniele SM, Ornella LA, Palatnik JF, Veggi LM. Reference genes for real-time PCR quantification of microRNAs and messenger RNAs in rat models of hepatotoxicity. PloS one. 2012;7:e36323. doi: 10.1371/journal.pone.0036323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski P, Pierzchala M, Goscik J, Pareek CS, Zwierzchowski L. Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. J Appl Genet. 2008;49:367–372. doi: 10.1007/BF03195635. [DOI] [PubMed] [Google Scholar]
- Mehta R, Birerdinc A, Hossain N, Afendy A, Chandhoke V, Younossi Z, Baranova A. Validation of endogenous reference genes for qRT-PCR analysis of human visceral adipose samples. BMC Mol Biol. 2010;11:39. doi: 10.1186/1471-2199-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moioli B, D’Andrea M, Pilla F. Candidate genes affecting sheep and goat milk quality. Small Rumin Res. 2007;68:179–192. [Google Scholar]
- Nesvadbova M, Knoll A. Evaluation of reference genes for gene expression studies in pig muscle tissue by real-time PCR. Czech J Anim Sci. 2011;56:213–216. [Google Scholar]
- Novinscak A, Gadkar VJ, Filion M. Optimization of RNA isolation and qRT-PCR strategies to monitor microbial gene expression in soil. Phytopathology. 2011;101:S130–S130. [Google Scholar]
- Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67. doi: 10.1186/1471-2199-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez R, Tupac-Yupanqui I, Dunner S. Evaluation of suitable reference genes for gene expression studies in bovine muscular tissue. BMC Mol Biol. 2008;9:79. doi: 10.1186/1471-2199-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Pierzchala M, Pareek CS, Urbanski P, Goluch D, Kamyczek M, Rozycki M, Kuryl J. Selection of reference genes for gene expression studies in porcine hepatic tissue using quantitative real-time polymerase chain reaction. Anim Sci Pap Rep. 2011;29:53–63. [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Rivera-Vega L, Mamidala P, Koch JL, Mason ME, Mittapalli O. Evaluation of Reference Genes for Expression Studies in Ash (Fraxinus spp.) Plant Mol Biol Rep. 2012;30:242–245. [Google Scholar]
- Schmid H, Cohen CD, Henger A, Irrgang S, Schlondorff D, Kretzler M. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003;64:356–360. doi: 10.1046/j.1523-1755.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- Spalenza V, Girolami F, Bevilacqua C, Riondato F, Rasero R, Nebbia C, Sacchi P, Martin P. Identification of internal control genes for quantitative expression analysis by real-time PCR in bovine peripheral lymphocytes. Vet J. 2011;189:278–283. doi: 10.1016/j.tvjl.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veys K, Labro AJ, De Schutter E, Snyders DJ. Quantitative single-cell ion-channel gene expression profiling through an improved qRT-PCR technique combined with whole cell patch clamp. J Neurosci Methods. 2012;209:227–234. doi: 10.1016/j.jneumeth.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Wang GP, Xu CS. Reference gene selection for real-time RT-PCR in eight kinds of rat regenerating hepatic cells. Mol Biotechnol. 2010;46:49–57. doi: 10.1007/s12033-010-9274-5. [DOI] [PubMed] [Google Scholar]
- Yoon JR, Laible PD, Gu M, Scott HN, Collart FR. Express primer tool for high-throughput gene cloning and expression. Biotechniques. 2002;33:1328–1333. doi: 10.2144/02336bc03. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tang ZL, Wang N, Long LQ, Li K. Evaluating a set of reference genes for expression normalization in multiple tissues and skeletal muscle at different development stages in pigs using quantitative real-time polymerase chain reaction. DNA Cell Biol. 2012;31:106–113. doi: 10.1089/dna.2011.1249. [DOI] [PubMed] [Google Scholar]
- Zi XD, Xu HW, Wang Y. Variation in sequences and mRNA expression levels of inhibin subunits alpha (INHA) and beta A (INHBA) genes between prolific and nonprolific goat breeds. Mol Reprod Dev. 2012;79:238–238. doi: 10.1002/mrd.22001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Result of the gel electrophoresis of total RNA isolated from goat tissue samples. 1: heart, 2: liver, 3: spleen, 4: lung, 5: kidney, 6: stomach, 7: small intestine, 8: uterus, 9; ovary, 10: muscle. RNA samples were run on a 1% agarose gel at 100 V for 30 min. All the samples show high-quality RNA because there is a clear appearance of the 28S, 18S and 5S rRNA bands. The results indicated that total RNA was isolated successfully and with no degradation and could be used for further research.


