Abstract
During embryo implantation in pigs, the uterine endometrium undergoes dramatic morphological and functional changes accompanied with dynamic gene expression. Since the greatest amount of embryonic losses occur during this period, it is essential to understand the expression and function of genes in the uterine endometrium. Although many reports have studied gene expression in the uterine endometrium during the estrous cycle and pregnancy, the pattern of global gene expression in the uterine endometrium in response to the presence of a conceptus (embryo/fetus and associated extraembryonic membranes) has not been completely determined. To better understand the expression of pregnancy-specific genes in the endometrium during the implantation period, we analyzed global gene expression in the endometrium on day (D) 12 and D15 of pregnancy and the estrous cycle using a microarray technique in order to identify differentially expressed endometrial genes between D12 of pregnancy and D12 of the estrous cycle and between D15 of pregnancy and D15 of the estrous cycle. Results showed that the global pattern of gene expression varied with pregnancy status. Among 23,937 genes analyzed, 99 and 213 up-regulated genes and 92 and 231 down-regulated genes were identified as differentially expressed genes (DEGs) in the uterine endometrium on D12 and D15 of pregnancy compared to D12 and D15 of the estrous cycle, respectively. Functional annotation clustering analysis showed that those DEGs included genes involved in immunity, steroidogenesis, cell-to-cell interaction, and tissue remodeling. These findings suggest that the implantation process regulates differential endometrial gene expression to support the establishment of pregnancy in pigs. Further analysis of the genes identified in this study will provide insight into the cellular and molecular bases of the implantation process in pigs.
Keywords: Pig, Uterus, Endometrium, Implantation, Microarray)
INTRODUCTION
Embryonic loss reaches up to approximately 20% by day (D) 20 of pregnancy in pigs (Bennett and Leymaster, 1989). The most critical period in pigs is considered to be D12 of pregnancy, the time when implantation of the conceptus (embryo/fetus and associated extraembryonic membranes) begins and maternal recognition of pregnancy occurs via estrogen secretion by the conceptus (Bazer and Thatcher, 1977; Geisert et al., 1982). It has been suggested that asynchrony of embryo development and inappropriate interactions between the conceptus and the uterine endometrium are major reasons for embryonic mortality during early pregnancy (Geisert and Yelich, 1997). Thus, to increase the rate of successful pregnancies, it is essential to understand the mechanism of the establishment of pregnancy.
The implantation process is a well-coordinated interaction between the developing conceptus and the maternal uterus (Geisert and Yelich, 1997). At the time of implantation, the conceptus produces estrogen, which acts as a signal for maternal recognition of pregnancy in pigs (Bazer et al., 1986). It has been suggested that estrogen reorients prostaglandin F2α (PGF2α) secretion from an endocrine to an exocrine manner and exerts an anti-luteolytic action (Spencer and Bazer, 2004). In addition to estrogen, the conceptus produces cytokines, such as interleukin (IL)-1β, interferon (IFN)-γ and IFN-δ, growth factors, and proteases (Tuo et al., 1996; Geisert and Yelich, 1997; Lefevre et al., 1998; Ross et al., 2003a). In response to these factors, the uterine endometrium undergoes dramatic morphological and functional changes to become receptive to the developing conceptus.
In the presence of a conceptus during early pregnancy, the expressions of several genes in the uterine endometrium are known to change. For example, fibroblast growth factor 7 (FGF7) is abundantly expressed in uterine endometrial epithelial cells during early pregnancy and is increased by estrogen treatment in endometrial explant cultures (Ka et al., 2001). Secreted phosphoprotein 1 (SPP1, or osteopontin), which binds to integrin receptors and functions in cell-to-cell attachment, is localized to endometrial luminal epithelial cells during early pregnancy and is induced by estrogen (Garlow et al., 2002; White et al., 2005). In addition, lysophosphatidic acid receptor 3 (L PAR3), which is a receptor for lysophosphatidic acid that possesses a growth factor-like activity, is expressed in uterine endometrial epithelial cells during early pregnancy and is induced by estrogen (Seo et al., 2008). Although it has been studied on the expression of many genes in the uterine endometrium during the estrous cycle and pregnancy, the pattern of global gene expression in the uterine endometrium in response to the presence of the conceptus has not been completely determined in pigs.
There have been many efforts to understand the expression and function of the uterine endometrial genes responsible for the establishment and maintenance of pregnancy using various gene expression analysis techniques in pigs. Suppression subtractive hybridization (SHH) has been used to determine novel gene expressions in Meishan-Landrace conceptuses and endometrial tissues at D15 of pregnancy (Vallee et al., 2003). In a recent study, we characterized several differentially expressed genes in the uterine endometrium on D12 of the estrous cycle and pregnancy using an annealing control primer (ACP)-based PCR (ACP-PCR) technique (Ka et al., 2009). Microarray technology has also been used to analyze gene expression in uterine endometrial tissues on D30 (Ka et al., 2008) or in cyclic uterine endometrium in pigs (Lee et al., 2005; Whitworth et al., 2005; Green et al., 2006). However, there are limited reports that have measured the global expression of genes in the uterine endometrium during embryo implantation in pigs.
Therefore, this study aimed to determine the global expression of uterine genes at the time of embryo implantation using a microarray technique in order to identify differentially expressed genes in the uterine endometrium on D12 and D15 of both the estrous cycle and of pregnancy. We compared microarray data from D12 and D15 of pregnancy to those from D12 and D15 of the estrous cycle and identified pregnancy-related variations in the patterns of global gene expression.
MATERIALS AND METHODS
Animals and tissue preparation
All experimental procedures involving animals were conducted in accordance with the Guide for Care and Use of Research Animals in Teaching and Research and approved by the Institutional Animal Care and Use Committee of Yonsei University. Crossbred sexually matured gilts that were either not mated or were mated with fertile boars after the onset of estrus were assigned to the D12 or D15 estrous cycle groups or the D12 or D15 pregnancy groups, respectively (n = 3 per d/status). Pregnancy was confirmed by the presence of an apparently normal conceptus in uterine flushings. Endometrium dissected from the myometrium was collected from the middle portion of the uterine horn. Endometrial tissues were snap-frozen in liquid nitrogen and stored at −80°C for RNA extraction.
RNA isolation
Total RNA from endometrial tissues was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) and the RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The purity and integrity of the total RNA were checked using NanoDrop (NanoDrop Technologies, Wilmington, DE, USA) and Experion (Bio-rad, Hercules, CA, USA), respectively.
Microarray analysis
Five micrograms of total RNA were used for labeling. Probe synthesis from the total RNA samples, hybridization, detection, and scanning were performed according to standard protocols from Affymetrix Inc. (Santa Clara, CA, USA). Briefly, cDNA was synthesized from total RNA using the One-Cycle cDNA Synthesis Kit (Affymetrix). Single-stranded cDNA was synthesized using Superscript II reverse transcriptase and T7-oligo primers at 42°C for 1 h Double-stranded (ds)-cDNA was obtained using DNA ligase, DNA polymerase I, and RNase H at 16°C for 2 h, followed by T4 DNA polymerase at 16°C for 5 min for gap filling. After clean-up with a Sample Cleanup Module (Affymetrix), ds-cDNA was used for in vitro transcription (IVT). cDNA was transcribed using the GeneChip IVT Labeling Kit (Affymetrix) in the presence of biotin-labeled CTP and UTP. Ten micrograms of labeled cRNA were fragmented to 35–200 bp using fragmentation buffer (Affymetrix). Fragmented cRNA was hybridized to the porcine genome (Affymetrix) at 45°C for 16 h according to the Affymetrix standard protocol. After hybridization, the arrays were washed in a GeneChip Fluidics Station 450 with a non-stringent wash buffer at 25°C followed by a stringent wash buffer at 50°C. After washing, the arrays were stained with a streptavidin-phycoerythrin complex. After staining, intensities were determined with a GeneChip scanner 3000 (Affymetrix) controlled by GeneChip Operating Software (Affymetrix).
Data analysis
Expression profiles were analyzed using GeneChip Operating Software (Affymetrix) to identify changes in expression levels in the uterine endometrium during the estrous cycle and early pregnancy. The operating software determined absolute analysis metrics (Detection, Detection p-value) using the scanned probe array data and compared the results between the different treatment group signals to generate the Change, Change p-value, and Signal log ratio (fold change) metrics after pre-processing of the raw data using the MAS5 algorithm. For normalization, data from each expression array were scaled to normalize the overall fluorescence intensity across each chip (average target intensity set at 500). The one-sided Wilcoxon’s signed rank test was employed to generate the detection p-value (p< 0.05). Two sets of algorithms were generated and used to determine change significance and change quantity metrics for every probe set. The change algorithm generated a Change p-value and an associated fold-change value, while the second algorithm gave a quantitative estimate of the change in gene expression in the form of the Signal log ratio. In the present study, the level of gene expression was considered to be increased if the Change p-value was less than 0.0025 and was considered to be decreased if the Change p-value was greater than 0.9975. Only relative changes equal to or greater than that of a two-fold level of expression were considered. We considered genes that matched the established Change p-value and the fold-change value to be differentially expressed genes. Probe identification was obtained using the gene ontology mining tool NetAffx (http://www.affymetrix.com/analysis/index.affx) and human homologues (Tsai et al., 2006).
Functional annotation clustering analysis
To evaluate biological functions of differentially expressed genes affected by pregnancy status in the uterine endometrium, we conducted functional annotation clustering analysis throughout DAVID program (Database for Annotation, Visualization, and Integrated Discovery, http://david.niaid.nih.gov/david/version2/index.htm) (Glynn et al., 2003). Official gene symbols of differentially expressed genes were submitted to DAVID for analysis of functional annotation clusters. Functional annotation clusters were determined by gathering related Gene Ontology (GO) terms with respect to biological process, cellular component, and molecular function. Because of limited number of annotated genes for porcine genome in NetAffx resulting from the limited availability of full-length porcine cDNA sequence, we used human gene symbols annotated for the Affymetrix porcine genome microarray probe identifications for the genes whose annotation was not available in NetAffx, as described in Tsai et al. (2006). Functional annotation clustering analysis using porcine and human gene symbols in DAVID program was conducted at highest stringency to clarify biological function of DEGs in the uterine endometrium. Functional groups were ordered by their overall enrichment score for the group based on the EASE scores of each term annotated.
Quantitative real-time RT-PCR
To analyze levels of selected differentially expressed gene mRNAs in the uterine endometrium on D12 and D15 of the estrous cycle and pregnancy, real-time RT-PCR was performed using the Applied Biosystems StepOnePlus System (Applied Biosystems, Foster City, CA, USA) using the SYBR Green method. Complementary DNAs were synthesized from 4 μg of total RNA isolated from different uterine endometrial tissues, and newly synthesized cDNAs (total volume of 21 μl) were diluted 1:4 with sterile water and then used for PCR. Specific primers based on porcine MUC5AC (GenBank accession number U10281.1; forward, 5′- CCA TCA TTT ACG AAG AGA CCG ACC-3′; reverse, 5′- GCC AGG TTT CAC CCT TCA TTC T-3′), NRIP1 (GenBank accession number BV726910.1; forward, 5′-TCA AAA CTC CCC TGA GTC CTC CTT TC-3′; reverse, 5′- ACG TCC TTG TCT TGT GTT TCT CGA CT-3′), RBP7 (GenBank accession number NM_001145222.1; forward, 5′- GAG CAA AAT GGG GAT TCT TTT ACC A-3′; reverse, 5′- ACA CTT GGC CTT CAC AGA ACA TCT C-3′), ADAMTS20 (GenBank accession number XM_003126621.1; forward, 5′- GTT GCT GAT GGT ACT CCT TGT GGA A-3′; reverse, 5′- CGA TAA CGC CAG GTA ATT GTC ATC T-3′), and porcine ribosomal protein L7 (RPL7), (GenBank accession number NM_001113217; forward, 5′-AAG CCA AGC ACT ATC ACA AGG AAT ACA-3′; reverse, 5′-TGC AAC ACC TTT CTG ACC TTT GG-3′) were designed to amplify cDNAs of 216 bp, 250 bp, 250 bp, 285 bp, and 172 bp, respectively. The Power SYBR Green PCR Master Mix (Applied Biosystems) was used for PCR reactions. Final reaction volume was 20 μl including 2 μl of cDNA, 10 μl of 2X Master mix, 2 μl of each primer, and 4 μl of dH2O. PCR conditions were 95°C for 15 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The results are reported as the expression relative to the level detected on D12 of the estrous cycle after normalization of the transcript amount to the endogenous RPL7 control by the 2−∆∆CT method (Livak and Schmittgen, 2001).
Statistic analysis
Data from real-time RT-PCR analysis for comparison of expression levels of selected genes in endometrium on D12 and D15 of the estrous cycle and pregnancy were subjected to the Student’s t test procedure of SAS, and are presented as means with standard errors.
RESULTS
Comparison of gene expression in the uterine endometrium between D12 and D15 of pregnancy and D12 and D15 of the estrous cycle
To determine global gene expression profiles in the uterine endometrium on D12 and D15 of the estrous cycle and of pregnancy in pigs, microarray analyses were performed using a total of 12 Affymetrix porcine genome microarray chips. To understand pregnancy-specific changes in endometrial gene expression, we identified differentially expressed genes with at least a two-fold increase or decrease in the uterine endometrium between D12 of pregnancy and D12 of the estrous cycle, as well as those with at least a two-fold increase or decrease between D15 of pregnancy and D15 of the estrous cycle.
After analyzing the raw data using the MAS5 algorithm of the GeneChip Operating Software, differentially expressed genes among the compared pairs were obtained. We considered genes that were below a Detection p-value of 0.05 and a two-fold change to be differentially expressed genes. Among 23,937 genes, 99 and 213 genes were up-regulated in the uterine endometrium on D12 and D15 of pregnancy compared to D12 and D15 of the estrous cycle, respectively. In addition, 92 and 231 genes were down-regulated in the endometrium on D12 and D15 of pregnancy compared to D12 and D15 of the estrous cycle, respectively. Volcano plot images showing comparative analyses of endometrial gene expression between the estrous cycle and pregnancy are shown in Figure 1.
Figure 1.
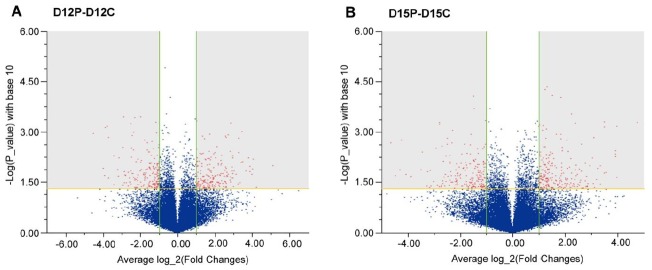
Volcano plot images for the comparative analysis of endometrial gene expression on day (D) 12 of pregnancy to D12 of the estrous cycle (A) and on D15 of pregnancy to D15 of the estrous cycle (B) in pigs. The X axis represents the average log_2 (fold-change), and the Y axis represents the -log (p-value) with base 10. Location in the upper left shaded area implies down-regulation; location in the upper right shaded area implies up-regulation. Cutoff values were below p<0.05 (horizontal line) and above an average two-fold change (vertical lines).
Genes differentially expressed on D12 of pregnancy compared to those of D12 of the estrous cycle
Genes that had higher expression in the uterine endometrium on D12 of pregnancy compared to those of D12 of the estrous cycle included sulfotransferase family 1E (SULT1E1), LPAR3, caspase 3 (CASP3), interleukin 1 receptor accessory protein (IL1RAP), and fibroblast growth factor 7 (FGF7) (Supplementary Table 1). Genes that had lower expression in the endometrium on D12 of pregnancy compared to those of D12 of the estrous cycle included glutathione S-transferase (GST), mucin (MUC5AC), solute carrier family 2 (SLC2A2), cytochrome P450 3A46 (CYP3A46), keratin 7 (KRT7), chemokine ligand 12 (CXCL12), and interferon regulatory factor 7 (IRF7) (Supplementary Table 2).
Genes differentially expressed on D15 of pregnancy compared to those of D15 of the estrous cycle
Genes that had higher expression in the uterine endometrium on D15 of pregnancy compared to those of D15 of the estrous cycle included endoplasmic reticulum degradation enhancer mannosidase alpha-like 3 (EDEM3), neurotrimin (NTM), UDP-glucose ceramide glucosyltransferase (UCGC), nuclear receptor interacting protein 1 (NRIP1), and SMAD family member 2 (SMAD2) (Supplementary Table 3). Genes that had lower expression in the endometrium on D15 of pregnancy compared to those of D15 of the estrous cycle included cellular retinol binding protein 7 (RBP7), parathyroid hormone (PTH), and myosin VI (MYO6) (Supplementary Table 4).
Functional annotation clustering analysis
To identify putative biological functions regulated by pregnancy in the uterine endometrium, we performed functional annotation clustering analysis of differentially expressed genes by the DAVID program. As a result, we obtained enriched functional groups in each cluster and clarified gene ontology (GO) terms or biological features of DEGs in the uterine endometrium (Figure 2). In genes up-regulated in the uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle, functional annotation clustering analysis showed that functional groups of membrane fraction and endoplasmic reticulum were enriched biological feature (Supplementary Table 5). Biological functions of cell adhesion and immune cell activation were enriched in genes down-regulated in uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle (Supplementary Table 6). RNA splicing and mRNA metabolism were enriched in genes up-regulated in the uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle (Supplementary Table 7), whereas cytoskeloton, microtubule, actin dynein related functions were enriched in genes down-regulated in the uterine endometrium on D15 of pregnancy compared to those on D15 of the estrous cycle (Supplementary Table 8).
Figure 2.
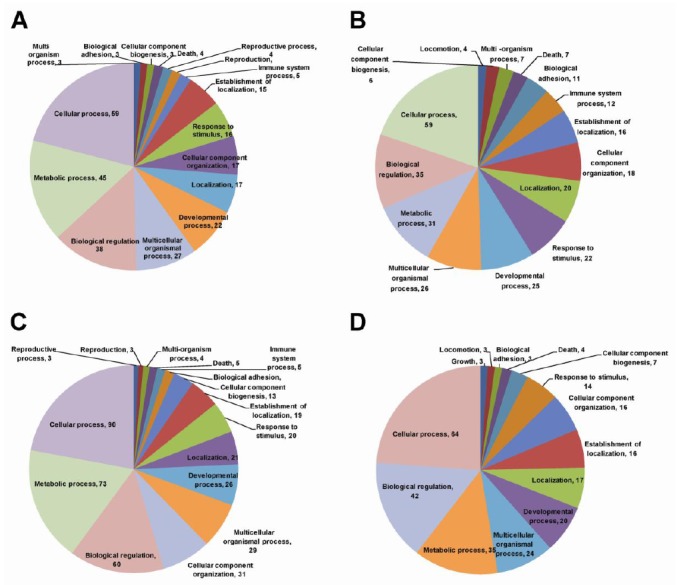
Functional annotation charts showing distribution of ontological analysis of up-regulated (A) and down-regulated genes (B) in the uterine endometrium on day (D) 12 of pregnancy compared with those on D12 of the estrous cycle, and up-regulated (C) and down-regulated genes (D) on D15 of pregnancy compared with those on D15 of the estrous cycle. Biological terms were identified by genes with respect to biological process at level one through DAVID. Gene ontology terms or biological features of differently expressed genes and the number of genes in each category are indicated.
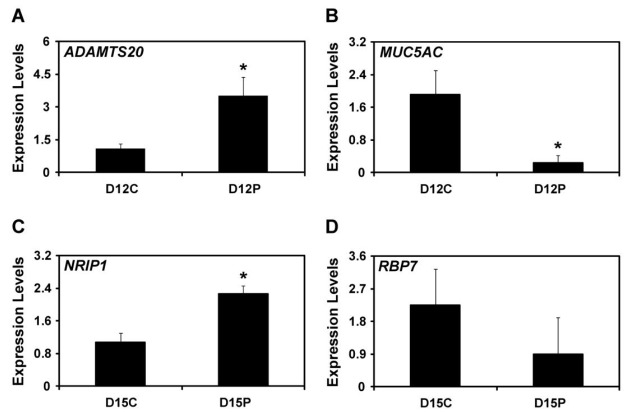
Real-time RT-PCR analysis of ADAMTS20, MUC5AC, NRIP1, and RBP7 in the uterine endometria on D12 and D15 of the estrous cycle and pregnancy
To determine validity of our microanalysis data, we selected ADAMTS20, MUC5AC, NRIP1, and RBP7 from genes up-regulated or down-regulated in comparison groups between D12 and D15 of the estrous cycle and pregnancy, and analyzed expression levels of those genes using realtime RT-PCR analysis. Levels of ADAMTS20 expression were significantly higher in the uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle (p<0.05). Expression levels of MUC5AC were significantly lower in the uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle (p<0.05). Levels of NRIP1 expression was significantly higher in the uterine endometrium on D15 of the pregnancy compared with those on D15 of the estrous cycle (p<0.05). However, levels of RBP7 expression were not different in the uterine endometrium between D15 of the estrous cycle and pregnancy (p>0.05).
DISCUSSION
The establishment of pregnancy is a complex process that requires well-coordinated interactions between the maternal uterus and the developing conceptus. In pigs, the implantation process begins around D12 of pregnancy and completes around D18 (Geisert and Yelich, 1997). During this period, the presence of the conceptus in the uterus induces changes in endometrial gene expression in preparation for conceptus attachment to the endometrial epithelial cells and extension of the life span of the corpus luteum in the ovaries (Spencer and Bazer, 2004). To better understand the implantation process, this study aimed to identify changes in endometrial gene expression on D12 and D15 of pregnancy in comparison to expression on D12 and D15 of the estrous cycle using a microarray technique. In this study, we identified genes that were up-regulated or down-regulated in the uterine endometrium during the implantation period. These genes that were differentially expressed according to pregnancy status included genes involved in immunity, steroidogenesis, cell-to-cell interaction, and tissue remodeling. Furthermore, functional annotation functional clustering analysis of these genes showed that biological functions of cell-to-cell interaction and immune cell activation were decreased in the uterine endometrium on D12 of pregnancy compared to those on D12 of the estrous cycle and RNA processing was increased in the uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle.
On around D12 of pregnancy in pigs, maternal recognition of pregnancy occurs through a mechanism explained by the endocrine-exocrine theory (Bazer and Thatcher, 1977). The elongating conceptus secretes estrogen into the uterine lumen, which, in turn, acts on the uterine endometrium and redirects the secretion of PGF2α toward the uterine lumen (exocrine) so that the corpus luteum (CL) function is maintained in the ovary. On the other hand, the absence of a conceptus in cyclic pigs on D12 causes secretion of PGF2α into the uterine vasculature (endocrine), which exerts luteolytic action on the CL. Thus, it is likely that the expression of endometrial genes on D12 of pregnancy is affected by the action of the estrogen originating from the conceptus. Indeed, it has been shown that the expressions of FGF7 and LPAR3 are up-regulated following estrogen treatment in uterine endometrium (Ka et al., 2008; Seo et al., 2008). In addition to estrogen, the conceptus also produces cytokines such as IL-1β, IFN-γ and IFN-δ (Spencer and Bazer, 2004), and the uterine environment during this period is also influenced by progesterone of ovarian origin. Therefore, expression of endometrial genes on D12 of pregnancy would be affected by a coordinated interaction of these factors.
Genes up-regulated in the endometrium of pregnant pigs on D12 of pregnancy compared to genes of D12 of the estrous cycle include SULT1E1, LPAR3, IL1RAP, and FGF7. Previous studies in pigs have demonstrated that these genes are expressed in the endometrium at significantly higher levels on D12 of pregnancy than on D12 of the estrous cycle (Ka et al., 2000; Kim et al., 2002; Ross et al., 2003b; Seo et al., 2008), indicating the validity of our microarray approach in analyzing gene expression. In addition, many other genes, including ADAMTS17, ADAMTS20, CASP3, CCNA2, and CYP19, were also identified as being up-regulated in the endometrium of pregnant pigs on D12 of pregnancy compared with genes on D12 of the estrous cycle. In functional annotation clustering analysis using genes up-regulated in the uterine endometrium on D12 of pregnancy compared with genes on D12 of the estrous cycle, any notable biological function was not found due to low relatedness of biological terms and low numbers of genes having similar function.
Down-regulated genes in the endometrium of pigs on D12 of pregnancy compared with genes on D12 of the estrous cycle include genes representing biological functions of leukocyte activation (CD86, CD8A, CXCL12, KLRK1, MLL5, PTPN22, and UNC13D), cell adhesion (AJAP1, CNTN5, CXCL12, CYFIP2, EGFL6, FAT1, ITGB4, ITGB7, MUC5AC, PKP2, and ROBO1), and cell junction (AJAP1, CYFIP2, GABRA5, GRIP1, PKP2, SHROOM2, and TNS3). During the implantation period in pigs, immune cell population changes and effector T cell population decreases in the subepithelial regions of the uterine endometrium (Croy et al., 1987; Bischof et al., 1996; Kaeoket et al., 2003), suggesting that maternal immune suppression contributes to the establishment of pregnancy. CD86, together with CD80, regulates T cell function as a ligand for CD28 and cytotoxic lymphocyte antigen-4 (CTLA-4), and expression of CD86 is detected in macrophages and dendritic cells at the feto-maternal interface in human (Banchereau et al., 2000) and mouse (Repnik et al., 2008). Interestingly, decreased expression of CD86 in the uterine endometrium with the conceptus on D12 of pregnancy compared D12 of the estrous cycle indicate that regulation of maternal uterine T cell function may also be important for the establishment of pregnancy in pigs. CXCL12 plays a key role in lymphocyte trafficking, cellular proliferation, organogenesis, vascularization, and embryogenesis (Murdoch, 2000; Wu et al., 2004). CXCL12 has been shown to be expressed in the uterine endometrium of humans and bovines (Hanna et al., 2003; Mansouri-Attia et al., 2009). In human trophoblasts and decidual cells, CXCL12 increases the invasiveness of trophoblasts and matrix metalloproteinase (MMP) 9 and MMP2 activity (Zhou et al., 2008) and has also been shown to be associated with endovascular invasion of CD16-NK cells in human placenta (Hanna et al., 2003). In this study, expression of CXCL12 decreased in pregnant uterine endometrium. Although its function in the uterine endometrium needs to be further elucidated during early pregnancy in pigs, modulation of CXCL12 action may also be important for embryo implantation.
During the implantation period in rodents, humans, and ruminants, it has been shown that expression of MUC1, a type of transmembrane mucin, is decreased in uterine endometrial epithelial cells to allow for adhesion of the conceptus trophectoderm to endometrial epithelial cells (Carson et al., 1998; Spencer et al., 2004). MUC5AC is a secreted, large, gel-forming type of mucin (Gendler and Spicer, 1995). Its expression has been measured in human uterine endocervical epithelial cells as a major form of gel-forming mucin in the menstrual cycle (Gipson et al., 1997), and it is associated with a change in eosinophilic cells in endometrial carcinomas (Moritani et al., 2005). Decreased expression of MUC5AC in pregnant uterine endometrium suggests that MUC5AC, in addition to MUC1, may block adhesion of the conceptus trophectoderm and must be removed for the trophectoderm to undergo epithelial cell attachment.
We also compared the genes expressed in the endometrium on D15 of pregnancy to those expressed on D15 of the estrous cycle. On D15 of the estrous cycle, the period of late diestrus in pigs, luteolysis is initiated by extended exposure of the endometrium to progesterone for 10 to 12 d, followed by increased pulsatile production of PGF2α (Ziecik, 2002). According to a study in sheep, the initial exposure to progesterone down-regulates the endometrial epithelial expressions of ESR1 and PGR, and the loss of the action of progesterone permits estrogen to increase ESR1 and OXTR in the uterine endometrium (Spencer and Bazer, 2004). Pulsatile release of oxytocin induces subluteolytic pulses of PGF2α from the uterine endometrium. PGF2α causes a supplemental release of oxytocin from the CL, which in turn stimulates the secretion of a luteolytic pulse of PGF2α from the uterine endometrium through a positive feedback mechanism. This positive feedback loop continues until the CL is depleted of oxytocin (Spencer and Bazer, 2004). On the other hand, on D15 of pregnancy, maternal recognition of pregnancy is established, and interdigitation between the chorion and endometrial epithelial cells develops to form the epitheliochorial placenta (Dantzer, 1985). During this time, the uterine environment is under the influence of progesterone, which is continuously produced from the ovaries, and cytokines originating from the conceptus, such as IFN-γ and IFN-δ (Cencic et al., 2003). It has been shown that a large amount of IFN-γ is secreted by the pig conceptus between d 12 and 20 of pregnancy (up to 250 mg per uterine horn), with the highest levels of synthesis on D15 and D16 (La Bonnardiere et al., 1991). At this time, ESR1 expression is decreased in the endometrium, and PGR is only detectable in stromal cells (Geisert et al., 1993; 1994).
In this study, many novel genes whose roles in the uterus have not been fully studied were identified as being differentially expressed in the endometrium between D15 of pregnancy and D15 of the estrous cycle. Among genes that were up-regulated in the endometrium on D15 of pregnancy compared to those on D15 of the estrous cycle were AKAP1, AKAP11, AKAP13, CLCN3, EDEM3, NRIP1, and SMAD2; down-regulated genes in the endometrium on D15 of pregnancy compared to those on D15 of the estrous cycle included ATP2C2, EFHA2, FOXN3, MYO6, PER1, PTH and RBP7. NRIP1, formerly known as receptor interacting protein 140 (RIP140), is a widely expressed corepressor that has the potential to inhibit the transcriptional activities of most nuclear receptors, including ESR1, PGR, and GR. Studies have shown that NRIP1 is expressed in metabolic tissues and plays a critical role in lipid metabolism (Fritah, 2009). The importance of NRIP1 in reproduction has been shown in a study of Nrip1-null mice (White et al., 2000), which are viable, but whose females are infertile due to a failure to release mature oocytes (White et al., 2000). The expression and function of NRIP1 in reproductive tissues have not been heavily studied, but some data in mice and rats have shown that Nrip1 is expressed in the uterus (Nephew et al., 2000; Leonardsson et al., 2002). In mice, Nrip1 expression is localized to glandular epithelial cells, stromal cells, and decidual cells, but is not observed in luminal epithelial cells in the uterine endometrium (Leonardsson et al., 2002). In women, mutation of the NRIP1 gene is associated with endometriosis, suggesting that mutation of this gene may be a predisposing factor for endometriosis (Caballero et al., 2005). There is no information available on the expression and function of NRIP1 in the porcine uterus. However, NRIP1 may play an important role in regulating lipid hormonal actions for the establishment of pregnancy, because the uterus is continuously influenced by steroid hormones (mainly estrogen and progesterone) during early pregnancy. Further characterization of the mechanism of NRIP1 action in the uterus during early pregnancy is needed.
SMAD2 is one of the key modulators in the TGF-β signaling pathway. When the TGF receptor is activated by TGF-β binding, SMAD2 binds to the TGF-β-receptor complex and is activated by C-terminal phosphorylation. Phosphorylated SMAD2 pairs with SMAD4 and relocates to the nucleus, where it acts as a transcription factor to induce target gene expression (ten Dijke and Hill, 2004). In rodents, SMAD2 expression in the uterus has been well characterized. During the estrous cycle, Smad2 expression is localized to luminal and glandular epithelial cells, but its expression is predominantly localized to the subluminal stroma surrounding the implanting embryo during the peri-implantation period (Liu et al., 2004). In the porcine uterus, phosphorylated SMAD2/3 was localized to endometrial luminal and glandular epithelilal cells, stromal cells, and endothelial cells (as well as the conceptus trophectoderm) on D13 of pregnancy, and activation of SMAD2/3 in endometrial luminal epithelial cells by TGF-β infusion into the uterine lumen has been demonstrated (Massuto et al., 2010). In pigs, it has been suggested that TGF-β, which is present at the maternal-fetal interface during the implantation period (Gupta et al., 1998), acts in trophectodermal cell-to-luminal epithelial cell adhesion (Massuto et al., 2010). Thus, increased expression of SMAD2 in the endometrium of pregnant pigs on D15 may indicate that TGF-β signaling is activated in conceptus implantation.
Recently, microarray analysis has been applied in determining DEGs in the uterine endometrium in pregnant pigs compared with genes expressed in non-pregnant pigs, showing that DEGs are involved in gene function of developmental process, transporter activity, calcium ion binding, apoptosis, cell motility, enzyme-linked protein receptor signaling pathway, positive regulation of cell proliferation, ion homeostasis, and hormone activity (Østrup et al., 2010). Microarray analysis has also been applied to determine the pattern of global gene expression during the implantation period in several species, including humans (Kao et al., 2002), mice (Reese et al., 2001), bovine (Ushizawa et al., 2004), and ovine (Satterfield et al., 2009). A direct comparison of genes expressed in the uterine endometrium during the implantation period in pigs to genes in other species is not possible because of varying experimental settings between studies and limited information on annotated genes in the porcine genome. However, it seems that very few genes expressed in the porcine uterine endometrium match the genes expressed in other species, which may result from different mechanisms of implantation and placentation in pigs.
The data presented in this study provides information on the genes expressed in the uterine endometrium during the implantation period in pigs. Expression of a gene during this period does not necessarily mean that the gene is critical for conceptus implantation; as such, further functional characterization of those genes is essential. Nevertheless, data from this study will provide insight into the mechanism of the implantation process for the establishment of pregnancy in pigs. In addition, the experimental setting used in this study may be applicable to the screening of candidate genes related to prolificacy among pig breeds and to low implantation rates in the production of somatic cell nuclear transfer cloned pigs.
Figure 3.
Real-time RT-PCR analysis of selected differentially expressed genes, ADAMTS20 (A), MUC5AC (B), NRIP1 (C), and RBP7 (D), in the uterine endometrium during the implantation period in pigs. Levels of ADAMTS20 and MUC5AC mRNA were significantly different in the endometrium on D12 of the estrous cycle and pregnancy (p<0.05). Levels of NRIP1 mRNA were significantly higher in the endometrium on D15 of pregnancy than D15 of the estrous cycle (p<0.05), but levels of RBP7 were not different (p>0.05). Abundance of mRNA is presented as the expression relative to the levels of ADAMTS20, MUC5AC, NRIP1 and RBP7 mRNAs measured in the uterine endometrium after normalization of the transcript amount to RPL7 mRNA. Data are presented as means with standard error.
ACKNOWLEDGEMENTS
This work was supported by the Yonsei University Research Fund of 2009, and the Next Generation BioGreen 21 Program (PJ007997), Rural Development Administration, and, in part, by the National Research Foundation Grant (NRF-2010-413-B00024) funded by the Korean Government, Republic of Korea.
Supplementary Data
Table 1.
List of top 30 genes up-regulated in the uterine endometrium on day (D) 12 of pregnancy compared to genes on D12 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.12.1.S1_at | 5.1138 | SULT1E1 | Sulfotransferase family 1E, estrogen-preferring, member 1 | NM_213992 |
| Ssc.31161.1.A1_at | 4.2407 | ADAMTS20 | ADAM metallopeptidase with thrombospondin type 1 motif, 20 | NM_025003 |
| Ssc.7637.1.A1_at | 4.0102 | LIMCH1 | LIM and calponin homology domains 1 | NM_001112717 |
| Ssc.17508.2.A1_at | 3.9781 | RCAN2 | Regulator of calcineurin 2 | NM_005822 |
| Ssc.3102.2.A1_at | 3.8907 | LPAR3 | Lysophosphatidic acid receptor 3 | NM_012152 |
| Ssc.3102.1.S1_at | 3.8637 | LPAR3 | Lysophosphatidic acid receptor 3 | NM_001162402 |
| Ssc.9019.1.A1_at | 3.8489 | NPR3 | Natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | NM_000908 |
| Ssc.7890.1.S1_at | 3.3443 | C5orf23 | Chromosome 5 open reading frame 23 | NM_024563 |
| Ssc.27625.1.S1_at | 2.9314 | GJB5 | Gap junction protein, beta 5, 31.1kDa | NM_005268 |
| Ssc.14085.1.A1_at | 2.8601 | LRRTM3 | Leucine rich repeat transmembrane neuronal 3 | NM_178011 |
| Ssc.18819.1.A1_at | 2.8266 | SYT13 | Synaptotagmin XIII | NM_020826 |
| Ssc.29244.1.A1_at | 2.6149 | SORCS3 | Sortilin-related VPS10 domain containing receptor 3 | NM_014978 |
| Ssc.12902.1.A1_at | 2.3709 | SLC22A2 | Solute carrier family 22 (organic cation transporter), member 2 | NM_003058 |
| Ssc.21585.2.S1_at | 2.3324 | UBR7 | Ubiquitin protein ligase E3 component n-recognin 7 (putative) | NM_001100417 |
| Ssc.12523.1.A1_at | 2.3272 | CLIC4 | Chloride intracellular channel 4 | NM_013943 |
| Ssc.15886.1.S1_at | 2.3001 | CASP3 | Caspase 3, apoptosis-related cysteine peptidase | NM_214131 |
| Ssc.7864.1.A1_at | 2.1904 | IL1RAP | Interleukin 1 receptor accessory protein | NM_002182 |
| Ssc.18027.1.S1_at | 2.1855 | BTBD3 | BTB (POZ) domain containing 3 | NM_014962 |
| Ssc.6441.2.S1_at | 2.1576 | DHRS7 | Dehydrogenase/reductase (SDR family) member 7 | NM_016029 |
| Ssc.8552.3.S1_a_at | 2.1220 | GGTA1 | Glycoprotein, alpha-galactosyltransferase 1 | NM_213810 |
| Ssc.21460.1.A1_at | 1.9430 | CREG1 | Cellular repressor of E1A-stimulated genes 1 | NM_003851 |
| Ssc.13221.1.A1_at | 1.8729 | PC | Pyruvate carboxylase | NM_000920 |
| Ssc.4999.1.S1_at | 1.8630 | AVPI1 | Arginine vasopressin-induced 1 | NM_021732 |
| Ssc.14304.1.A1_at | 1.8514 | CLSTN2 | Calsyntenin 2 | NM_022131 |
| Ssc.15923.1.A1_at | 1.8044 | FGF7 | Fibroblast growth factor 7 | AF217463.1 |
| Ssc.30862.1.S1_at | 1.8001 | DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | NM_012328 |
| Ssc.31013.2.S1_at | 1.7794 | KCTD3 | Potassium channel tetramerisation domain containing 3 | NM_016121 |
| Ssc.12056.1.A1_at | 1.7746 | LOXL4 | Lysyl oxidase-like 4 | NM_032211 |
| Ssc.24450.1.S1_at | 1.7716 | F13B | Coagulation factor XIII, B polypeptide | NM_001994 |
| Ssc.12412.1.A1_at | 1.7315 | SDF2L1 | Stromal cell-derived factor 2-like 1 | NM_022044 |
Table 2.
List of top 30 genes down-regulated in the uterine endometrium on day (D) 12 of pregnancy compared to genes on D12 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.10837.1.A1_at | −3.994277 | ROBO1 | Roundabout, axon guidance receptor, homolog 1 (Drosophila) | NM_002941 |
| Ssc.15850.1.S1_a_at | −3.892519 | TRA@ | T cell receptor alpha locus | NW_001838110.1 |
| Ssc.16377.2.A1_at | −3.697358 | GSTA3 | Glutathione S-transferase alpha 3 | NM_000847 |
| Ssc.3904.1.S1_at | −2.781738 | RPS6KA6 | Ribosomal protein S6 kinase, 90kDa, polypeptide 6 | NM_014496 |
| Ssc.16767.1.S1_at | −2.76431 | HNRNPL | Heterogeneous nuclear ribonucleoprotein L-like | NM_001005335 |
| Ssc.338.1.S1_at | −2.656394 | MUC5AC | Mucin 5AC, oligomeric mucus/gel-forming | NM_017511.1 |
| Ssc.22190.1.S1_at | −2.632552 | AJAP1 | Adherens junctions associated protein 1 | NM_001042478 |
| Ssc.8621.1.A1_at | −2.490124 | PRKACB | Protein kinase, cAMP-dependent, catalytic, Beta | NM_002731 |
| Ssc.23849.1.A1_at | −2.457561 | SLC2A2 | Solute carrier family 2 (facilitated glucose transporter), member 2 | NM_000340 |
| Ssc.25254.1.S1_at | −2.378661 | RHOJ | Ras homolog gene family, member J | NM_020663 |
| Ssc.21224.1.A1_at | −2.352619 | KRT7 | Keratin 7 | NM_005556 |
| Ssc.6556.2.A1_at | −2.344349 | GABRA5 | Gamma-aminobutyric acid (GABA) A receptor, alpha 5 | NM_000810 |
| Ssc.27912.1.S1_at | −2.205287 | ZNF608 | Zinc finger protein 608 | NM_020747 |
| Ssc.24434.1.S1_at | −2.16435 | C14orf145 | Chromosome 14 open reading frame 145 | NM_152446.3 |
| Ssc.26326.1.A1_at | −2.134067 | CYP3A46 | Cytochrome P450 3A46 | NM_001134824 |
| Ssc.11260.1.A1_at | −2.124412 | GSTA1 | Glutathione S-transferase alpha 1 | NM_145740 |
| Ssc.8053.1.S1_at | −1.953481 | POLL | Polymerase (DNA directed), lambda | NM_013274 |
| Ssc.2618.1.S1_at | −1.827725 | MAMDC4 | MAM domain containing 4 | NM_206920 |
| Ssc.5641.1.S1_at | −1.80541 | ANK2 | Ankyrin 2, neuronal | NM_001127493 |
| Ssc.18992.1.A1_at | −1.784956 | ANKRD6 | Ankyrin repeat domain 6 | NM_014942 |
| Ssc.30445.1.A1_at | −1.761477 | ALK | Anaplastic lymphoma receptor tyrosine Kinase | NM_004304 |
| Ssc.27318.1.S1_at | −1.757688 | KRT7 | Keratin 7 | NM_005556 |
| Ssc.30532.1.A1_at | −1.725776 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | NM_005431 |
| Ssc.27626.1.S1_at | −1.719731 | EGFL6 | EGF-like-domain, multiple 6 | NM_015507 |
| Ssc.12223.1.A1_at | −1.690582 | PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | NM_012411 |
| Ssc.336.1.S1_at | −1.631299 | USP18 | Ubiquitin specific peptidase 18 | NM_213826 |
| Ssc.15708.1.S1_at | −1.621459 | HBZ | Hemoglobin, zeta | NM_005332 |
| Ssc.7243.1.A1_at | −1.619644 | CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | NM_001009580 |
| Ssc.21162.1.S1_s_at | −1.609334 | IRF7 | Interferon regulatory factor 7 | NM_001097428 |
| Ssc.286.1.S1_s_at | −1.553267 | IRG6 | Inflammatory response protein 6 | NM_213817 |
Table 3.
List of top 30 genes up-regulated in the uterine endometrium on day (D) 15 of pregnancy compared to genes on D15 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.28494.1.S1_at | 3.9383217 | RAB5B | RAB5B, member RAS oncogene family | NM_002868 |
| Ssc.18273.1.A1_at | 3.9374428 | MAGI1 | Membrane associated guanylate kinase, WW and PDZ domain containing 1 | NM_173515 |
| Ssc.22734.1.A1_at | 3.505833 | EDEM3 | ER degradation enhancer, mannosidase alpha- like 3 | NM_025191 |
| Ssc.6392.1.S1_at | 3.3603669 | NTM | Neurotrimin | NM_001144058.1 |
| Ssc.25740.1.A1_at | 3.3474057 | AKAP1 | A kinase (PRKA) anchor protein 1 | NM_003488 |
| Ssc.9329.1.A1_at | 3.0693123 | PTPLAD2 | Protein tyrosine phosphatase-like A domain containing 2 | NM_001010915 |
| Ssc.10593.2.A1_at | 3.0355328 | IFI44L | Interferon-induced protein 44-like | NM_006820 |
| Ssc.11925.1.A1_at | 2.8589389 | SEL1L | Sel-1 suppressor of lin-12-like (C. Elegans) | NM_005065 |
| Ssc.25678.1.S1_at | 2.8213114 | SYT1 | Synaptotagmin I | NM_005639 |
| Ssc.12809.5.A1_at | 2.7832047 | HBA2 | Hemoglobin, alpha 2; hemoglobin, alpha 1 | NM_000517 |
| Ssc.7455.1.A1_at | 2.7595911 | AASDHPPT | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | NM_015423 |
| Ssc.7682.1.A1_at | 2.6842858 | OSBPL9 | Oxysterol binding protein-like 9 | NM_024586 |
| AFFX-Ss_18SrRNA_at | 2.6236143 | AFFX-Ss_18SrRNA | AFFX-Ss_18SrRNA | Null |
| Ssc.21585.2.S1_at | 2.5961034 | UBR7 | Ubiquitin protein ligase E3 component n-recognin 7 (putative) | NM_001100417 |
| Ssc.13473.1.A1_at | 2.5538991 | UGCG | UDP-glucose ceramide glucosyltransferase | NM_003358 |
| Ssc.27407.1.A1_at | 2.5424531 | NRIP1 | Nuclear receptor interacting protein 1 | NM_003489 |
| Ssc.3975.2.A1_at | 2.4775446 | STK38L | Serine/threonine kinase 38 like | NM_015000.3 |
| Ssc.28192.1.A1_at | 2.4555041 | FAM153A | Family with sequence similarity 153, member A | NM_173663 |
| Ssc.20353.1.S1_at | 2.3855645 | MALT1 | Metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) | NR_002819.2 |
| Ssc.11870.1.A1_at | 2.2433352 | NRIP1 | Nuclear receptor interacting protein 1 | NM_003489 |
| Ssc.17508.2.A1_at | 2.2266604 | RCAN2 | Regulator of calcineurin 2 | NM_005822 |
| Ssc.8143.2.A1_at | 2.1943771 | RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 | NM_014247 |
| Ssc.17152.2.A1_at | 2.1849978 | COQ2 | Coenzyme Q2 homolog, prenyltransferase (Yeast) | NM_015697 |
| Ssc.27130.1.A1_at | 2.1615496 | SLC25A36 | Solute carrier family 25, member 36 | NM_001104647 |
| Ssc.3325.3.S1_at | 2.1184436 | EXT1 | Exostoses (multiple) 1 | NM_000127 |
| Ssc.22645.1.S1_at | 2.0685056 | DDX46 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | NM_014829 |
| Ssc.6175.1.S1_at | 2.0623846 | LCOR | Ligand dependent nuclear receptor corepressor | NM_032440 |
| Ssc.13601.1.A1_at | 1.8954884 | MYO1B | Myosin IB | NM_001130158 |
| Ssc.24402.1.S1_at | 1.8920959 | CCDC76 | Coiled-coil domain containing 76 | NM_019083 |
| Ssc.27133.1.A1_at | 1.8789606 | TMEM68 | Transmembrane protein 68 | NM_152417 |
Table 4.
List of top 30 genes down-regulated in the uterine endometrium on day (D) 15 of pregnancy compared to genes on D15 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.25087.1.S1_at | −4.665941 | DNAJC13 | DnaJ (Hsp40) homolog, subfamily C, member 13 | NM_015268 |
| Ssc.2015.2.S1_at | −4.469763 | ZNF449 | Zinc finger protein 449 | NM_152695 |
| Ssc.7817.1.A1_at | −4.302148 | NETO1 | Neuropilin (NRP) and tolloid (TLL)-like 1 | NM_138966 |
| Ssc.23424.1.A1_at | −4.072875 | DTNA | Dystrobrevin, alpha | NM_001128175 |
| Ssc.8989.1.A1_at | −3.882042 | PPHLN1 | Periphilin 1 | NM_016488 |
| Ssc.21088.2.S1_at | −3.009555 | CSTF1 | Cleavage stimulation factor, 3′ pre-RNA, subunit 1, 50kDa | NM_001033521 |
| Ssc.9834.2.S1_at | −2.875995 | Null | Sn36_F05.f sn Sus scrofa cDNA | CB471302.1 |
| Ssc.1600.1.A1_at | −2.684489 | RIMKLB | Ribosomal modification protein rimK-like family member B | NM_020734.2 |
| Ssc.1600.1.A1_a_at | −2.616954 | RIMKLB | Ribosomal modification protein rimK-like family member B | NM_020734.2 |
| Ssc.27385.1.S1_at | −2.580643 | RBP7 | Retinol binding protein 7, cellular | NM_001145222 |
| Ssc.668.1.S1_at | −2.543521 | PTH | Parathyroid hormone | NM_214401 |
| Ssc.958.1.S1_at | −2.543376 | ALDOB | Aldolase B, fructose-bisphosphate | NM_000035 |
| Ssc.27953.1.S1_at | −2.289442 | MLL | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | NM_005933 |
| Ssc.21468.2.A1_at | −2.194019 | LARP1 | La ribonucleoprotein domain family, member 1 | NM_015315 |
| Ssc.30456.1.A1_at | −2.155805 | CCDC135 | Coiled-coil domain containing 135 | NM_032269 |
| Ssc.27345.1.S1_at | −2.107692 | Null | 940760 MARC 4PIG Sus scrofa cDNA | CN153625.1 |
| Ssc.7219.1.A1_at | −1.926105 | ERC2 | ELKS/RAB6-interacting/CAST family member 2 | NM_015576 |
| Ssc.22791.1.A1_at | −1.922899 | EEA1 | Early endosome antigen 1 | NM_003566 |
| Ssc.7672.1.A1_at | −1.847485 | CAPZB | Capping protein (actin filament) muscle Z-line, beta | NM_004930 |
| Ssc.25130.1.A1_at | −1.83943 | C6orf98 | Chromosome 6 open reading frame 98 | NM_001099267 |
| Ssc.30379.1.A1_at | −1.831398 | ARMC3 | Armadillo repeat containing 3 | NM_173081 |
| Ssc.7191.1.A1_at | −1.805 | LSM14A | LSM14A, SCD6 homolog A (S. Cerevisiae) | NM_001114093 |
| Ssc.2589.1.S1_at | −1.790171 | TNNI3 | Troponin I type 3 (cardiac) | NM_001098599 |
| Ssc.23948.1.A1_at | −1.727051 | DYNC1I2 | Dynein, cytoplasmic 1, intermediate chain 2 | NM_001378 |
| Ssc.8133.1.A1_at | −1.725898 | GPM6B | Glycoprotein M6B | NM_001001994 |
| Ssc.28842.1.A1_at | −1.685047 | CCDC113 | Coiled-coil domain containing 113 | NM_014157 |
| Ssc.7917.1.A1_at | −1.680845 | Null | MI-P-E6-ahn-e-09-1-UM.s1 MI-P-E6 Sus scrofa cDNA clone | BQ599768.1 |
| Ssc.29910.1.A1_at | −1.633574 | MTMR2 | Myotubularin related protein 2 | NM_016156 |
| Ssc.6616.1.S1_at | −1.629585 | C1orf92 | Chromosome 1 open reading frame 92 | NM_144702 |
| Ssc.28950.1.A1_at | −1.627009 | EXOC6 | Exocyst complex component 6 | NM_019053.4 |
Table 5.
Functional annotation clusters of up-regulated genes in uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 2.56 | Membrane fraction (12), cell fraction (14), insoluble fraction (12) |
| 2 | 1.04 | Endoplasmic reticulum part (6), endoplasmic reticulum membrane (5), endoplasmic reticulum (10), nuclear envelope-endoplasmic reticulum network (5), microsome (4), vesicular fraction (4), organelle membrane (9), endomembrane system (6) |
| 3 | 0.88 | Blood vessel morphogenesis (4), blood vessel development (4), vasculature development (4) |
| 4 | 0.86 | Cellular macromolecule catabolic process (9), proteolysis (11), macromolecule catabolic process (9), proteolysis involved in cellular protein catabolic process (7), cellular protein catabolic process (7), protein catabolic process (7), ubiquitin- protein ligase activity (3), small conjugating protein ligase activity (3), ligase activity, forming carbon-nitrogen bonds (3), ubiquitin-dependent protein catabolic process (3), modification-dependent macromolecule catabolic process (6), acid- amino acid ligase activity (3), modification-dependent protein catabolic process (6) |
| 5 | 0.80 | Intracellular organelle lumen (14), organelle lumen (14), nuclear lumen (10), membrane-enclosed lumen (14), nucleoplasm (8) |
| 6 | 0.72 | Proteolysis (11), metalloendopeptidase activity (3), endopeptidase activity (5), negative regulation of apoptosis (5), negative regulation of programmed cell death(5), negative regulation of cell death (5), metallopeptidase activity (3), peptidase activity, acting on L-amino acid peptides (5), peptidase activity (5), regulation of apoptosis (6), regulation of programmed cell death (6), regulation of cell death (6) |
| 7 | 0.72 | Phosphatase activity (5), phosphoprotein phosphatase activity (4), phosphate metabolic process (9), dephosphorylation (4), phosphorus metabolic process (9), protein amino acid dephosphorylation (3), protein serine/threonine kinase activity(3), phosphorylation (5), protein amino acid phosphorylation (5), protein kinase activity (3) |
| 8 | 0.71 | Positive regulation of multicellular organismal process (4), positive regulation of cell proliferation (5), regulation of growth (4) |
| 9 | 0.65 | Cell cycle (8), mitotic cell cycle (5), cell cycle process (5), cell cycle phase (4) |
| 10 | 0.65 | Glycosaminoglycan binding (3), polysaccharide binding (3), pattern binding (3), carbohydrate binding (3) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D12 of pregnancy compared with those on D12 of the estrous cycle through DAVID.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 6.
Functional annotation clusters of down-regulated genes in uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 2.04 | Plasma membrane part (22), integral to plasma membrane (11), intrinsic to plasma membrane (11) |
| 2 | 2.03 | Cell adhesion (11), biological adhesion (11), cell-cell adhesion (4) |
| 3 | 1.50 | Leukocyte activation (7), cell activation (7), lymphocyte activation (6), T cell activation (4), immune effector process (4), regulation of leukocyte activation (4), regulation of cell activation (4), leukocyte mediated immunity (3), cell surface (5), positive regulation of response to stimulus (4), positive regulation of immune system process (4), immune response (7), external side of plasma membrane (3) |
| 4 | 1.48 | Immune system development (7), hemopoiesis (6), hemopoietic or lymphoid organ development (6), myeloid cell differentiation (3), protein homodimerization activity (4), nucleoplasm part (3) |
| 5 | 1.46 | Cell junction (7), adherens junction (4), anchoring junction 4) |
| 6 | 1.45 | Biopolymer glycosylation (4), glycoprotein biosynthetic process (4), glycoprotein metabolic process (4), glycosylation (4), protein amino acid glycosylation (4) |
| 7 | 0.98 | Apoptosis (7), programmed cell death (7), cell death (7), death (7), negative regulation of cell proliferation (4) |
| 8 | 0.83 | DNA metabolic process (8), response to DNA damage stimulus (5), DNA repair (4), cellular response to stress (5), DNA binding (7) |
| 9 | 0.83 | Induction of apoptosis (5), induction of programmed cell death (5), positive regulation of apoptosis (5), positive regulation of programmed cell death (5), positive regulation of cell death (5), regulation of apoptosis (7), regulation of programmed cell death (7), regulation of cell death (7) |
| 10 | 0.76 | Response to ionizing radiation (3), response to radiation (3), response to abiotic stimulus (3) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D15 of pregnancy compared with those on D15 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 7.
Functional annotation clusters of up-regulated genes in uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 3.46 | Nuclear speck (7), nuclear body (8), RNA splicing (9) |
| 2 | 2.21 | Nucleotide binding (30), purine ribonucleotide binding (25), AT P binding (21), ribonucleotide binding (25), purine nucleotide binding (25), adenyl ribonucleotide binding (21), adenyl nucleotide binding (21), purine nucleoside binding (21), nucleoside binding (21) |
| 3 | 2.12 | Nuclear body (8), nucleoplasm part (12), nucleoplasm (15), membrane-enclosed lumen (24), nuclear lumen (20), intracellular organelle lumen (23), organelle lumen (23), nucleolus (9) |
| 4 | 1.51 | RNA splicing (9), mrna metabolic process (9), RNA processing (11), mrna processing (8), spliceosome assembly (3), RNA splicing, via transesterification reactions (5), RNA splicing, via transesterification reactions with bulged adenosine as nucleophile (5), nuclear mrna splicing, via spliceosome (5), RNA binding (11), ribonucleoprotein complex assembly (3), ribonucleoprotein complex biogenesis (4), cellular macromolecular complex assembly (5), cellular macromolecular complex subunit organization (5) |
| 5 | 1.15 | Nuclear body (8), nuclear export (4), nucleocytoplasmic transport (4), nuclear transport (4), mrna transport (3), establishment of RNA localization (3), nucleic acid transport (3), RNA transport (3), RNA localization (3), nucleobase, nucleoside, nucleotide and nucleic acid transport (3), intracellular transport (8) |
| 6 | 1.06 | Receptor-mediated endocytosis (4), membrane organization (8), membrane invagination (4), endocytosis (4), vesicle-mediated transport (6) |
| 7 | 0.96 | Helicase activity (6), ATP-dependent helicase activity (3), purine NTP-dependent helicase activity (3), atpase activity, coupled (4), atpase activity (4) |
| 8 | 0.95 | Enzyme binding (8), protein kinase binding (4), kinase binding(4) |
| 9 | 0.94 | Macromolecular complex assembly (11), macromolecular complex subunit organization (11), protein complex assembly (8), protein complex biogenesis (8), cellular macromolecular complex assembly (5), cellular macromolecular complex subunit organization (5), protein oligomerization (3) |
| 10 | 0.75 | Protein serine/threonine kinase activity (7), enzyme linked receptor protein signaling pathway (6), phosphate metabolic process (11), phosphorus metabolic process (11), protein kinase activity (8), protein amino acid phosphorylation (8), phosphorylation (9) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D12 of pregnancy compared with those on D12 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 8.
Functional annotation clusters of down-regulated genes in uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 1.68 | Adenyl ribonucleotide binding (15), purine ribonucleotide binding (17), purine nucleotide binding (17), ribonucleotide binding (17), nucleoside binding (15), adenyl nucleotide binding (15), purine nucleoside binding (15), ATP binding (14), nucleotide binding (18) |
| 2 | 1.50 | Cytoskeletal part (11), microtubule cytoskeleton (7), intracellular non-membrane- bounded organelle (18), non-membrane- bounded organelle (18), centrosome (4), cytoskeleton (13), microtubule organizing center (4) |
| 3 | 1.50 | Cytoskeletal part (11), dynein complex (3), microtubule associated complex (4), microtubule-based movement (4), microtubule cytoskeleton (7), motor activity (3), microtubule motor activity (3), microtubule (4), microtubule-based process (4) |
| 4 | 1.20 | Cytoskeletal part (11), regulation of actin filament length (3), regulation of cellular component size (5), regulation of actin cytoskeleton organization (3), regulation of actin filament-based process (3), regulation of organelle organization (4), actin cytoskeleton (4), cytoskeletal protein binding (6), regulation of cytoskeleton organization (3), actin binding (4) |
| 5 | 1.17 | Regulation of neuron differentiation (4), regulation of neurogenesis (4), regulation of nervous system development (4), regulation of cell development (4), neuron differentiation (4) |
| 6 | 0.92 | Protein amino acid dephosphorylation (4), dephosphorylation (4), phosphorus metabolic process (9), phosphoprotein phosphatase activity (4), phosphate metabolic process (9), protein tyrosine phosphatase activity (3), phosphatase activity (4), protein kinase activity (5), protein serine/threonine kinase activity (4), protein amino acid phosphorylation (5), phosphorylation (5) |
| 7 | 0.84 | Central nervous system neuron axonogenesis (3), central nervous system neuron development (3), central nervous system neuron differentiation (3), cell projection organization (5), cell motion (5), axonogenesis (3), cell morphogenesis involved in neuron differentiation (3), neuron projection morphogenesis (3), cell projection morphogenesis (3), cell morphogenesis involved in differentiation (3), cell part morphogenesis (3), neuron differentiation (4), neuron projection development (3), cell morphogenesis (3), cellular component morphogenesis (3), neuron development (3) |
| 8 | 0.82 | Regulation of synaptic transmission (3), regulation of transmission of nerve impulse (3), regulation of neurological system process (3), regulation of system process (4) |
| 9 | 0.75 | Cell projection organization (5), cytoskeletal protein binding (6), cytoskeleton organization (3) |
| 10 | 0.71 | Learning or memory (3), neurological system process (10), behavior (5), cognition (4) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D15 of pregnancy compared with those on D15 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
REFERENCES
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Thatcher WW. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F2alpha by the uterine endometrium. Prostaglandins. 1977;14:397–400. doi: 10.1016/0090-6980(77)90185-x. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Vallet JL, Roberts RM, Sharp DC, Thatcher WW. Role of conceptus secretory products in establishment of pregnancy. J Reprod Fertil. 1986;76:841–850. doi: 10.1530/jrf.0.0760841. [DOI] [PubMed] [Google Scholar]
- Bennett GL, Leymaster KA. Integration of ovulation rate, potential embryonic viability and uterine capacity into a model of litter size in swine. J Anim Sci. 1989;67:1230–1241. doi: 10.2527/jas1989.6751230x. [DOI] [PubMed] [Google Scholar]
- Bischof RJ, Lee R, Lee CS, Meeusen E. Dynamic changes in the lymphocyte subpopulations of pig uterine lymph nodes. Vet Immunol Immunopathol. 1996;51:315–324. doi: 10.1016/0165-2427(95)05529-0. [DOI] [PubMed] [Google Scholar]
- Caballero V, Ruiz R, Sainz JA, Cruz M, Lopez-Nevot MA, Galan JJ, Real LM, de Castro F, Lopez-Villaverde V, Ruiz A. Preliminary molecular genetic analysis of the Receptor Interacting Protein 140 (RIP140) in women affected by endometriosis. J Exp Clin Assist Reprod. 2005;2:11. doi: 10.1186/1743-1050-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson DD, DeSouza MM, Regisford EG. Mucin and proteoglycan functions in embryo implantation. Bioessays. 1998;20:577–583. doi: 10.1002/(SICI)1521-1878(199807)20:7<577::AID-BIES9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Cencic A, Guillomot M, Koren S, La Bonnardiere C. Trophoblastic interferons: do they modulate uterine cellular markers at the time of conceptus attachment in the pig? Placenta. 2003;24:862–869. doi: 10.1016/s0143-4004(03)00135-8. [DOI] [PubMed] [Google Scholar]
- Croy BA, Wood W, King GJ. Evaluation of intrauterine immune suppression during pregnancy in a species with epitheliochorial placentation. J Immunol. 1987;139:1088–1095. [PubMed] [Google Scholar]
- Dantzer V. Electron microscopy of the initial stages of placentation in the pig. Anat Embryol (Berl) 1985;172:281–293. doi: 10.1007/BF00318976. [DOI] [PubMed] [Google Scholar]
- Fritah A. Control of skeletal muscle metabolic properties by the nuclear receptor corepressor RIP140. Appl Physiol Nutr Metab. 2009;34:362–367. doi: 10.1139/H09-026. [DOI] [PubMed] [Google Scholar]
- Garlow JE, Ka H, Johnson GA, Burghardt RC, Jaeger LA, Bazer FW. Analysis of osteopontin at the maternal-placental interface in pigs. Biol Reprod. 2002;66:718–725. doi: 10.1095/biolreprod66.3.718. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Renegar RH, Thatcher WW, Roberts RM, Bazer FW. Establishment of pregnancy in the pig: I. Interrelationships between preimplantation development of the pig blastocyst and uterine endometrial secretions. Biol Reprod. 1982;27:925–939. doi: 10.1095/biolreprod27.4.925. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Brenner RM, Moffatt RJ, Harney JP, Yellin T, Bazer FW. Changes in oestrogen receptor protein, mRNA expression and localization in the endometrium of cyclic and pregnant gilts. Reprod Fertil Dev. 1993;5:247–260. doi: 10.1071/rd9930247. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Pratt TN, Bazer FW, Mayes JS, Watson GH. Immunocytochemical localization and changes in endometrial progestin receptor protein during the porcine oestrous cycle and early pregnancy. Reprod Fertil Dev. 1994;6:749–760. doi: 10.1071/rd9940749. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Yelich JV. Regulation of conceptus development and attachment in pigs. J Reprod Fertil Suppl. 1997;52:133–149. [PubMed] [Google Scholar]
- Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- Gipson IK, Ho SB, Spurr-Michaud SJ, Tisdale AS, Zhan Q, Torlakovic E, Pudney J, Anderson DJ, Toribara NW, Hill JA., 3rd Mucin genes expressed by human female reproductive tract epithelia. Biol Reprod. 1997;56:999–1011. doi: 10.1095/biolreprod56.4.999. [DOI] [PubMed] [Google Scholar]
- Glynn DJ, Sherman BT, Hosack DA, Yang J, Baseler MW, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Green JA, Kim JG, Whitworth KM, Agca C, Prather RS. The use of microarrays to define functionally-related genes that are differentially expressed in the cycling pig uterus. Soc Reprod Fertil Suppl. 2006;62:163–176. [PubMed] [Google Scholar]
- Gupta A, Dekaney CM, Bazer FW, Madrigal MM, Jaeger JA. Beta transforming growth factors (TGFbeta) at the porcine conceptus-maternal interface. Part II: uterine TGFbeta bioactivity and expression of immunoreactive TGFbetas (TGFbeta1, TGFbeta2, and TGFbeta3) and their receptors (type I and type II) Biol Reprod. 1998;59:911–917. doi: 10.1095/biolreprod59.4.911. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, Katz G, Haimov-Kochman R, Fujii N, Yagel S, Peled A, Mandelboim O. CXCL12 expression by invasive trophoblasts induces the specific migration of CD16-human natural killer cells. Blood. 2003;102:1569–1577. doi: 10.1182/blood-2003-02-0517. [DOI] [PubMed] [Google Scholar]
- Ka H, Seo H, Kim M, Moon S, Kim H, Lee CK. Gene expression profiling of the uterus with embryos cloned by somatic cell nuclear transfer on day 30 of pregnancy. Anim Reprod Sci. 2008;108:79–91. doi: 10.1016/j.anireprosci.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Ka H, Seo H, Kim M, Choi Y, Lee CK. Identification of differentially expressed genes in the uterine endometrium on day 12 of the estrous cycle and pregnancy in pigs. Mol Reprod Dev. 2009;76:75–84. doi: 10.1002/mrd.20935. [DOI] [PubMed] [Google Scholar]
- Ka H, Jaeger LA, Johnson GA, Spencer TE, Bazer FW. Keratinocyte growth factor is up-regulated by estrogen in the porcine uterine endometrium and functions in trophectoderm cell proliferation and differentiation. Endocrinology. 2001;142:2303–2310. doi: 10.1210/endo.142.6.8194. [DOI] [PubMed] [Google Scholar]
- Ka H, Spencer TE, Johnson GA, Bazer FW. Keratinocyte growth factor: expression by endometrial epithelia of the porcine uterus. Biol Reprod. 2000;62:1772–1778. doi: 10.1095/biolreprod62.6.1772. [DOI] [PubMed] [Google Scholar]
- Kaeoket K, Persson E, Dalin AM. Influence of pre-ovulatory insemination and early pregnancy on the infiltration by cells of the immune system in the sow endometrium. Anim Reprod Sci. 2003;75:55–71. doi: 10.1016/s0378-4320(02)00230-0. [DOI] [PubMed] [Google Scholar]
- Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC. Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002;143:2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- Kim JG, Vallet JL, Rohrer GA, Christenson RK. Characterization of porcine uterine estrogen sulfotransferase. Domest Anim Endocrinol. 2002;23:493–506. doi: 10.1016/s0739-7240(02)00172-8. [DOI] [PubMed] [Google Scholar]
- La Bonnardiere C, Martinat-Botte F, Terqui M, Lefevre F, Zouari K, Martal J, Bazer FW. Production of two species of interferon by Large White and Meishan pig conceptuses during the peri-attachment period. J Reprod Fertil. 1991;91:469–478. doi: 10.1530/jrf.0.0910469. [DOI] [PubMed] [Google Scholar]
- Lee SH, Zhao SH, Recknor JC, Nettleton D, Orley S, Kang SK, Lee BC, Hwang WS, Tuggle CK. Transcriptional profiling using a novel cDNA array identifies differential gene expression during porcine embryo elongation. Mol Reprod Dev. 2005;71:129–139. doi: 10.1002/mrd.20291. [DOI] [PubMed] [Google Scholar]
- Lefevre F, Guillomot M, D’Andrea S, Battegay S, La Bonnardiere C. Interferon-delta: the first member of a novel type I interferon family. Biochimie. 1998;80:779–788. doi: 10.1016/s0300-9084(99)80030-3. [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Jacobs MA, White R, Jeffery R, Poulsom R, Milligan S, Parker M. Embryo transfer experiments and ovarian transplantation identify the ovary as the only site in which nuclear receptor interacting protein 1/RIP140 action is crucial for female fertility. Endocrinology. 2002;143:700–707. doi: 10.1210/endo.143.2.8656. [DOI] [PubMed] [Google Scholar]
- Liu G, Lin H, Zhang X, Li Q, Wang H, Qian D, Ni J, Zhu C. Expression of Smad2 and Smad4 in mouse uterus during the oestrous cycle and early pregnancy. Placenta. 2004;25:530–537. doi: 10.1016/j.placenta.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mansouri-Attia N, Aubert J, Reinaud P, Giraud-Delville C, Taghouti G, Galio L, Everts RE, Degrelle S, Richard C, Hue I, Yang X, Tian XC, Lewin HA, Renard JP, Sandra O. Gene expression profiles of bovine caruncular and intercaruncular endometrium at implantation. Physiol Genomics. 2009;39:14–27. doi: 10.1152/physiolgenomics.90404.2008. [DOI] [PubMed] [Google Scholar]
- Massuto DA, Kneese EC, Johnson GA, Burghardt RC, Hooper RN, Ing NH, Jaeger LA. Transforming growth factor beta (TGFB) signaling is activated during porcine implantation: proposed role for latency-associated peptide interactions with integrins at the conceptus-maternal interface. Reproduction. 2010;139:465–478. doi: 10.1530/REP-09-0447. [DOI] [PubMed] [Google Scholar]
- Moritani S, Kushima R, Ichihara S, Okabe H, Hattori T, Kobayashi TK, Silverberg SG. Eosinophilic cell change of the endometrium: a possible relationship to mucinous differentiation. Mod Pathol. 2005;18:1243–1248. doi: 10.1038/modpathol.3800412. [DOI] [PubMed] [Google Scholar]
- Murdoch C. CXCR4: chemokine receptor extraordinaire. Immunol Rev. 2000;177:175–184. doi: 10.1034/j.1600-065x.2000.17715.x. [DOI] [PubMed] [Google Scholar]
- Nephew KP, Long X, Osborne E, Burke KA, Ahluwalia A, Bigsby RM. Effect of estradiol on estrogen receptor expression in rat uterine cell types. Biol Reprod. 2000;62:168–177. doi: 10.1095/biolreprod62.1.168. [DOI] [PubMed] [Google Scholar]
- Østrup E, Bauersachs S, Blum H, Wolf E, Hyttel P. Differential endometrial gene expression in pregnant and nonpregnant sows. Biol Reprod. 2010;83:277–285. doi: 10.1095/biolreprod.109.082321. [DOI] [PubMed] [Google Scholar]
- Reese J, Das SK, Paria BC, Lim H, Song H, Matsumoto H, Knudtson KL, DuBois RN, Dey SK. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001;276:44137–44145. doi: 10.1074/jbc.M107563200. [DOI] [PubMed] [Google Scholar]
- Repnik U, Tilburgs T, Roelen DL, van der Mast BJ, Kanhai HH, Scherjon S, Claas FH. Comparison of macrophage phenotype between decidua basalis and decidua parietalis by flow cytometry. Placenta. 2008;29:405–412. doi: 10.1016/j.placenta.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Ross JW, Malayer JR, Ritchey JW, Geisert RD. Characterization of the interleukin-1beta system during porcine trophoblastic elongation and early placental attachment. Biol Reprod. 2003a;69:1251–1259. doi: 10.1095/biolreprod.103.015842. [DOI] [PubMed] [Google Scholar]
- Ross JW, Ashworth MD, Hurst AG, Malayer JR, Geisert RD. Analysis and characterization of differential gene expression during rapid trophoblastic elongation in the pig using suppression subtractive hybridization. Reprod Biol Endocrinol. 2003b;1:23. doi: 10.1186/1477-7827-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield MC, Song G, Kochan KJ, Riggs PK, Simmons RM, Elsik CG, Adelson DL, Bazer FW, Zhou H, Spencer TE. Discovery of candidate genes and pathways in the endometrium regulating ovine blastocyst growth and conceptus elongation. Physiol Genomics. 2009;39:85–99. doi: 10.1152/physiolgenomics.00001.2009. [DOI] [PubMed] [Google Scholar]
- Seo H, Choi M, Kim Y, Lee CK, Ka H. Analysis of lysophosphatidic acid (LPA) receptor and LPA-induced endometrial prostaglandin-endoperoxide synthase 2 expression in the porcine uterus. Endocrinology. 2008;149:6166–6175. doi: 10.1210/en.2008-0354. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Bazer FW. Conceptus signals for establishment and maintenance of pregnancy. Reprod Biol Endocrinol. 2004;2:49. doi: 10.1186/1477-7827-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Johnson GA, Bazer FW, Burghardt RC. Implantation mechanisms: insights from the sheep. Reproduction. 2004;128:657–668. doi: 10.1530/rep.1.00398. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Tsai S, Cassady JP, Freking BA, Nonneman DJ, Rohrer GA, Piedrahita JA. Annotation of the Affymetrix porcine genome microarray. Anim Genet. 2006;37:423–424. doi: 10.1111/j.1365-2052.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- Tuo W, Harney JP, Bazer FW. Developmentally regulated expression of interleukin-1 beta by peri-implantation conceptuses in swine. J Reprod Immunol. 1996;31:185–198. doi: 10.1016/0165-0378(96)00975-8. [DOI] [PubMed] [Google Scholar]
- Ushizawa K, Herath CB, Kaneyama K, Shiojima S, Hirasawa A, Takahashi T, Imai K, Ochiai K, Tokunaga T, Tsunoda Y, Tsujimoto G, Hashizume K. cDNA microarray analysis of bovine embryo gene expression profiles during the pre-implantation period. Reprod Biol Endocrinol. 2004;2:77. doi: 10.1186/1477-7827-2-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee M, Beaudry D, Roberge C, Matte JJ, Blouin R, Palin MF. Isolation of differentially expressed genes in conceptuses and endometrial tissue of sows in early gestation. Biol Reprod. 2003;69:1697–1706. doi: 10.1095/biolreprod.103.019307. [DOI] [PubMed] [Google Scholar]
- White FJ, Ross JW, Joyce MM, Geisert RD, Burghardt RC, Johnson GA. Steroid regulation of cell specific secreted phosphoprotein 1 (osteopontin) expression in the pregnant porcine uterus. Biol Reprod. 2005;73:1294–1301. doi: 10.1095/biolreprod.105.045153. [DOI] [PubMed] [Google Scholar]
- White R, Leonardsson G, Rosewell I, Ann Jacobs M, Milligan S, Parker M. The nuclear receptor corepressor nrip1 (RIP140) is essential for female fertility. Nat Med. 2000;6:1368–1374. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, Bivens NJ, Forrester LJ, Mathialagan N, Green JA, Prather RS. Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod. 2005;72:1437–1451. doi: 10.1095/biolreprod.104.037952. [DOI] [PubMed] [Google Scholar]
- Wu X, Li DJ, Yuan MM, Zhu Y, Wang MY. The expression of CXCR4/CXCL12 in first-trimester human trophoblast cells. Biol Reprod. 2004;70:1877–1885. doi: 10.1095/biolreprod.103.024729. [DOI] [PubMed] [Google Scholar]
- Zhou WH, Du MR, Dong L, Yu J, Li DJ. Chemokine CXCL12 promotes the cross-talk between trophoblasts and decidual stromal cells in human first-trimester pregnancy. Hum Reprod. 2008;23:2669–2679. doi: 10.1093/humrep/den308. [DOI] [PubMed] [Google Scholar]
- Ziecik AJ. Old, new and the newest concepts of inhibition of luteolysis during early pregnancy in pig. Domest Anim Endocrinol. 2002;23:265–275. doi: 10.1016/s0739-7240(02)00162-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1.
List of top 30 genes up-regulated in the uterine endometrium on day (D) 12 of pregnancy compared to genes on D12 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.12.1.S1_at | 5.1138 | SULT1E1 | Sulfotransferase family 1E, estrogen-preferring, member 1 | NM_213992 |
| Ssc.31161.1.A1_at | 4.2407 | ADAMTS20 | ADAM metallopeptidase with thrombospondin type 1 motif, 20 | NM_025003 |
| Ssc.7637.1.A1_at | 4.0102 | LIMCH1 | LIM and calponin homology domains 1 | NM_001112717 |
| Ssc.17508.2.A1_at | 3.9781 | RCAN2 | Regulator of calcineurin 2 | NM_005822 |
| Ssc.3102.2.A1_at | 3.8907 | LPAR3 | Lysophosphatidic acid receptor 3 | NM_012152 |
| Ssc.3102.1.S1_at | 3.8637 | LPAR3 | Lysophosphatidic acid receptor 3 | NM_001162402 |
| Ssc.9019.1.A1_at | 3.8489 | NPR3 | Natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | NM_000908 |
| Ssc.7890.1.S1_at | 3.3443 | C5orf23 | Chromosome 5 open reading frame 23 | NM_024563 |
| Ssc.27625.1.S1_at | 2.9314 | GJB5 | Gap junction protein, beta 5, 31.1kDa | NM_005268 |
| Ssc.14085.1.A1_at | 2.8601 | LRRTM3 | Leucine rich repeat transmembrane neuronal 3 | NM_178011 |
| Ssc.18819.1.A1_at | 2.8266 | SYT13 | Synaptotagmin XIII | NM_020826 |
| Ssc.29244.1.A1_at | 2.6149 | SORCS3 | Sortilin-related VPS10 domain containing receptor 3 | NM_014978 |
| Ssc.12902.1.A1_at | 2.3709 | SLC22A2 | Solute carrier family 22 (organic cation transporter), member 2 | NM_003058 |
| Ssc.21585.2.S1_at | 2.3324 | UBR7 | Ubiquitin protein ligase E3 component n-recognin 7 (putative) | NM_001100417 |
| Ssc.12523.1.A1_at | 2.3272 | CLIC4 | Chloride intracellular channel 4 | NM_013943 |
| Ssc.15886.1.S1_at | 2.3001 | CASP3 | Caspase 3, apoptosis-related cysteine peptidase | NM_214131 |
| Ssc.7864.1.A1_at | 2.1904 | IL1RAP | Interleukin 1 receptor accessory protein | NM_002182 |
| Ssc.18027.1.S1_at | 2.1855 | BTBD3 | BTB (POZ) domain containing 3 | NM_014962 |
| Ssc.6441.2.S1_at | 2.1576 | DHRS7 | Dehydrogenase/reductase (SDR family) member 7 | NM_016029 |
| Ssc.8552.3.S1_a_at | 2.1220 | GGTA1 | Glycoprotein, alpha-galactosyltransferase 1 | NM_213810 |
| Ssc.21460.1.A1_at | 1.9430 | CREG1 | Cellular repressor of E1A-stimulated genes 1 | NM_003851 |
| Ssc.13221.1.A1_at | 1.8729 | PC | Pyruvate carboxylase | NM_000920 |
| Ssc.4999.1.S1_at | 1.8630 | AVPI1 | Arginine vasopressin-induced 1 | NM_021732 |
| Ssc.14304.1.A1_at | 1.8514 | CLSTN2 | Calsyntenin 2 | NM_022131 |
| Ssc.15923.1.A1_at | 1.8044 | FGF7 | Fibroblast growth factor 7 | AF217463.1 |
| Ssc.30862.1.S1_at | 1.8001 | DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | NM_012328 |
| Ssc.31013.2.S1_at | 1.7794 | KCTD3 | Potassium channel tetramerisation domain containing 3 | NM_016121 |
| Ssc.12056.1.A1_at | 1.7746 | LOXL4 | Lysyl oxidase-like 4 | NM_032211 |
| Ssc.24450.1.S1_at | 1.7716 | F13B | Coagulation factor XIII, B polypeptide | NM_001994 |
| Ssc.12412.1.A1_at | 1.7315 | SDF2L1 | Stromal cell-derived factor 2-like 1 | NM_022044 |
Table 2.
List of top 30 genes down-regulated in the uterine endometrium on day (D) 12 of pregnancy compared to genes on D12 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.10837.1.A1_at | −3.994277 | ROBO1 | Roundabout, axon guidance receptor, homolog 1 (Drosophila) | NM_002941 |
| Ssc.15850.1.S1_a_at | −3.892519 | TRA@ | T cell receptor alpha locus | NW_001838110.1 |
| Ssc.16377.2.A1_at | −3.697358 | GSTA3 | Glutathione S-transferase alpha 3 | NM_000847 |
| Ssc.3904.1.S1_at | −2.781738 | RPS6KA6 | Ribosomal protein S6 kinase, 90kDa, polypeptide 6 | NM_014496 |
| Ssc.16767.1.S1_at | −2.76431 | HNRNPL | Heterogeneous nuclear ribonucleoprotein L-like | NM_001005335 |
| Ssc.338.1.S1_at | −2.656394 | MUC5AC | Mucin 5AC, oligomeric mucus/gel-forming | NM_017511.1 |
| Ssc.22190.1.S1_at | −2.632552 | AJAP1 | Adherens junctions associated protein 1 | NM_001042478 |
| Ssc.8621.1.A1_at | −2.490124 | PRKACB | Protein kinase, cAMP-dependent, catalytic, Beta | NM_002731 |
| Ssc.23849.1.A1_at | −2.457561 | SLC2A2 | Solute carrier family 2 (facilitated glucose transporter), member 2 | NM_000340 |
| Ssc.25254.1.S1_at | −2.378661 | RHOJ | Ras homolog gene family, member J | NM_020663 |
| Ssc.21224.1.A1_at | −2.352619 | KRT7 | Keratin 7 | NM_005556 |
| Ssc.6556.2.A1_at | −2.344349 | GABRA5 | Gamma-aminobutyric acid (GABA) A receptor, alpha 5 | NM_000810 |
| Ssc.27912.1.S1_at | −2.205287 | ZNF608 | Zinc finger protein 608 | NM_020747 |
| Ssc.24434.1.S1_at | −2.16435 | C14orf145 | Chromosome 14 open reading frame 145 | NM_152446.3 |
| Ssc.26326.1.A1_at | −2.134067 | CYP3A46 | Cytochrome P450 3A46 | NM_001134824 |
| Ssc.11260.1.A1_at | −2.124412 | GSTA1 | Glutathione S-transferase alpha 1 | NM_145740 |
| Ssc.8053.1.S1_at | −1.953481 | POLL | Polymerase (DNA directed), lambda | NM_013274 |
| Ssc.2618.1.S1_at | −1.827725 | MAMDC4 | MAM domain containing 4 | NM_206920 |
| Ssc.5641.1.S1_at | −1.80541 | ANK2 | Ankyrin 2, neuronal | NM_001127493 |
| Ssc.18992.1.A1_at | −1.784956 | ANKRD6 | Ankyrin repeat domain 6 | NM_014942 |
| Ssc.30445.1.A1_at | −1.761477 | ALK | Anaplastic lymphoma receptor tyrosine Kinase | NM_004304 |
| Ssc.27318.1.S1_at | −1.757688 | KRT7 | Keratin 7 | NM_005556 |
| Ssc.30532.1.A1_at | −1.725776 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | NM_005431 |
| Ssc.27626.1.S1_at | −1.719731 | EGFL6 | EGF-like-domain, multiple 6 | NM_015507 |
| Ssc.12223.1.A1_at | −1.690582 | PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | NM_012411 |
| Ssc.336.1.S1_at | −1.631299 | USP18 | Ubiquitin specific peptidase 18 | NM_213826 |
| Ssc.15708.1.S1_at | −1.621459 | HBZ | Hemoglobin, zeta | NM_005332 |
| Ssc.7243.1.A1_at | −1.619644 | CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | NM_001009580 |
| Ssc.21162.1.S1_s_at | −1.609334 | IRF7 | Interferon regulatory factor 7 | NM_001097428 |
| Ssc.286.1.S1_s_at | −1.553267 | IRG6 | Inflammatory response protein 6 | NM_213817 |
Table 3.
List of top 30 genes up-regulated in the uterine endometrium on day (D) 15 of pregnancy compared to genes on D15 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.28494.1.S1_at | 3.9383217 | RAB5B | RAB5B, member RAS oncogene family | NM_002868 |
| Ssc.18273.1.A1_at | 3.9374428 | MAGI1 | Membrane associated guanylate kinase, WW and PDZ domain containing 1 | NM_173515 |
| Ssc.22734.1.A1_at | 3.505833 | EDEM3 | ER degradation enhancer, mannosidase alpha- like 3 | NM_025191 |
| Ssc.6392.1.S1_at | 3.3603669 | NTM | Neurotrimin | NM_001144058.1 |
| Ssc.25740.1.A1_at | 3.3474057 | AKAP1 | A kinase (PRKA) anchor protein 1 | NM_003488 |
| Ssc.9329.1.A1_at | 3.0693123 | PTPLAD2 | Protein tyrosine phosphatase-like A domain containing 2 | NM_001010915 |
| Ssc.10593.2.A1_at | 3.0355328 | IFI44L | Interferon-induced protein 44-like | NM_006820 |
| Ssc.11925.1.A1_at | 2.8589389 | SEL1L | Sel-1 suppressor of lin-12-like (C. Elegans) | NM_005065 |
| Ssc.25678.1.S1_at | 2.8213114 | SYT1 | Synaptotagmin I | NM_005639 |
| Ssc.12809.5.A1_at | 2.7832047 | HBA2 | Hemoglobin, alpha 2; hemoglobin, alpha 1 | NM_000517 |
| Ssc.7455.1.A1_at | 2.7595911 | AASDHPPT | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | NM_015423 |
| Ssc.7682.1.A1_at | 2.6842858 | OSBPL9 | Oxysterol binding protein-like 9 | NM_024586 |
| AFFX-Ss_18SrRNA_at | 2.6236143 | AFFX-Ss_18SrRNA | AFFX-Ss_18SrRNA | Null |
| Ssc.21585.2.S1_at | 2.5961034 | UBR7 | Ubiquitin protein ligase E3 component n-recognin 7 (putative) | NM_001100417 |
| Ssc.13473.1.A1_at | 2.5538991 | UGCG | UDP-glucose ceramide glucosyltransferase | NM_003358 |
| Ssc.27407.1.A1_at | 2.5424531 | NRIP1 | Nuclear receptor interacting protein 1 | NM_003489 |
| Ssc.3975.2.A1_at | 2.4775446 | STK38L | Serine/threonine kinase 38 like | NM_015000.3 |
| Ssc.28192.1.A1_at | 2.4555041 | FAM153A | Family with sequence similarity 153, member A | NM_173663 |
| Ssc.20353.1.S1_at | 2.3855645 | MALT1 | Metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) | NR_002819.2 |
| Ssc.11870.1.A1_at | 2.2433352 | NRIP1 | Nuclear receptor interacting protein 1 | NM_003489 |
| Ssc.17508.2.A1_at | 2.2266604 | RCAN2 | Regulator of calcineurin 2 | NM_005822 |
| Ssc.8143.2.A1_at | 2.1943771 | RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 | NM_014247 |
| Ssc.17152.2.A1_at | 2.1849978 | COQ2 | Coenzyme Q2 homolog, prenyltransferase (Yeast) | NM_015697 |
| Ssc.27130.1.A1_at | 2.1615496 | SLC25A36 | Solute carrier family 25, member 36 | NM_001104647 |
| Ssc.3325.3.S1_at | 2.1184436 | EXT1 | Exostoses (multiple) 1 | NM_000127 |
| Ssc.22645.1.S1_at | 2.0685056 | DDX46 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | NM_014829 |
| Ssc.6175.1.S1_at | 2.0623846 | LCOR | Ligand dependent nuclear receptor corepressor | NM_032440 |
| Ssc.13601.1.A1_at | 1.8954884 | MYO1B | Myosin IB | NM_001130158 |
| Ssc.24402.1.S1_at | 1.8920959 | CCDC76 | Coiled-coil domain containing 76 | NM_019083 |
| Ssc.27133.1.A1_at | 1.8789606 | TMEM68 | Transmembrane protein 68 | NM_152417 |
Table 4.
List of top 30 genes down-regulated in the uterine endometrium on day (D) 15 of pregnancy compared to genes on D15 of the estrous cycle by average fold change
| Probe set ID | Average log_2(Fold change) | Gene symbol | Gene title | RefSeq transcript ID |
|---|---|---|---|---|
| Ssc.25087.1.S1_at | −4.665941 | DNAJC13 | DnaJ (Hsp40) homolog, subfamily C, member 13 | NM_015268 |
| Ssc.2015.2.S1_at | −4.469763 | ZNF449 | Zinc finger protein 449 | NM_152695 |
| Ssc.7817.1.A1_at | −4.302148 | NETO1 | Neuropilin (NRP) and tolloid (TLL)-like 1 | NM_138966 |
| Ssc.23424.1.A1_at | −4.072875 | DTNA | Dystrobrevin, alpha | NM_001128175 |
| Ssc.8989.1.A1_at | −3.882042 | PPHLN1 | Periphilin 1 | NM_016488 |
| Ssc.21088.2.S1_at | −3.009555 | CSTF1 | Cleavage stimulation factor, 3′ pre-RNA, subunit 1, 50kDa | NM_001033521 |
| Ssc.9834.2.S1_at | −2.875995 | Null | Sn36_F05.f sn Sus scrofa cDNA | CB471302.1 |
| Ssc.1600.1.A1_at | −2.684489 | RIMKLB | Ribosomal modification protein rimK-like family member B | NM_020734.2 |
| Ssc.1600.1.A1_a_at | −2.616954 | RIMKLB | Ribosomal modification protein rimK-like family member B | NM_020734.2 |
| Ssc.27385.1.S1_at | −2.580643 | RBP7 | Retinol binding protein 7, cellular | NM_001145222 |
| Ssc.668.1.S1_at | −2.543521 | PTH | Parathyroid hormone | NM_214401 |
| Ssc.958.1.S1_at | −2.543376 | ALDOB | Aldolase B, fructose-bisphosphate | NM_000035 |
| Ssc.27953.1.S1_at | −2.289442 | MLL | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | NM_005933 |
| Ssc.21468.2.A1_at | −2.194019 | LARP1 | La ribonucleoprotein domain family, member 1 | NM_015315 |
| Ssc.30456.1.A1_at | −2.155805 | CCDC135 | Coiled-coil domain containing 135 | NM_032269 |
| Ssc.27345.1.S1_at | −2.107692 | Null | 940760 MARC 4PIG Sus scrofa cDNA | CN153625.1 |
| Ssc.7219.1.A1_at | −1.926105 | ERC2 | ELKS/RAB6-interacting/CAST family member 2 | NM_015576 |
| Ssc.22791.1.A1_at | −1.922899 | EEA1 | Early endosome antigen 1 | NM_003566 |
| Ssc.7672.1.A1_at | −1.847485 | CAPZB | Capping protein (actin filament) muscle Z-line, beta | NM_004930 |
| Ssc.25130.1.A1_at | −1.83943 | C6orf98 | Chromosome 6 open reading frame 98 | NM_001099267 |
| Ssc.30379.1.A1_at | −1.831398 | ARMC3 | Armadillo repeat containing 3 | NM_173081 |
| Ssc.7191.1.A1_at | −1.805 | LSM14A | LSM14A, SCD6 homolog A (S. Cerevisiae) | NM_001114093 |
| Ssc.2589.1.S1_at | −1.790171 | TNNI3 | Troponin I type 3 (cardiac) | NM_001098599 |
| Ssc.23948.1.A1_at | −1.727051 | DYNC1I2 | Dynein, cytoplasmic 1, intermediate chain 2 | NM_001378 |
| Ssc.8133.1.A1_at | −1.725898 | GPM6B | Glycoprotein M6B | NM_001001994 |
| Ssc.28842.1.A1_at | −1.685047 | CCDC113 | Coiled-coil domain containing 113 | NM_014157 |
| Ssc.7917.1.A1_at | −1.680845 | Null | MI-P-E6-ahn-e-09-1-UM.s1 MI-P-E6 Sus scrofa cDNA clone | BQ599768.1 |
| Ssc.29910.1.A1_at | −1.633574 | MTMR2 | Myotubularin related protein 2 | NM_016156 |
| Ssc.6616.1.S1_at | −1.629585 | C1orf92 | Chromosome 1 open reading frame 92 | NM_144702 |
| Ssc.28950.1.A1_at | −1.627009 | EXOC6 | Exocyst complex component 6 | NM_019053.4 |
Table 5.
Functional annotation clusters of up-regulated genes in uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 2.56 | Membrane fraction (12), cell fraction (14), insoluble fraction (12) |
| 2 | 1.04 | Endoplasmic reticulum part (6), endoplasmic reticulum membrane (5), endoplasmic reticulum (10), nuclear envelope-endoplasmic reticulum network (5), microsome (4), vesicular fraction (4), organelle membrane (9), endomembrane system (6) |
| 3 | 0.88 | Blood vessel morphogenesis (4), blood vessel development (4), vasculature development (4) |
| 4 | 0.86 | Cellular macromolecule catabolic process (9), proteolysis (11), macromolecule catabolic process (9), proteolysis involved in cellular protein catabolic process (7), cellular protein catabolic process (7), protein catabolic process (7), ubiquitin- protein ligase activity (3), small conjugating protein ligase activity (3), ligase activity, forming carbon-nitrogen bonds (3), ubiquitin-dependent protein catabolic process (3), modification-dependent macromolecule catabolic process (6), acid- amino acid ligase activity (3), modification-dependent protein catabolic process (6) |
| 5 | 0.80 | Intracellular organelle lumen (14), organelle lumen (14), nuclear lumen (10), membrane-enclosed lumen (14), nucleoplasm (8) |
| 6 | 0.72 | Proteolysis (11), metalloendopeptidase activity (3), endopeptidase activity (5), negative regulation of apoptosis (5), negative regulation of programmed cell death(5), negative regulation of cell death (5), metallopeptidase activity (3), peptidase activity, acting on L-amino acid peptides (5), peptidase activity (5), regulation of apoptosis (6), regulation of programmed cell death (6), regulation of cell death (6) |
| 7 | 0.72 | Phosphatase activity (5), phosphoprotein phosphatase activity (4), phosphate metabolic process (9), dephosphorylation (4), phosphorus metabolic process (9), protein amino acid dephosphorylation (3), protein serine/threonine kinase activity(3), phosphorylation (5), protein amino acid phosphorylation (5), protein kinase activity (3) |
| 8 | 0.71 | Positive regulation of multicellular organismal process (4), positive regulation of cell proliferation (5), regulation of growth (4) |
| 9 | 0.65 | Cell cycle (8), mitotic cell cycle (5), cell cycle process (5), cell cycle phase (4) |
| 10 | 0.65 | Glycosaminoglycan binding (3), polysaccharide binding (3), pattern binding (3), carbohydrate binding (3) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D12 of pregnancy compared with those on D12 of the estrous cycle through DAVID.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 6.
Functional annotation clusters of down-regulated genes in uterine endometrium on D12 of pregnancy compared with those on D12 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 2.04 | Plasma membrane part (22), integral to plasma membrane (11), intrinsic to plasma membrane (11) |
| 2 | 2.03 | Cell adhesion (11), biological adhesion (11), cell-cell adhesion (4) |
| 3 | 1.50 | Leukocyte activation (7), cell activation (7), lymphocyte activation (6), T cell activation (4), immune effector process (4), regulation of leukocyte activation (4), regulation of cell activation (4), leukocyte mediated immunity (3), cell surface (5), positive regulation of response to stimulus (4), positive regulation of immune system process (4), immune response (7), external side of plasma membrane (3) |
| 4 | 1.48 | Immune system development (7), hemopoiesis (6), hemopoietic or lymphoid organ development (6), myeloid cell differentiation (3), protein homodimerization activity (4), nucleoplasm part (3) |
| 5 | 1.46 | Cell junction (7), adherens junction (4), anchoring junction 4) |
| 6 | 1.45 | Biopolymer glycosylation (4), glycoprotein biosynthetic process (4), glycoprotein metabolic process (4), glycosylation (4), protein amino acid glycosylation (4) |
| 7 | 0.98 | Apoptosis (7), programmed cell death (7), cell death (7), death (7), negative regulation of cell proliferation (4) |
| 8 | 0.83 | DNA metabolic process (8), response to DNA damage stimulus (5), DNA repair (4), cellular response to stress (5), DNA binding (7) |
| 9 | 0.83 | Induction of apoptosis (5), induction of programmed cell death (5), positive regulation of apoptosis (5), positive regulation of programmed cell death (5), positive regulation of cell death (5), regulation of apoptosis (7), regulation of programmed cell death (7), regulation of cell death (7) |
| 10 | 0.76 | Response to ionizing radiation (3), response to radiation (3), response to abiotic stimulus (3) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D15 of pregnancy compared with those on D15 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 7.
Functional annotation clusters of up-regulated genes in uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 3.46 | Nuclear speck (7), nuclear body (8), RNA splicing (9) |
| 2 | 2.21 | Nucleotide binding (30), purine ribonucleotide binding (25), AT P binding (21), ribonucleotide binding (25), purine nucleotide binding (25), adenyl ribonucleotide binding (21), adenyl nucleotide binding (21), purine nucleoside binding (21), nucleoside binding (21) |
| 3 | 2.12 | Nuclear body (8), nucleoplasm part (12), nucleoplasm (15), membrane-enclosed lumen (24), nuclear lumen (20), intracellular organelle lumen (23), organelle lumen (23), nucleolus (9) |
| 4 | 1.51 | RNA splicing (9), mrna metabolic process (9), RNA processing (11), mrna processing (8), spliceosome assembly (3), RNA splicing, via transesterification reactions (5), RNA splicing, via transesterification reactions with bulged adenosine as nucleophile (5), nuclear mrna splicing, via spliceosome (5), RNA binding (11), ribonucleoprotein complex assembly (3), ribonucleoprotein complex biogenesis (4), cellular macromolecular complex assembly (5), cellular macromolecular complex subunit organization (5) |
| 5 | 1.15 | Nuclear body (8), nuclear export (4), nucleocytoplasmic transport (4), nuclear transport (4), mrna transport (3), establishment of RNA localization (3), nucleic acid transport (3), RNA transport (3), RNA localization (3), nucleobase, nucleoside, nucleotide and nucleic acid transport (3), intracellular transport (8) |
| 6 | 1.06 | Receptor-mediated endocytosis (4), membrane organization (8), membrane invagination (4), endocytosis (4), vesicle-mediated transport (6) |
| 7 | 0.96 | Helicase activity (6), ATP-dependent helicase activity (3), purine NTP-dependent helicase activity (3), atpase activity, coupled (4), atpase activity (4) |
| 8 | 0.95 | Enzyme binding (8), protein kinase binding (4), kinase binding(4) |
| 9 | 0.94 | Macromolecular complex assembly (11), macromolecular complex subunit organization (11), protein complex assembly (8), protein complex biogenesis (8), cellular macromolecular complex assembly (5), cellular macromolecular complex subunit organization (5), protein oligomerization (3) |
| 10 | 0.75 | Protein serine/threonine kinase activity (7), enzyme linked receptor protein signaling pathway (6), phosphate metabolic process (11), phosphorus metabolic process (11), protein kinase activity (8), protein amino acid phosphorylation (8), phosphorylation (9) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D12 of pregnancy compared with those on D12 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.
Table 8.
Functional annotation clusters of down-regulated genes in uterine endometrium on D15 of pregnancy compared with those on D15 of the estrous cycle
| Annotation clustera | Enrichment scoreb | Biological termsc |
|---|---|---|
| 1 | 1.68 | Adenyl ribonucleotide binding (15), purine ribonucleotide binding (17), purine nucleotide binding (17), ribonucleotide binding (17), nucleoside binding (15), adenyl nucleotide binding (15), purine nucleoside binding (15), ATP binding (14), nucleotide binding (18) |
| 2 | 1.50 | Cytoskeletal part (11), microtubule cytoskeleton (7), intracellular non-membrane- bounded organelle (18), non-membrane- bounded organelle (18), centrosome (4), cytoskeleton (13), microtubule organizing center (4) |
| 3 | 1.50 | Cytoskeletal part (11), dynein complex (3), microtubule associated complex (4), microtubule-based movement (4), microtubule cytoskeleton (7), motor activity (3), microtubule motor activity (3), microtubule (4), microtubule-based process (4) |
| 4 | 1.20 | Cytoskeletal part (11), regulation of actin filament length (3), regulation of cellular component size (5), regulation of actin cytoskeleton organization (3), regulation of actin filament-based process (3), regulation of organelle organization (4), actin cytoskeleton (4), cytoskeletal protein binding (6), regulation of cytoskeleton organization (3), actin binding (4) |
| 5 | 1.17 | Regulation of neuron differentiation (4), regulation of neurogenesis (4), regulation of nervous system development (4), regulation of cell development (4), neuron differentiation (4) |
| 6 | 0.92 | Protein amino acid dephosphorylation (4), dephosphorylation (4), phosphorus metabolic process (9), phosphoprotein phosphatase activity (4), phosphate metabolic process (9), protein tyrosine phosphatase activity (3), phosphatase activity (4), protein kinase activity (5), protein serine/threonine kinase activity (4), protein amino acid phosphorylation (5), phosphorylation (5) |
| 7 | 0.84 | Central nervous system neuron axonogenesis (3), central nervous system neuron development (3), central nervous system neuron differentiation (3), cell projection organization (5), cell motion (5), axonogenesis (3), cell morphogenesis involved in neuron differentiation (3), neuron projection morphogenesis (3), cell projection morphogenesis (3), cell morphogenesis involved in differentiation (3), cell part morphogenesis (3), neuron differentiation (4), neuron projection development (3), cell morphogenesis (3), cellular component morphogenesis (3), neuron development (3) |
| 8 | 0.82 | Regulation of synaptic transmission (3), regulation of transmission of nerve impulse (3), regulation of neurological system process (3), regulation of system process (4) |
| 9 | 0.75 | Cell projection organization (5), cytoskeletal protein binding (6), cytoskeleton organization (3) |
| 10 | 0.71 | Learning or memory (3), neurological system process (10), behavior (5), cognition (4) |
Top ten of annotation clusters identified by official gene symbols of up-regulated gene list on D15 of pregnancy compared with those on D15 of the estrous cycle.
Enrichment score represents significance of each cluster annotation and the relatedness of the terms and the genes associated with terms. Enrichment score is calculated by overall EASE scores (the modified Fisher extract p value) of each term member.
The gene ontology (GO) terms are group of terms having similar biological meaning with respect to biological process, cellular component, and molecular function. GO functional annotation term (FAT) was used for the analysis because GO term cannot universally define the specificity of a given term. The number in parenthesis indicate the number of differentially expressed genes contribute to the clustered term.