Abstract
In this study, we investigated the relationship between unsaturated fatty acids influencing beef flavor and four types of SNPs (c.280A>G, c.388G>A, c.408G>C and c.456A>G) located at exon 2, 3 and 4 of the FABP4 gene, which is a fatty acid binding protein 4 in Korean cattle (n = 513). When analyzing the relationship between single genotype, fatty acids and carcass trait, individuals of GG, GG, CC and GG genotypes that are homozygotes, had a higher content of unsaturated fatty acids and marbling scores than other genotypes (p<0.05). Then, haplotype block showed strong significant relationships not only with unsaturated fatty acids (54.73%), but also with marbling scores (5.82) in ht1×ht1 group (p<0.05). This ht1×ht1 group showed significant differences with unsaturated fatty acids and marbling scores that affected beef flavor in Korean cattle. Therefore, it can be inferred that the ht1×ht1 types might be valuable new markers for use in the improvement of Korean cattle.
Keywords: Fatty Acid Binding Protein 4 (FABP4), Korean Cattle, Unsaturated Fatty Acid, Haplotype, Beef Flavor
INTRODUCTION
Consumer taste depends on the quality, health and flavor of beef. The quality of the fat is determined by its fatty acid composition, and beef quality depends not only on the marbling score but also on fatty acid composition (Cameron et al., 1991; Matsubara et al., 1998; Laborde et al., 2001).
Fatty acid composition of intramuscular tissue in cattle has become important in the beef industry because fat tissue containing abundant mono-unsaturated fatty acid reflects a lower melting point, which contributes positively to beef flavor and tenderness (Dryden et al., 1970; Jeremiah et al., 1996; Melton et al., 1982; Sturdivant et al., 1992). Oleic acid within fatty acid, which accounts for the highest percentage of fatty acid in bovine adipose tissue, makes a significant contribution to beef flavor (Dryden et al., 1970; Yosimura et al., 1983; Tsuji et al., 2008). These factors are also of great interest because of their implications for human health. Saturated fatty acids are considered to have the most harmful cardiovascular effects (Mozaffarian et al., 2005). In contrast, mono-unsaturated fatty acids increase hepatic low density lipoprotein (LDL) receptor activity, thereby decreasing the circulating concentration of LDL-cholesterol (Woollet et al., 1992; Rudel et al., 1995). Therefore, juicier beef has been reported to show higher unsaturated fatty acid content and higher beef quality, and it was estimated that a higher content of oleic acid suggested an already deep relationship between several factors, along with unsaturated fatty acid, that influence beef flavor (Waldman et al., 1965). Nevertheless, knowledge about the genetic variation of fatty acid composition is limited.
The fatty acid binding protein 4 (FABP4) gene plays an important role in lipid hydrolysis and intracellular fatty acid trafficking in different tissues (Damcoot et al., 2004; Chmurzynska et al., 2006). The uptake of intracellular fatty acid is essential as a signal molecule to trigger the process of preadipocyte differentiation and terminal differentiation-related gene expression (Amri et al., 1994; Duplus et al., 2 000).
Recently, several reports have shown the relationship of single nucleotide polymorphisms (SNPs) of the bovine FABP4 to fatty acid composition. Two SNPs of FABP4 have been identified that positively affect marbling score and subcutaneous fat depth, and fall into a significant QTL interval for beef marbling associated with BTA 14 (Michal et al., 2006; Pannier et al., 2010). The variation of c.220 A>G (I74V) associated with palmitoleic acid in adipose tissue of beef cattle has been described in crosses between Japanese Black Cattle and Holstein (p<0.05) (Hoashi et al., 2008; Mannen et al., 2011; Narukami et al., 2011). In addition, two SNPs in FABP4, g.2774 G>C (intron1) and g.3631 G>A (exon3) affect carcass weight and marbling score in Korean cattle (Lee et al., 2010). Another study showed associations of carcass weight and back fat thickness with c.220 A>G (I74V) and c.328 G>A (V110M) (p<0.05) (Cho et al., 2007).
FABP4, which is highly associated with oleic acid, is an important factor for beef flavor, and has been considered a valuable factor for use in improvement of beef quality. Therefore, this study was conducted to obtain genetic information on exonic SNPs of FABP4 and their association with unsaturated fatty acids which could make them important factors in higher beef quality and for application of these factors for improvement of beef flavor in Korean cattle.
MATERIALS AND METHODS
Sampling and phenotypic data
Korean cattle (called Hanwoo in Korea) population (n = 513) were bred in Gyeungbuk, Korea. Steers in Gyeonbuk province were fed according to the feeding program of each farmer. They were the progeny of 18 sires. The pedigree records of 513 steers were collected by the Charmpoom Hanwoo Research Institution. In general, they were weaned at 6 months of age, castrated at 6 months of age, fed with growth stage feed for 18 months, and given a high concentration diet in the last 6 months. Twenty-four hours after slaughter, carcass weight was measured and the carcass traits were dissected at the last rib and the first lumber vertebra according to the Animal Product Grading System of Korea for measurement of carcass traits. All steers were slaughtered at 941 days of age (mean±SD: 941.36±71.51), with an average carcass weight of 427.25 kg. Back fat thickness and marbling score were measured or scored in the left carcass cut across the vertebra between the last thoracic vertebra and the first lumbar vertebra. The marbling degree was scored from 1 to 9 with a mean of 5.43 where a larger score meant more abundant intramuscular fat (Table 1).
Table 1.
Phenotypic mean and standard deviation (SD) of Korean cattle used in the current genetic association analysis
| Trait | Description | Mean | SD |
|---|---|---|---|
| Carcass trait | |||
| CW (kg) | Carcass weight | 427.25 | 43.28 |
| BFT (cm) | Backfat thickness | 13.22 | 5.13 |
| MS | Marbling score | 5.43 | 1.93 |
| Fatty acid composition (%) | |||
| C14:0 | Myristic acid | 3.63 | 0.65 |
| C16:0 | Palmitic acid | 25.65 | 1.96 |
| C18:0 | Stearic acid | 10.48 | 1.39 |
| C14:1 | Myristoleic acid | 1.24 | 0.38 |
| C16:1 | Palmitoleic acid | 6.61 | 0.97 |
| C18:1 | Oleic acid | 44.3 | 2.66 |
| C18:2n6 | Linoleic acid | 3.02 | 0.76 |
| C18:3n3 | Linolenic acid | 0.35 | 0.2 |
| SFA | Saturated fatty acid | 40.6 | 2.86 |
| MUFA | Monounsaturated fatty acid | 53.5 | 2.96 |
| Fatty acid index | |||
| M/S3 | MUFA/SFA | 1.32 | 0.16 |
| IC14 | (C14:1/(C14:0+C14:1))×100 | 25.37 | 6.37 |
| IC16 | (C16:1/(C16:0+C16:1))×100 | 20.5 | 2.81 |
| IC18 | (C18:1/(C18:0+C18:1))×100 | 80.86 | 2.43 |
Fatty acid analysis
Total lipids were extracted from approximately 500 mg of m. longissimus dorsi muscle with chloroform: methanol (2:1, vol/vol) according to the methods of Folch (Folch et al., 1957). Extracted lipids were then methylated by the method of O’Keefe (O’Keefe et al., 1968) with sodium methylate. The extract was then filtered through filter paper in a water bath (40°C). The filtrate was mixed with distilled water, from which a layer of methanol and water was removed. Then, following removal of the chloroform and lipid layers using nitrogen gas, the sample was treated with BF3-methanol (14%) and then subjected to transmethylation at 65°C. Total composition of fatty acids was analyzed using gas-chromatography (Perkin-Elmer CO., USA). These included myristic acid, palmitic acid, stearic acid, myristoleic acid, palmitoleic acid, oleic acid, linoleic acid, and linolenic acid. The components and their functions were used as phenotypes for genetic association analyses (Table 1).
Sequencing of FABP4 and SNP genotyping
Total genomic DNA was extracted from m. longissimus dorsi muscle using the COSMO GENETECH kit (COSMO co, Ltd. Korea). Exonic SNPs in FABP4 were selected based on the nucleotide sequence of the bovine FABP4 in GenBank (Accession no. NM_174314.2), and genotypes at 9 SNPs were preliminary analyzed. Primers for amplification and extension were designed for single-base extension (Vreeland et al., 2002) using forward, reverse, and extension primer sequences (Table 2).
Table 2.
The information of primer sequence for SNPs of FABP4 (Accession number: NM_174314.2) gene of BTA 14 in Korean cattle
| SNP | Exon | Sequences | Product size | |
|---|---|---|---|---|
| c.181T>G | 2 | F1 | TTGACATTTTTCTTTTCCCAAC | 200 |
| R2 | TGACTTTCCTGTCATCTGGAG | |||
| E3 | CTGGCATGGCCAAACCCACT | |||
| c.193T>G | 2 | F | TTGACATTTTTCTTTTCCCAAC | 200 |
| R | TGACTTTCCTGTCATCTGGAG | |||
| E | AACCCACTTTGATCATCAGT | |||
| c.212C>A | 2 | F | TATAGGCGTGGGCTTTGCTA | 202 |
| R | GCTCCAGTTCTTTATTTCTCACC | |||
| E | TTTGAATGGGGGTGTGGTCA | |||
| c.280A>G | 2 | F | GGGGTGTGGTCACCATTAAA | 178 |
| R | TGGTTTGCTATAGGCAGCAG | |||
| E | TGGGCCAGGAATTTGATGAA | |||
| c.352T>G | 3 | F | CAAGGGCGATTGTCTATTTCTC | 208 |
| R | ATTATCCCCACAGAGCATCG | |||
| E | CTCTGGTACAAGTACAAAAC | |||
| c.364T>C | 3 | F | TTATGGGATGACCTAGCACTAAAA | 201 |
| R | TTTTTCCCTCCATCATTGTAATC | |||
| E | TACAAAACTGGGATGGAAAA | |||
| c.388G>A | 3 | F | ATTATCCCCACAGAGCATCG | 219 |
| R | TTTTCATTCTACAAGGGCGATT | |||
| E | CCACCATAAAGAGAAAACTC | |||
| c.408G>C | 3 | F | CATCGTAAACTTAGATGAAGGTGCT | 200 |
| R | CATTCTACAAGGGCGATTGTCT | |||
| E | GTGGATGATAAGATGGTGCT | |||
| c.456A>G | 4 | F | TTCCTGCTCAACATTGAAGG | 190 |
| R | GTCATGGAGTTCGATGCAAA | |||
| E | AACAGAGTTTATGAGAGAGC |
Forward sequence.
Reverse sequence.
Extension sequence.
Reactions of primer extension were performed using the SNaPshot ddNTP Primer Extension Kit (Applied Biosystems, Foster City, CA, USA). To purify the reactions of primer extension, mixtures containing exonuclase 1 and shrimp alkaline phosphatase were added to the reaction mixtures. Samples were cultured at 37°C for 1 h and then inactivated at 72°C for 15 min. PCR products were well mixed by addition of Genescan 120 LIZ standard and HiDi formamide (Applied Biosystems, Foster City, CA, USA), followed by denaturation at 95 °C for 5 min. Electrophoresis was then performed using an ABI PRISM 3130XL Genetic Analyzer, followed by an assay of electrophoresis products using GeneMapper v.4.0 software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
For statistical analysis, heterozygosity (H), minor allele frequency (MAF) and haplotype block were tested using Haploviewer v4.2 (Barret et al., 2005). Hardy-Weinberg equilibrium (HWE) in the examined population was tested by comparing expected and observed genotype frequencies using the Chi-square test.
Associations between individual SNPs and carcass traits and fatty acid composition in the 513 animals were analyzed by mixed ANCOVA (analysis of covariance) linear model using SPSS v19.0 software (SPSS Inc., Chicago, IL, USA). The following model was used: Yijk = (μ+Si+Pj+Gk+β×age+eijk in which Yijk is the phenotypic observations of traits, μ is the overall mean of each trait, Si is the random effect of sire, Pj is the fixed effect of the place at the calving (14 classes), Gk is fixed effect of the SNP genotypes, β is a regression coefficient, age is a covariate for age in days at slaughter, and eijk is a random residual. Significant differences among mean values of different genotypes were calculated using Duncan’s multiple range test and determined by p<0.05.
This genetic association model was applied to haplotype analysis as well as individual SNP analysis by replacing genotype effect with haplotype effect. For haplotype analysis, pairwise linkage (r2) was estimated, and linkage disequilibrium blocks were identified by the 4-gamete rule using Haploviewer v4.2 (Wang et al., 2002). Also, phase probabilities for each site were calculated for each individual using this software (PHASE) (input option: ignoring families). Using this software, phase probabilities of all polymorphic sites for haplotypes were calculated for each individual.
RESULTS
Nine candidate SNPs of exons in FABP4 were statistically analyzed to determine the genotypic and allelic frequencies, heterozygote, MAF (minor allele frequency) and HWE (Hardy-Weinberg equilibrium), shown in Table 3.
Table 3.
Genotype frequency distribution of exonic variants in FABP4 (Accession number: NM_174314.2) in Korean cattle
| SNP | Exon | AA change | Genotype frequency (N) | H1 | MAF2 | HWE3 | ||
|---|---|---|---|---|---|---|---|---|
| c.181T>G | 2 | L41V | TT 1.000 (44) |
TG 0.000 (0) |
GG 0.000 (0) |
0.00 | 0.00 | - |
| c.193T>G | 2 | L45V | TT 1.000 (44) |
TG 0.000 (0) |
GG 0.000 (0) |
0.00 | 0.00 | - |
| c.212C>A | 2 | T51N | CC 1.000 (44) |
CA 0.000 (0) |
AA 0.000 (0) |
0.00 | 0.00 | - |
| c.280A>G | 2 | I74V | AA 0.386 (17) |
AG 0.409 (18) |
GG 0.205 (9) |
0.48 | 0.41 | 0.429 |
| c.352T>G | 3 | W98G | TT 1.000 (44) |
TG 0.000 (0) |
GG 0.000 (0) |
0.00 | 0.00 | - |
| c.364T>C | 3 | S102P | TT 1.000 (44) |
TC 0.000 (0) |
CC 0.000 (0) |
0.00 | 0.00 | - |
| c.388G>A | 3 | V110M | GG 0.591 (26) |
GA 0.341 (15) |
AA 0.068 (3) |
0.36 | 0.28 | 0.681 |
| c.408G>C | 3 | L116L | GG 0.363 (16) |
GC 0.477 (21) |
CC 0.160 (8) |
0.47 | 0.44 | 0.808 |
| c.456A>G | 4 | A132A | AA 0.386 (17) |
AG 0.409 (18) |
GG 0.205 (9) |
0.48 | 0.41 | 0.429 |
Heterozygosity.
Minor allele frequency.
p-value for testing Hardy-Weinberg equilibrium.
Table 3 shows five monomorphic loci in Korean cattle were excluded in the following genetic association analysis. MAF for the other loci ranged from 0.28 to 0.44, and their genotypes did not deviate from the Hardy-Weinberg equilibrium (p>0.05, Table 3). A total of four selected SNPs were associated with a variety of fatty acid composition and carcass traits (p<0.05, Table 4). First, comparison between these results for carcass weight and the final selected four SNPs did not affect Korean cattle. However, for back fat thickness, GG genotypes were the lowest at 11.91 in c.280A>G SNP (p<0.04), for marbling score, c.388G>A SNPs were the highest at 5.66 in GG genotypes, and there were statistically significant differences (p<0.004). Then, for palmitic acid which has a high proportion of saturated fatty acids, c.408G>C SNP was the highest at 25.61, 25.79 in GG, GC genotypes (p<0.02) and the content of SFA had a similar tendency at 40.75, 40.77 and 40.75 in AG, GC and AG genotypes (p<0.05).
Table 4.
Genotypic effects of exonic variants in FABP4 (Accession number: NM_174314.2) on fatty acid composition of intramuscular fat
| SNP | c.280A>G | c.388G>A | c.408G>C | c.456A>G | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | AA | AG | GG | p-value | GG | GA | AA | p-value | GG | GC | CC | p-value | AA | AG | GG | p-value | |
| N | 155 | 277 | 81 | 286 | 209 | 18 | 178 | 294 | 41 | 155 | 277 | 81 | |||||
| Carcass trait | |||||||||||||||||
| CW (kg) | 430.81±3.44 | 425.48±2.61 | 426.48±4.87 | 0.465 | 431.05±2.61 | 423.25±2.90 | 413.17±9.19 | 0.052 | 428.65±3.09 | 425.44±2.61 | 434.10±6.27 | 0.423 | 429.23±3.38 | 425.48±2.60 | 429.49±5.06 | 0.605 | |
| BFT (cm) | 13.61±0.42b | 13.38±0.31b | 11.91±0.45a | 0.040 | 12.97±0.28 | 13.67±0.38 | 12.06±1.14 | 0.202 | 13.20±0.40 | 13.29±0.29 | 12.78±0.65 | 0.835 | 13.08±0.42 | 13.38±0.31 | 12.93±0.49 | 0.719 | |
| MS | 5.43±0.15 | 5.32±0.12 | 5.78±0.21 | 0.181 | 5.66±0.11b | 5.17±0.13ab | 4.67±0.41a | 0.004 | 5.36±0.14 | 5.34±0.11 | 5.93±0.30 | 0.182 | 5.47±0.15 | 5.32±0.12 | 5.69±0.21 | 0.309 | |
| Fatty acid composition (%) | |||||||||||||||||
| C14:0 | 3.67±0.05 | 3.66±0.04 | 3.52±0.06 | 0.193 | 3.63±0.04 | 3.64±0.04 | 3.80±0.16 | 0.572 | 3.64±0.05 | 3.65±0.04 | 3.47±0.09 | 0.226 | 3.67±0.05 | 3.66±0.04 | 3.50±0.07 | 0.130 | |
| C16:0 | 25.75±0.15 | 25.66±0.11 | 25.42±0.23 | 0.496 | 25.59±0.12 | 25.65±0.13 | 26.58±0.51 | 0.119 | 25.61±0.15b | 25.79±0.11b | 24.88±0.27a | 0.020 | 25.75±0.15 | 25.66±0.12 | 25.42±0.23 | 0.478 | |
| C18:0 | 10.40±0.10 | 10.59±0.08 | 10.26±0.15 | 0.125 | 10.40±0.08 | 10.57±0.09 | 10.73±0.35 | 0.329 | 10.43±0.10 | 10.55±0.08 | 10.21±0.18 | 0.277 | 10.37±0.10 | 10.59±0.08 | 10.32±0.15 | 0.159 | |
| C14:1 | 1.24±0.03 | 1.22±0.02 | 1.31±0.04 | 0.234 | 1.24±0.02 | 1.25±0.03 | 1.15±0.09 | 0.587 | 1.24±0.03 | 1.24±0.02 | 1.28±0.06 | 0.801 | 1.25±0.03 | 1.22±0.02 | 1.29±0.04 | 0.373 | |
| C16:1 | 6.69±0.07 | 6.54±0.06 | 6.70±0.11 | 0.216 | 6.65±0.06 | 6.56±0.06 | 6.49±0.29 | 0.554 | 6.65±0.07 | 6.58±0.06 | 6.60±0.12 | 0.811 | 6.70±0.08 | 6.54±0.06 | 6.67±0.11 | 0.213 | |
| C18:1 | 44.16±0.20a | 44.15±0.16a | 45.08±0.30b | 0.016 | 44.56±0.16b | 44.04±0.17ab | 43.16±0.51a | 0.019 | 44.31±0.19a | 44.09±0.15a | 45.71±0.38b | 0.001 | 44.16±0.20a | 44.15±0.16a | 45.06±0.30b | 0.020 | |
| C18:2n6 | 3.08±0.06b | 3.06±0.04b | 2.78±0.08a | 0.009 | 2.94±0.05 | 3.13±0.05 | 3.02±0.22 | 0.053 | 3.06±0.06b | 3.05±0.04b | 2.65±0.11a | 0.005 | 3.07±0.06b | 3.06±0.04b | 2.81±0.08a | 0.021 | |
| C18:3n3 | 0.36±0.01 | 0.34±0.01 | 0.38±0.02 | 0.276 | 0.36±0.01 | 0.34±0.01 | 0.33±0.04 | 0.666 | 0.36±0.01 | 0.34±0.01 | 0.38±0.03 | 0.337 | 0.36±0.01 | 0.34±0.01 | 0.38±0.02 | 0.247 | |
| SFA | 40.65±0.22ab | 40.75±0.17b | 40.02±0.32a | 0.038 | 40.42±0.17 | 40.76±0.19 | 41.59±0.63 | 0.137 | 40.56±0.21b | 40.77±0.16b | 39.62±0.41a | 0.047 | 40.65±0.22ab | 40.75±0.17b | 40.02±0.32a | 0.038 | |
| MUFA | 53.49±0.22a | 53.24±0.18a | 54.43±0.33b | 0.006 | 53.84±0.17b | 53.16±0.19ab | 52.21±0.65a | 0.007 | 53.58±0.22a | 53.28±0.17a | 54.74±0.39b | 0.012 | 53.51±0.23a | 53.24±0.17a | 54.40±0.33b | 0.008 | |
| Fatty acid index | |||||||||||||||||
| M/S3 | 1.33±0.01a | 1.32±0.01a | 1.37±0.01b | 0.024 | 1.34±0.01b | 1.31±0.01ab | 1.26±0.03a | 0.030 | 1.33±0.01a | 1.32±0.01a | 1.39±0.02b | 0.020 | 1.33±0.01a | 1.32±0.01a | 1.37±0.02b | 0.026 | |
| IC14 | 25.31±0.50 | 24.92±0.37 | 27.03±0.76 | 0.232 | 25.52±0.38 | 25.37±0.43 | 23.02±1.39 | 0.272 | 25.32±0.45 | 25.21±0.38 | 26.77±0.90 | 0.337 | 25.39±0.50 | 24.92±0.37 | 26.88±0.76 | 0.056 | |
| IC16 | 20.63±0.21 | 20.32±0.17 | 20.87±0.35 | 0.229 | 20.65±0.16 | 20.38±0.19 | 19.55±0.66 | 0.200 | 20.62±0.20 | 20.35±0.16 | 21.01±0.37 | 0.289 | 20.66±0.22 | 20.32±0.17 | 20.83±0.34 | 0.253 | |
| IC18 | 80.92±0.19ab | 80.65±0.15a | 81.45±0.25b | 0.030 | 81.05±0.14 | 80.65±0.17 | 80.09±0.62 | 0.073 | 80.93±0.17 | 80.69±0.14 | 81.74±0.28 | 0.078 | 80.97±0.18 | 80.65±0.15 | 81.36±0.26 | 0.055 | |
Means with different superscripts in the same row within each SNP indicate statistical difference with p<0.05.
SFASaturated fatty acid, MUFA Mono unsaturated fatty acid, M/S Mono unsaturated fatty acid/Saturated fatty acid, IC14 (C14:1/(C14:0 +C14:1))×100, IC16(C16:1/(C16:0+C16:1))×100, IC18 (C18:1/(C18:0+C18:1))×100, CW carcass weight, BFT backfat thickness, MS marbling score from 1 to 9 indicating that a larger score means more abundant intramuscular fat.
On the other hand, there was a statistically significant difference in unsaturated fatty acids, oleic acid and MUFA, because they were the highest at 45.08 and 54.43 in GG genotype of c.280A>G SNP (p<0.05) and 44.56 and 53.84 in GG genotype of c.388G>A SNP (p<0.05). Also, they had higher contents of oleic acid and MUFA than individuals of other genotypes, since their composition in the CC genotype of c.408G>C SNP, GG genotype of c.456A>G SNP were 45.71, 54.74 and 45.06, 54.40, respectively (p<0.05).
Four polymorphic SNPs located in exons showed a significant difference in content of unsaturated fatty acids by analysis of linkage disequilibrium. Thus, the haplotype block was prepared and the results are shown in Table 5 and Figure 1.
Table 5.
Haplotype frequency distribution of exonic variants by linkage disequilibrium (LD) block of FABP4 in Korean cattle
| Block | c.280A>G | c.388G>A | c.408G>C | c.456A>G | Frequency |
|---|---|---|---|---|---|
| ht1 | G | G | C | G | 0.351646 |
| ht2 | A | G | G | A | 0.328343 |
| ht3 | A | A | G | A | 0.227501 |
| G | G | G | G | 0.061786 | |
| Except | G | G | G | A | 0.006100 |
| A | G | C | G | 0.005441 | |
| A | G | G | G | 0.004699 | |
| G | A | G | A | 0.003970 | |
| G | A | C | G | 0.003161 | |
| A | A | C | A | 0.002993 | |
| A | G | C | A | 0.002945 | |
| G | A | G | G | 0.000939 |
Figure 1.
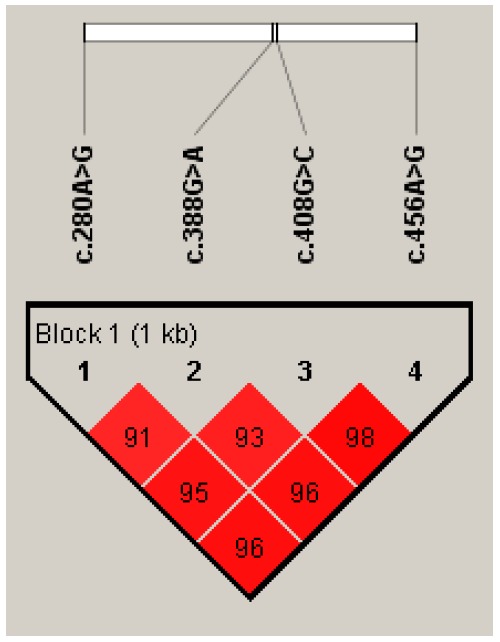
Pairwise linkage estimates between exonic SNPs in FABP4 gene.
Haplotype block included c.280A>G in exon 2 and c.388G>A, c.408G>C in exon 3, and c.456A>G in exon 4. Haplotypic associations of block were found with fatty acid compositions (p<0.05, Table 6). The haplotypes were further associated with marbling score (p<0.038), but not with carcass weight and back fat thickness (p>0.05, Table 6) as shown in the single locus analysis.
Table 6.
Haplotypic effects within each linkage disequilibrium (LD) block in FABP4 on fatty acid composition of intramuscular fat
| Trait | LD Block
|
||||||
|---|---|---|---|---|---|---|---|
| ht1×ht1 (N = 38) |
ht1×ht2 (N = 136) |
ht1×ht3 (N = 151) |
ht2×ht2 (N = 56) |
ht2×ht3 (N = 87) |
ht3×ht3 (N = 31) |
p-value | |
| Carcass trait | |||||||
| CW (kg) | 431.63±6.55 | 430.62±3.74 | 419.54±3.62 | 435.95±6.14 | 428.64±4.03 | 419.19±7.31 | 0.089 |
| BFT (cm) | 12.39±0.65 | 13.17±0.42 | 13.11±0.42 | 13.20±0.71 | 13.44±0.53 | 13.77±1.11 | 0.899 |
| MS | 5.82±0.31b | 5.51±0.17ab | 5.18±0.15ab | 5.80±0.24b | 5.49±0.21ab | 4.68±0.29a | 0.038 |
| Fatty acid composition (%) | |||||||
| C14:0 | 3.46±0.09a | 3.77±0.05b | 3.56±0.05ab | 3.46±0.07a | 3.72±0.07b | 3.78±0.10b | 0.004 |
| C16:0 | 24.84±0.29a | 25.90±0.15b | 25.67±0.16ab | 25.22±0.28ab | 25.73±0.20ab | 25.97±0.38b | 0.031 |
| C18:0 | 10.19±0.18 | 10.42±0.11 | 10.63±0.12 | 10.21±0.17 | 10.51±0.13 | 10.52±0.27 | 0.338 |
| C14:1 | 1.27±0.06 | 1.23±0.03 | 1.25±0.03 | 1.23±0.04 | 1.25±0.04 | 1.19±0.07 | 0.956 |
| C16:1 | 6.64±0.12 | 6.68±0.07 | 6.51±0.08 | 6.66±0.11 | 6.64±0.10 | 6.63±0.18 | 0.796 |
| C18:1 | 45.68±0.40c | 43.99±0.23a | 44.19±0.21ab | 45.17±0.34bc | 44.00±0.28a | 43.61±0.43a | 0.001 |
| C18:2n6 | 2.67±0.12a | 3.07±0.06bc | 3.04±0.05bc | 2.87±0.12ab | 3.22±0.08c | 3.03±0.16bc | 0.005 |
| C18:3n3 | 0.38±0.03ab | 0.32±0.01a | 0.36±0.02ab | 0.42±0.03b | 0.34±0.02ab | 0.32±0.03a | 0.038 |
| SFA | 39.56±0.43a | 40.85±0.22bc | 40.66±0.24bc | 39.84±0.40ab | 40.85±0.28bc | 41.02±0.52c | 0.044 |
| MUFA | 54.73±0.42c | 53.29±0.24ab | 53.27±0.25ab | 54.35±0.38bc | 53.32±0.29ab | 52.80±0.53a | 0.011 |
| Fatty acid index | |||||||
| M/S | 1.39±0.02c | 1.31±0.01a | 1.32±0.01ab | 1.38±0.02bc | 1.32±0.01a | 1.29±0.03a | 0.013 |
| IC14 | 26.65±0.95 | 24.54±0.55 | 25.79±0.52 | 26.24±0.75 | 25.07±0.67 | 23.71±1.01 | 0.169 |
| IC16 | 21.13±0.39 | 20.50±0.22 | 20.25±0.25 | 20.95±0.36 | 20.52±0.29 | 20.35±0.49 | 0.466 |
| IC18 | 81.75±0.29c | 80.83±0.19ab | 80.61±0.21ab | 81.52±0.31bc | 80.69±0.24ab | 80.55±0.51a | 0.037 |
Means with different superscripts in the same row within each linkage disequilibrium block indicate statistical difference with p<0.05.
SFA = Saturated fatty acid, MUFA = Mono unsaturated fatty acid, M/S = Mono unsaturated fatty acid/Saturated fatty acid, IC14 = (C14:1/(C14:0+C14:1))×100, IC16= (C16:1/(C16:0+C16:1))×100, IC18 = (C18:1/(C18:0+C18:1))×100, CW = Carcass weight, BFT = Backfat thickness, MS = Marbling score from 1 to 9 indicating that a larger score means more abundant intramuscular fat.
DISCUSSION
In this study, we investigated the association between exonic SNPs of FABP4, fatty acid and carcass traits in Korean cattle. Table 4 shows that the selected four SNPs had significant association with oleic acid and MUFA among the fatty acids. In particular, c.280A>G SNP had an association with back fat thickness, and in case of c.388G>A SNP, with marbling score. Also, it has been reported that c.220 A>G (I74V) SNP has an association with back fat thickness (Cho et al., 2007), and g.3631 G>A (V110M) SNP has an association with marbling score (Lee et al., 2010). The results drawn by the two studies are remarkably consistent.
On the other hand, I74V SNP only associated with palmitoleic acid in Holstein Cattle (Narukami et al., 2011), and V110M SNP did not have any association with fatty acid in Japanese Black Cattle (Hoashi et al., 2008; Mannen et al., 2011). These results can be considered to be different from our results because the breeds are different. Also, Michal et al. (2006) reported that bovine FABP4 genotype significantly affected the marbling score and the depth of subcutaneous fat in Wagyu×Limousine F2 population. In case of SNPs that Michal et al. (2006) studied, the fact that they are located in the part of 3’UTR in FABP4 is different from the positions of the four SNPs we investigated, so there is a slight difference in the conclusion. However, we suggest that there was a coincidence with the result that each SNP of FABP4 had associations not only with fatty acids but also with back fat thickness and marbling score.
Among these four SNPs, c.280A>G and c.388G>A are non-synonymous SNPs that have a change in the amino acid sequence caused by the nucleotide changes. Especially, in the case of c.280A>G SNP, as ATC nucleotide into GTC, changes the encoded amino acid from isoleucine to valine. c.388G>A SNP is also a non-synonymous SNP where valine replaces methionine as GTG nucleotide changes into ATG. On the other hand, c.408G>C and c.456A>G are synonymous SNPs, which do not have changes of amino acid, but do have nucleotide changes. These two novel SNPs (c.408G>C, c.456A>G) were not found to have an association with fatty acid type.
The LD coefficients (r2) between individual SNPs, which reached the values for 5% level of genome-wide significance for single-marker associations, were also calculated in order to assess LD, and haplotype blocks were identified using the so-called four-gamete rule (Wang et al., 2002). In the four-gamete rule approach, the population frequencies of the four possible two-marker haplotypes were computed. If all four were observed with frequency of at least 0.01, recombination is deemed to have taken place. Blocks were then formed by consecutive markers where only three gametes were observed. These analyses were carried out in Haploviewer v4.2 (Barret et al., 2005).
Table 6 shows that, homozygous genotypes with G, G, C and G allele at c.280A>G, c.388G>A, c.408G>C and c.456A>G had an increased proportion of oleic acid, the major content of monounsaturated fatty acids, and thus an increased proportion of monounsaturated fatty acids. Similarly, there was a decreased proportion of palmitic acid, the major content of saturated fatty acids, and thus decreased proportion of saturated fatty acids.
In regards to the relationship with carcass traits, the homozygous genotypes did not have any association with carcass weight and back fat thickness, but in marbling score, in the highest level of significance was in the ht1×ht1 group, which is the same as the result of fatty acid (p<0.05) (Table 6). Especially, there was no single gene association for palmitic acid and myristic acid. However, based on the conclusions which showed significance in the result of haplotype, it can be seen that the effect of haplotype genes were better than the effect of single gene. Simultaneous use of multiple markers would accelerate genetic improvement by such haplotypic effects or by epistatic effects (Ryu et al., 2011).
Accordingly, it is expected that haplotype blocks of c.280A>G c.388G>A, c.408G>C, and c.456A>G SNPs at exon 2, 3, and 4 are able to increase the contents of unsaturated fatty acids and marbling score associated with beef flavor, using significant relationships with the unsaturated fatty acids, and marbling score. Therefore, such genotypes as those of the ht1×ht1 groups, having a higher content of unsaturated fatty acids and intramuscular marbling scores associated with beef flavor, could make a positive contribution to the development of the Korean cattle industry.
ACKNOWLEDGEMENTS
This research was supported by the Yeungnam University research grants in 2010.
REFERENCES
- Amri EG, Ailhaud G, Grimaldi PA. Fatty acid as signal transducing molecules: involvement in the differentiation of preadipose to adipose cells. J Lipid Res. 1994;35:930–937. [PubMed] [Google Scholar]
- Barret JC, Fry B, Maller J, Daly MJ. Haploview: anaylsis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cameron ND, Enser MB. Fatty acid composition of lipid in longissimus dorsi muscle of Duroc and British landrace pigs and its relationship with eating quality. Meat Sci. 1991;29:295–307. doi: 10.1016/0309-1740(91)90009-F. [DOI] [PubMed] [Google Scholar]
- Chmurzynska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Cho S, Park TS, Yoon DH, Cheong HS, Namgoong S, Park BL, Lee HW, Han CS, Kim EM, Cheng Il-C, Kim H, Shin HD. Identification of genetic polymorphism in FABP3 and FABP4 and putative association with bak fat thickness in Korean native cattle. BMB Report. 2007;41:29–34. doi: 10.5483/bmbrep.2008.41.1.029. [DOI] [PubMed] [Google Scholar]
- Damcoot CM, Moffett SP, Feingold E, Barmade MM, Marshall JA, Hamman RF, Ferrel RF. Genetic variation in fatty acid-binding protein-4 and peroxisome proliferator-activated receptor gamma interactively influence insulin sensitivity and body composition in male. Metab Clin Exp. 2004;53:303–309. doi: 10.1016/j.metabol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Dryden FD, Marchello JA. Influence of total lipid and fatty acid composition upon the palatability of three bovine muscles. J Anim Sci. 1970;31:36–41. [Google Scholar]
- Duplus E, Glorian M, Forest C. Fatty acid regulation of gene transcription. J Biol Chem. 2000;267:30749–30752. doi: 10.1074/jbc.R000015200. [DOI] [PubMed] [Google Scholar]
- Folch J, Lee M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissue. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Hoashi S, Hinenoya T, Tanaka A, Ohsaki H, Sasazaki H, Taniguchi M, Oyama K, Mukai M, Mannen H. Association between fatty acid compositions and genotypes of FABP4 and LXR-alpha in Japanese Black cattle. BMC Genet. 2008;9:84. doi: 10.1186/1471-2156-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeremiah IE. The influence of subcutaneous fat thickness and marbling on beef. Food Res Int. 1996;29:513–520. [Google Scholar]
- Laborde FL, Mandell IB, Tosh JJ, Wilton JW, Buchanan-Smith JG. Breed effects on growth performance, carcass characteristics, fatty acid composition, and palatability attributes in finishing steers. J Anim Sci. 2001;79:355–365. doi: 10.2527/2001.792355x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Van der werf JHJ, Lee SH, Park EW, Oh SH, Gibson JP, Thompson JM. Genetic polymorphisms of the bovine fatty acid binding protein 4 gene are significantly associated with marbling and carcass weight in Hanwoo (Korean Cattle) Anim Genet. 2010;41:442–444. doi: 10.1111/j.1365-2052.2010.02024.x. [DOI] [PubMed] [Google Scholar]
- Mannen H. Identification and utilization of genes associated with beef qualities. J Anim Sci. 2011;82:1–7. doi: 10.1111/j.1740-0929.2010.00845.x. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Inoue Y, Tahara K, Nakagawa K. Quality of and consumer preference for marketing beef. Animal Husbandry. 1998;34:10–15. [Google Scholar]
- Melton SL, Amiri M, Davis GW, Backus WR. Flavor and chemical characteristics of ground beef from grass-, forage-, grain- and grain-finished steers. J Anim Sci. 1982;55:77–87. [Google Scholar]
- Michal JJ, Zhang ZW, Gaskins CT, Jiang Z. The bovine fatty acid binding protein 4 gene is significantly associated with marbling and subcutaneous fat depth in Wagyu×Limousin F2 crosses. Anim Genet. 2006;37:400–402. doi: 10.1111/j.1365-2052.2006.01464.x. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D, Ascherio A, Hu FB, Stampfer MJ, Willett WC. Interplay between different polyunsaturated fatty acids and risk of coronary heart disease in man. Circulation. 2005;111:257–164. doi: 10.1161/01.CIR.0000152099.87287.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narukami T, Sasazaki S, Oyama K, Nogi T, Taniguchi M, Mannen H. Effect of DNA polymorphism related to fatty acid composition in adipose tissue of Holstein cattle. J Anim Sci. 2011;82:406–411. doi: 10.1111/j.1740-0929.2010.00855.x. [DOI] [PubMed] [Google Scholar]
- O’Keefe PW, Wellington GH, Mattick LR, Stouffer JR. Composition of bovine muscle lipids at various carcass locations. J Food Sci. 1968;33:188–192. [Google Scholar]
- Pannier L, Mullen AM, Hanill RM, Stapleton PC, Sweeney T. Association analysis of single nucleotide polymorphism in DGAT1, TG and FABP4 genes and intramuscular fat in crossbred Bos Taurus cattle. Meat Sci. 2010;85:515–518. doi: 10.1016/j.meatsci.2010.02.025. [DOI] [PubMed] [Google Scholar]
- Rudel LL, Park JS, Sawyer JK. Compared with dietary monounsaturated and saturated fat, polyunsaturated fat protects African green monkeys from coronary artery atherosclerosis. Arterioscler Thromb Vasc Biol. 1995;15:2101–2110. doi: 10.1161/01.atv.15.12.2101. [DOI] [PubMed] [Google Scholar]
- Ryu J, Kim Y, Kim C, Kim J, Lee C. Association of bovine carcass phenotypes with genes in an adaptive thermogenesis pathway. Mol Biol Rep. 2011 doi: 10.1007/s11033-011-0880-5. [DOI] [PubMed] [Google Scholar]
- Sturdivant CA, Lunt DK, Smith GC, Smith SB. Fatty acid composition of subcutaneous and intramuscular adipose tissue and M. longissimus dorsi of Waygu cattle. Meat Sci. 1992;32:449–458. doi: 10.1016/0309-1740(92)90086-J. [DOI] [PubMed] [Google Scholar]
- Tsuji S. Kobe beef as international brand; Symposium of Gyeungbuk Hanwoo Cluster.2008. [Google Scholar]
- Vreeland WN, Meagher RJ, Barron AE. Multiplexed, highthroughput genotype by single_base extension and endlabeled free solution electrophoresis. Anal Chem. 2002;74:4328–4333. doi: 10.1021/ac0258094. [DOI] [PubMed] [Google Scholar]
- Waldman RC, Suess GG, Lewis RW, Bray RW, Brungardt VH. Certain fatty acids of bovine tissue and their association with carcass characteristics. J Anim Sci. 1965;24:869. [Google Scholar]
- Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollet LA, Spady DK, Dietschy JM. Saturated and unsaturated fatty acid independently regulate low density lipoprotein receptor activity and production rate. J Lipid Res. 1992;33:77–78. [PubMed] [Google Scholar]
- Yosimura T, Namikawa K. Influence of breed, sex and anatomical location on lipid and fatty acid composition of bovine subcutaneous fat. Jpn J Zootech Sci. 1983;54:97–100. [Google Scholar]


