Abstract
The swine leukocyte antigen class II molecules are possibly associated with the induction of protective immunity. The study described here was to investigate the relationship between polymorphisms in exon 2 of the swine DQA gene and piglet diarrhea. This study was carried out on 425 suckling piglets from three purebred pig strains (Large White, Landrace and Duroc). The genetic diversity of exon 2 in swine DQA was detected by PCR-SSCP and sequencing analysis, eight unique SSCP patterns (AB, BB, BC, CC, CD, BD, BE and DD) representing five specific allele (A to E) sequences were detected. Sequence analysis revealed 21 nucleotide variable sites and resulting in 12 amino acid substitutions in the populations. A moderate level polymorphism and significant deviations from Hardy-Weinberg equilibrium of the genotypes distribution were observed in the populations (p<0.01). The association analysis indicated that there was a statistically significant difference in the score of piglet diarrhea between different genotypes, individuals with genotype CC showed a lower diarrhea score than genotypes AB (0.98±0.09), BB (0.85±0.77) and BC (1.25±0.23) (p<0.05), and significantly low than genotype BE (1.19±0.19) (p<0.01), CC genotype may be a most resistance genotype for piglet diarrhea.
Keywords: SLA, DQA Gene, Genetic Polymorphism, Piglet Diarrhea
INTRODUCTION
The Large White, Landrace, and Duroc are three important commercial pig breeds in the intensive breeding pig farms and are well-known for their characteristics of high prolificacy, fast growth and high lean rates. Due to large scale intensive production systems, pigs are exposed on various pathogens that cause infectious diseases and pathological symptoms (Lunney et al., 2009).
Piglet diarrhea is one of the serious threats to economic success in the swine industry, which causes increased mortality rates, poor growth and additional medical costs. It is estimated that 50% of piglet mortality is due to diarrhea deaths (Morris et al., 2002). Generally, the incidence of this disease is controlled by the combined function of genetic factors and pathogens such as contagious and environmental pathogens and poor management practices. An effective stimulation of the adaptive immune system is based on the activation of T lymphocytes via their recognition of pathogen-derived peptides presented by major histocompatibility complex (MHC) molecules to the respective T-cell receptors. The swine MHC, which codes for a series of extremely polymorphic cell-surface glycoproteins, is referred to swine leukocyte antigens (SLA) which function is to capture antigen peptides as well as present them to T-lymphocytes, thereby influencing host challenge to infectious disease (Lunney et al., 2009; Patch et al., 2011).
The associations of porcine SLA complex with swine immune responses, as well as disease resistance and susceptibility, have led to a number of comprehensive studies. SLA alleles and haplotypes have been associated with difference in swine immune response to various infectious agents (Mallard et al., 1989; Renard and Vaiman, 1989; Shinkai et al., 2012). In the NIH miniature pigs, SLA haplotypes have been shown to affect antibody responses to sheep erythrocytes and cell-mediated immune responses to tuberculin-purified protein (Mallard et al., 1989). Tissot et al. (1989) also found a relationship of haplotype B with a higher susceptibility of swine cutaneous malignant melanoma in Sinclair miniature swine. A possible SLA influence on the inherent disease susceptibility and resistance in PRRS has also been reported (Lewis et al., 2007; Molina et al., 2008). Yang et al. (2012) reported that polymorphism of the SLA-DRA gene has significant influence on diarrhea of piglets. All of those works indicated that a detailed analysis of SLA genes is essential to advance our understanding of the processes implicated in immune responses and the effects of intensive selection, such as via strict breeding programs.
Generally, the extensive polymorphism of MHC genes is expected to broaden the array of antigenic peptides that available for T-cell immunological recognition in stimulating and regulating immune surveillance. This extremely polymorphism, is essentially concentrated in the first domain (exon 2), in a region which forms the floor and wall of the peptide-binding groove of the class II genes (Smith et al., 2005).
The SLA class II genes expressed on the surface of professional antigen presenting cells are noted as SLA-DRA, SLA-DRB1, SLA-DQA, and SLA-DQB1. SLA-DRB1 and SLA-DQB1 loci display a very high degree of polymorphism, although the SLA-DQA gene has been reported as a moderately polymorphic gene, and SLA-DRA was reported to exhibit only limited polymorphism (Lunney et al., 2009; Ho et al., 2009; Ho et al., 2010). Research utilizing SLA class II genes primarily focus on exon 2 of the DRB and DQB because these loci code for the peptide binding region (PBR) which is in direct contact with the processed peptide (Babik et al., 2005; Luetkemeier et al., 2009; Park et al., 2010). However, there is currently no study on the relationship between SLA-DQA gene and piglet diarrhea. The swine DQA gene encodes the α chain of DQ antigen which is closely linked to DRB. The full-length of the genome sequence of this gene contains 5.5 kb that encodes for a polypeptide of 255 amino acid residues, which include four exons, with exon 1 encoding signal peptide region, with exon 2 and 3 encoding the corresponding extracellular α1 and α2 domains, and exon 4 encoding both transmembrane and cytoplasmic domains (Lunney et al., 2009).
Consequently, in the present study we characterized the genetic variation of the exon 2 of SLA II DQA locus from Large White, Landrace and Duroc pig strains and investigate the correlation between genotypes and piglet diarrhea, to identify the suitable candidate markers correlated with animal resistance potentially for use in genetic strategies to breed disease resistance pigs.
MATERIALS AND METHODS
Samples and data collection
The animals used in this experiment were from Linze Xinghua Pig Breeding Farm (Zhangye, Gansu, China). A total of 425 pure piglets were examined, including Landrace (n = 146), Large White (n = 148) and Duroc (n = 131) pigs.
Experimental piglets were born in June 2010 and the experiment conducted under forage and feeding management conditions and disease control in accordance with the Guide for Chinese Feeding Standard of Swine and approved by the institute of Ministry of Agriculture Feed Industry Center, China Agricultural University (Beijing, China). All piglets were monitored two times a day during the entire suckling period (from birth to weaning, 0 to 28 d) and assigned a daily score based on visual analysis of symptoms traits as follows: 0- normal, solid feces; 1- slight diarrhea, soft and loose feces; 2- moderate diarrhea, semi-liquid feces; 3- severe diarrhea, liquid and unformed feces (Kelly et al., 1990). Genomic DNA was extracted from swine ear tissues samples by standard phenol-chloroform extraction procedure (Sambrook and Russell, 2001) and quantified by using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA).
Genotyping by polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP)
PCR was performed to amplify a 378 bp fragment of the SLA-DQA gene which encompassed 81 bp intron 1, 249 bp exon 2 and 48 bp intron 2 using forward primer SLA-DQAe2F (5′-AGTCAAGTTCTCTTGTCACT-3′) and SLA-DQAe2R (5′-TGTGAACGGGTAGATTCTGT-3′). The primers were designed according to the published SLA-DQA sequence in GenBank (accession No. AY303988). PCR analyses were carried out in total of a 25 μl volume with 100 ng genomic DNA, 10 pmol of each primer, 2.5 mM dNTPs, 5 U of Taq DNA polymerase (TaKaRa, Dalian, China), and 10×reaction buffer (including 15 mM Mg2+). PCR conditions included an initial denaturing of 2 min at 94°C, followed by 30 cycles of a three-step process of denaturation at 94°C for 20 s, 55°C for 30 s and extension at 72°C for 30 s, followed by a final extension at 72°C for 10 min. PCR products were detected on a 2% agarose gel electrophoresis.
All samples were screened using SSCP analysis to scan for polymorphism in the amplified region. About 2 μl PCR product was mixed with 6 μl of the denaturing solution (98% formamide, 0.025% bromophenol blue, 0.025% xylene cyanole, and 10 mM EDTA), denatured for 10 min at 98°C, and immediately chilled on ice for 10 min. Denatured DNA samples were placed on 17 cm×17 cm, 10% acrylamide: bisacrylamide gel (29: 1) gels. Electrophoresis was run in 1×TBE buffer of 30 min (250V) at 4°C and then 15 h (180V) at room temperture. Gels were silver-stained according to the method of Byun et al. (2009).
Cloning and sequencing
To determine the sequence variation in the swine DQA gene, PCR products of individuals that showed different SSCP patterns were purified using DNA Fragment Quick Recover Kit (TaKaRa, Dalian, China). The purified PCR products were ligated into pMD®19-T vectors (TaKaRa, Dalian, China) and transformed into the Escherichia coli DH 5α strain. Recombinant clones were detected by blue/white screening, between 10 to 15 insert-positive clones for each transformation were picked and incubated overnight in terrific Luria-Bertani broth at 37°C, in a vibrating rotary incubator (200 rpm). Clones were screened using the PCR-SSCP approach as described by Zhou et al. (2008), and only those clones for which the SSCP patterns matched those of the corresponding genomic DNA were selected and subsequently sequenced by Sangon Biotech (Shanghai, China) Co., Ltd.
Statistical analysis
Sequence alignment, translation and comparison were performed using MEGA5 (Tamura et al., 2011); genotype and allele frequencies, observed heterozygosity (He), and testing for deviation from Hardy-Weinberg equilibrium were calculated by the POPGENE version 1.32 (1997); polymorphism information content (PIC) was calculated by Botstein’s methods (1980).
The average rates of nonsynonymous (dN) and synonymous (dS) substitution for antigen-binding sites (ABS) and non-antigen binding sites (non-ABS) were calculated according to Nei and Gojobori (1986) method with Jukes-Cantor (1969) correction, using the package MEGA5 (Tamura et al., 2011). The porcine ABS was defined by assuming congruence with human MHC sites (Brown et al., 1993).
The effects of genotype on piglet diarrhea were estimated using the general linear model (GLM) procedure of SAS software (SAS, 1999) according to the following statistical model:
Where Yijkl is the observation value of piglet diarrhea score, μ is the overall population means, Bi is the fixed effect of breed, Sj is fixed effect of gender, Gk is the effect of genotype, and e is the random residual effect. Preliminary analysis also included the fixed interaction effects of breed, sex and genotype, however, these interaction effects were subsequently removed because it did not have a significant effect.
RESULTS
Polymorphism analysis in exon 2 of the SLA-DQA gene
A 378 bp amplified fragment of the SLA-DQA gene was obtained. Based on the PCR products, SSCP was used for initial screening of samples, a total of 8 unique SSCP patterns (AB, BB, BC, CC, CD, BD, BE and DD) were detected among 425 pigs (Figure 1). Genotypes AB, BB, BD, BE and BC were detected in all three pig strains, genotype CC was only detected in the Landrace breed and genotypes CD and DD were only detected in Duroc breed. Homozygous individuals were found for alleles B, C and D, but alleles A and E only appeared in heterozygous individuals. Allele B was the most common allele, with frequencies of 0.61, 077, and 0.69 in Large White, Landrace and Duroc pigs, respectively.
Figure 1.
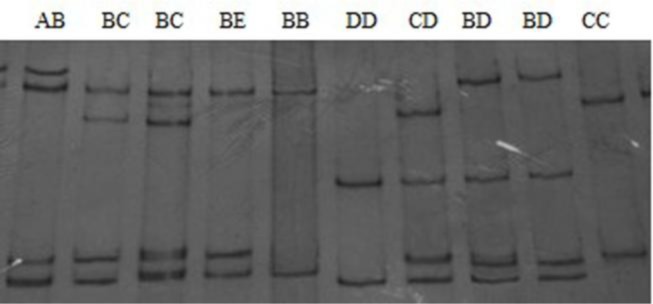
The genotyping results of the SLA-DQA gene exon 2 by PCR-SSCP.
Significant deviations from Hardy-Weinberg equilibrium were detected in the genotypes distribution in Large White, Landrace and Duroc pig populations (p<0.01). The observed levels of heterozygosity ranged from 0.39 to 0.54, and the PIC for the studied populations ranged from 0.37 to 0.47 (Table 1). According to the protocol of Vaiman et al. (1994), this locus is moderately polymorphic in the three populations (0.25<PIC<0.50).
Table 1.
Genotype and allele frequency, population genetic parameters of the SLA-DQA exon 2 in three pig strains
| Breed | Large White | Landrace | Duroc |
|---|---|---|---|
| n | 146 | 148 | 131 |
| Genotype frequency | |||
| AB | 0.60 (87) | 0.14 (20) | 0.16 (21) |
| BB | 0.22 (32) | 0.60 (89) | 0.46 (60) |
| BD | 0.09 (13) | 0.04(6) | 0.04 (5) |
| BE | 0.05 (8) | 0.10 (15) | 0.14 (18) |
| BC | 0.04 (6) | 0.07 (10) | 0.14 (18) |
| CC | 0.00 (0) | 0.06 (8) | 0.00 (0) |
| CD | 0.00 (0) | 0.00 (0) | 0.05 (6) |
| DD | 0.00 (0) | 0.00 (0) | 0.02 (3) |
| Allele frequency | |||
| A | 0.30 | 0.07 | 0.08 |
| B | 0.61 | 0.77 | 0.69 |
| C | 0.02 | 0.09 | 0.09 |
| D | 0.04 | 0.02 | 0.06 |
| E | 0.03 | 0.05 | 0.07 |
| x2 (HWE) | 59.29* | 57.37* | 43.74* |
| He | 0.54 | 0.39 | 0.49 |
| PIC | 0.47 | 0.37 | 0.46 |
x2 (HWE) = Chi-square test for Hardy-Weinberg equilibrium.
He = Observed heterozygosity. PIC = Polymorphism information content.
Hardy-Weinberg disequilibrium, p<0.01.
The nucleotide characterizations of these polymorphisms were revealed by cloning and sequencing. Comparison with the existing alleles in the Immuno Polymorphism Database (IPD)-MHC SLA sequence database (http://www.ebi.ac.uk/ipd/mhc/sla/) found two new mutations (c.3994T>A and c.4065G>A) for the first time in swine, which formed two new alleles. The new allele sequences have been deposited in GenBank database (http://www.ncbi.nlm.nih.gov/nuccore/) under accession number JX840404, JX840405.
Alignments of nucleotide and amino acid sequences showed that twenty-one of 249 (8.43%) nucleotide (Figure 2) and twelve of 82 (14.63%) amino acid variation sites (Figure 3) were identified in exon 2 of the swine DQA gene. We estimated the amino acid variation separately for PBR and non-PBR, the nonsynonymous substitutions detected cause changes at 12 amino acid sites, 6 (31.57%) were included in the 19 PBR (marked with+in Figure 3) sites, whereas only 6 (9.52%) of the 63 amino acid positions were polymorphic in the non-PBR. The rate of synonymous substitutions among DQA alleles occurred at higher frequencies than nonsynonymous substitutions at all sites as well as for the ABS and non-ABS in this study (dN/dS <1, purifying selection) (Table 2).
Figure 2.
Alignment of the SLA-DQA exon 2 allele nucleotide sequences refer to the sequence AY303988. Sequences correspond to positions 3898-4146 were the complete exon 2. Identical nucleotides are replaced by · and the substitutions are written as letters. Newly identified sequences are marked with * which have assigned GenBank accession numbers.
Figure 3.

Alignment of the SLA-DQA exon 2 allele amino acid sequences refer to the sequence AY30398. Putative antigen-binding sites (ABS) are marked with plus which have been defined according to Brown et al. (1993).
Table 2.
The average rates of synonymous substitutions (dS) and nonsynonymous substitutions (dN) among 5 SLA-DQA exon 2 alleles
| Positions | N | Nv | dN | dS | dN/dS |
|---|---|---|---|---|---|
| Combined | 82 | 12 | 0.034 (0.008) | 0.055 (0.009) | 0.618 |
| PBR | 19 | 6 | 0.085 (0.028) | 0.109 (0.023) | 0.800 |
| Non-PBR | 63 | 6 | 0.019 (0.003) | 0.041 (0.010) | 0.463 |
N = Numbers of encoded amino acid.
Nv = Numbers of variable amino acid.
Standard errors (in parentheses) were obtained through 1,000 bootstrap replicates.
Effects of genotypes on piglet diarrhea
Statistical results showed that a significantly difference at swine DQA exon 2 locus was observed between genotype effect and piglet diarrhea (p<0.05), whereas breed and sex showed little influence on piglet diarrhea (p>0.05). The least square means along with standard errors for piglet diarrhea score among different genotypes are given in Table 3. The genotypic linear contrasts suggested that diarrhea score of genotypes CC (0.36±0.19) was extremely lower than that of genotype BE (1.19±0.19) (p<0.01), and significantly lower than genotypes AB (0.98±0.09), BB (0.85±0.77) and BC (1.25±0.23) (p<0.05) (Table 3). The animals with genotype DD were excluded when association analysis was carried out because the sample size was less than three.
Table 3.
Association of different genotypes of the exon 2 of SLA-DQA gene with piglet diarrhea score
| Genotype | Samples number | Least square means±standard errors |
|---|---|---|
| AB | 130 | 0.98±0.09a |
| BB | 173 | 0.85±0.77a |
| BD | 43 | 0.80±0.15ac |
| BE | 26 | 1.19±0.19A |
| BC | 18 | 1.25±0.23a |
| CC | 27 | 0.36±0.19Bbc |
| CD | 5 | 1.12±0.43ac |
| DD | 3 | 1.23±0.56ac |
Values with different superscripts within the same line differ significantly at p<0.01.
Values with different superscripts within the same line differ significantly at p<0.05.
DISCUSSION
Genetic diversity of the SLA-DQA gene
One of the most remarkable features of the MHC genes is the extremely high degree of genetic polymorphism within loci. Extensive genetic studies have been carried out on the MHC II DQA gene of various species (Hickford et al., 2007; Janova et al., 2009); 24 previously DQA alleles and 13 new alleles were identified in the genus Equus by Janova et al. (2009), and 37 ovine DQA haplotypes were detected in 520 sheep by Hickford et al. (2007). Variation in swine the DQA gene is well characterized, 20 alleles have been identified with a moderate degree of polymorphism (Lunney et al., 2009; Ho et al., 2010). In this study, we have successfully characterized the coding sequence of swine DQA exon 2, a total of twenty-one nucleotide variation sites were identified, defining 5 alleles. In particular, with the contribution of two novel alleles was identified. These indicate that the SLA-DQA gene, which is similar to other vertebrates, is one of the most polymorphic genetic loci, with high allelic diversity and high levels of sequence divergence among alleles.
The distribution of all genotypes had significantly deviations from Hardy-Weinberg equilibrium (p>0.05) in Large White, Landrace and Duroc pig populations. Five heterozygous genotypes (AB, BC, BE, CD and BD) and three homozygous genotypes (BB, CC and DD) were present in this study. This may attributable to the difference of survival in an environment with selection pressure from antigens and increased other environmental influences acting to dilute the impact of genetic variation (Andersson et al., 2012). The advantage for heterozygous individuals may be due to their wider antigen recognition spectrum than homozygous against a wide variety of pathogens (Sommer et al., 2005), while missing homozygous genotypes may have been eliminated in the breeding process (Beuzen et al., 2000).
He and PIC are considered to be suitable parameters to estimate the genetic variation of populations. The average diversity in SLA II genes has been shown to have a high polymorphism with He (0.628) and PIC (0.581) (Yu et al., 2012). The He and PIC of our study were greater than 0.37 in all three populations, this concurs with the results of Lunney et al. (2009) that indicate SLA-DQA diplays a middle level of polymorphism, compared to previous swine SLA studies by Yu et al. (2012).
A popular explanation for the extensive polymorphic nature of MHC genes is that it is driven and maintained by balancing selection. The classical way to investigate whether an MHC gene has been subject to balancing selection is to investigate the ratio between non-synonymous and synonymous substitutions (dN/dS) in the PBR. Intriguingly, in the present investigation a low proportion of dN and dS at PBR suggests that this gene actually has been subject to purifying selection. This is in contrast to the previous DQA studies in other vertebrates species (Zhu et al., 2007; Goüy de Bellocq et al., 2009; Kamathand and Getz, 2011). Through purifying selection, non-synonymous substitutions that probably affect normal gene function will usually be eliminated (Hughes and Yeager, 1998).
Association between genotypes and piglet diarrhea
Previously studies have established that DQA gene is implicated in human atopic dermatitis, Puumala virus and Cowpox virus infections in bank voles (Deter et al., 2008; Kiyohara et al., 2008). In pigs, SLA alleles and haplotypes had been confirmed to be associated with some immune-related diseases (Tissot et al., 1989; Chen et al., 2005). Renard and Vaiman (1989) indicated that mortality rate before weaning caused by diarrhea was linked to SLA haplotypes.
The SLA class II DQA gene encodes cell-surface glycoproteins that preferentially recognize foreign antigens and, subsequently, alter disease and vaccine responsiveness (Smith et al., 2005). However, there is currently no study on the relationship between SLA-DQA gene and piglet diarrhea, however, a study by Yang et al. (2012) reported the diarrhea score of SLA class II DRA genotypes AA (1.10±0.19) and BB (1.05±0.14) significantly higher than AC (0.32±0.25) and CC (0.06±0.12). In the present study, we also observed a statistically significant difference in the score of piglet diarrhea between different genotypes, genotype CC (0.36±0.19) indicated a lower diarrhea score than genotypes AB (0.98±0.09), BB (0.85±0.77), BC (1.25±0.23), and BE (1.19±0.19). This finding supports the previous observations by Yang et al. (2012) and Renard and Vaiman (1989).
Amino acid mutations in the coding region have a significant impact on the stability of proteins, which largely depend on the chemical and physical properties of the mutated amino acid, these include mutation position (whether in functional domain) and charge properties (Wang and Moult, 2001). Six of the 12 polymorphic amino acid sites were included in the peptide binding region in our study, which might play a crucial role in immunological processes modulating disease pathogenesis. Especially, in the case of c.4086A>G and c.4087C>T, as linked ACG nucleotide variation into GTG of allele C, changes the encoded amino acid from threonine to valine; and c.4089A>G and c.4090C>T, as linked TTA nucleotide into GGA of allele C, changes the encoded amino acid from leucine to glycine. These are notable as both caused changes of amino acid properties from hydrophobic and polar to lyophobic and nonpolar and therefore may affect protein function or stability causing variation in genotype. Our statistical analysis revealed CC genotype showed a lower diarrhea score than other genotypes, indicating that those four nucleotide mutations are possible functional mutations related to piglet diarrhea and genotype CC may be a most resistance genotype for piglet diarrhea.
To our knowledge, this is the first report describing a significant association of the variation in swine DQA with piglet diarrhea. The results reported here provide a theoretical foundation for further pig anti-disease resistance breeding research. Considering piglet diarrhea is influenced by multiple factors, such as parasite disease biology, genetic background, and sample size and especially MHC typing method, further works (sample amplification and DNA typing) should be carried out to facilitate future understanding the associations with specific alleles and SNPs. This will provide more meaningful information with possible application in future pig disease resistance breeding programs.
Acknowledgments
This work is supported by Bio-Technology Special Program of Agriculture and Animal Husbandry of Gansu Province, China (Grant No.GNSW-2008-04, No.092 NKDA036 and No.0804NKCA 065).
REFERENCES
- Andersson LS, Swinbune EJ, Meadows JRS, Broström H, Eriksson S, Fikse WF, Frey R, Sundquist M, Tseng CT, Mikko S, Lindgren G. The same ELA class II risk factors confer equine insect bite hypersensitivity in two distinct populations. Immunogenetics. 2012;64:201–208. doi: 10.1007/s00251-011-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babik W, Durka W, Radwan J. Sequence diversity of the MHC-DRB gene in the Eurasian beaver (Castor fiber) Mol Ecol. 2005;14:4249–4257. doi: 10.1111/j.1365-294X.2005.02751.x. [DOI] [PubMed] [Google Scholar]
- Beuzen ND, Stear MJ, Chang KC. Molecular markers and their use in animal breeding. Vet J. 2000;160:42–52. doi: 10.1053/tvjl.2000.0468. [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Byun SO, Fang Q, Zhou H, Hickford JGH. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal Biochem. 2009;385:174–175. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Chen FX, Xie J, Li NL, Zhou Y, Xin LJ, Chou KY. Novel SLA-DQ alleles and their recombinant molecules in xenogeneic stimulation of human T cells. Transpl Immunol. 2005;14:83–89. doi: 10.1016/j.trim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Deter J, Bryja J, Chaval Y, Galan M, Henttonen H, Laakkonen J, Voutilainen L, Vapalahti O, Vaheri A, Salvador AR, Morand S, Cosson JF, Charbonnel N. Association between the DQA MHC class II gene and Puumala virus infection in Myodes glareolus, the bank vole. Infect Genet Evol. 2008;8:450–458. doi: 10.1016/j.meegid.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Goüy de Bellocq J, Suchentrunk F, Baird SJE, Schaschl E. Evolutionary history of an MHC gene in two leporid species: characterisation of Mhc-DQA in the European brown hare and comparison with the European rabbit. Immunogenetics. 2009;61:131–144. doi: 10.1007/s00251-008-0349-4. [DOI] [PubMed] [Google Scholar]
- Hickford JGH, Zhou HT, Fang Q. Haplotype analysis of the DQA genes in sheep: Evidence supporting recombination between the loci. J Anim Sci. 2007;85:577–582. doi: 10.2527/jas.2006-217. [DOI] [PubMed] [Google Scholar]
- Ho CS, Lunney JK, Ando A, Rogel-Gaillard C, Lee JH, Schook LB, Smith DM. Nomenclature for factors of the SLA system, update 2008. Tissue Antigens. 2009;73:307–315. doi: 10.1111/j.1399-0039.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- Ho CS, Lunney JK, Lee JH, Franzo-Romain MH, Martens GW, Rowland RR, Smith DM. Molecular characterization of swine leucocyte antigen class II genes in outbred pig populations. Anim Genet. 2010;41:428–432. doi: 10.1111/j.1365-2052.2010.02019.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–434. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- Janova E, Matiasovic J, Vahala J, Vodicka R, Dyk EV, Horin P. Polymorphism and selection in the major histocompatibility complex DRA and DQA genes in the family Equidae. Immunogenetics. 2009;61:513–527. doi: 10.1007/s00251-009-0380-0. [DOI] [PubMed] [Google Scholar]
- Kamathand PL, Getz WM. Adaptive molecular evolution of the major histocompatibility complex genes, DRA and DQA, in the genus Equus. BMC Evol Biol. 2011;11:128. doi: 10.1186/1471-2148-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DJ, O’Brien JJ, McCracken KJ. Effect of creep feeding on the incidence, duration and severity of post-weaning diarrhea in pigs. Res Vet Sci. 1990;49:223–228. [PubMed] [Google Scholar]
- Kiyohara C, Tanaka K, Miyake Y. Genetic susceptibility to atopic dermatitis. Allergol Int. 2008;57:39–56. doi: 10.2332/allergolint.R-07-150. [DOI] [PubMed] [Google Scholar]
- Lewis CR, Ait-Ali T, Clapperton M, Archibald AL, Bishop S. Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS) Viral Immunol. 2007;20:343–358. doi: 10.1089/vim.2007.0024. [DOI] [PubMed] [Google Scholar]
- Luetkemeier ES, Malhi RS, Beever JE, Schook LB. Diversification of porcine MHC class II genes: evidence for selective advantage. Immunogenetics. 2009;61:119–129. doi: 10.1007/s00251-008-0348-5. [DOI] [PubMed] [Google Scholar]
- Lunney JK, Ho CS. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev Comp Immunol. 2009;33:362–374. doi: 10.1016/j.dci.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Mallard BA, Wilkie BN, Kennedy BW. Genetic and other effects on antibody and cell mediated immune response in swine leucocyte antigen (SLA)-defined miniature pigs. Anim Genet. 1989;20:167–178. doi: 10.1111/j.1365-2052.1989.tb00854.x. [DOI] [PubMed] [Google Scholar]
- Molina RM, Cha SH, Chittick W, Lawson S, Murtaugh MP, Nelson EA, Christopher-Hennings J, Yoon KJ, Evans R, Rowland RRR, Zimmerman JJ. Immune response against porcine reproductive and respiratory syndrome virus during acute and chronic infection. Vet Immunol Immunopathol. 2008;126:283–292. doi: 10.1016/j.vetimm.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Morris RS, Davies PR, Lawton DE. Evolution of diseases in the world’s pig industry. 17th International Pig Veterinary Society Congress Proceedings; Ames, Iowa. 2002. pp. 1–10. In. [Google Scholar]
- Park K, Choi H, Thong LM, Kwon OJ, Kim JH, Lee HT, Kim YB, Park SB, Park C. Simple and comprehensive SLA-DQB1 genotyping using genomic PCR and direct sequencing. Tissue Antigens. 2010;76:301–310. doi: 10.1111/j.1399-0039.2010.01522.x. [DOI] [PubMed] [Google Scholar]
- Patch JR, Pedersen LE, Toka FN, Moraes M, Grubman MJ, Nielsen M, Jungersen G, Buus S, Golde WT. Induction of foot-and-mouth disease virus-specific cytotoxic T cell killing by vaccination. Clin Vaccine Immunol. 2011;18:280–288. doi: 10.1128/CVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POPGENE Version 1.32. For Molecular Biology and Biotechnology Centre. University of Alberta and Center for International Forestry Research; Alberta: 1997. [Google Scholar]
- Renard C, Vaiman M. Possible relationships between SLA and porcine reproduction. Reprod Nutr Dev. 1989;29:569–576. doi: 10.1051/rnd:19890506. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2001. [Google Scholar]
- SAS . SAS user’s guide: Statistics (Version 8.01 Ed.) SAS Inst Inc; Cary, NC, USA: 1999. [Google Scholar]
- Shinkai H, Arakawa A, Tanaka-Matsuda M, Ide-Okumura H, Terada K, Chikyu M, Kawarasaki T, Ando A, Uenishi H. Genetic variability in swine leukocyte antigen class II and Toll-like receptors affects immune responses to vaccination for bacterial infections in pigs. Comp Immunol Microbiol Infect Dis. 2012;35:523–532. doi: 10.1016/j.cimid.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Smith DM, Lunney JK, Martens GW, Ando A, Lee JH, Ho CS. Nomenclature for factors of the SLA class-I system, 2004. Tissue Antigens. 2005;65:136–149. doi: 10.1111/j.1399-0039.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot RG, Beattie CW, Amoss MS. The swine leucocyte antigen (SLA) complex and Sinclair swine cutaneous malignant melanoma. Anim Genet. 1989;20:51–57. doi: 10.1111/j.1365-2052.1989.tb00841.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, proteins structer, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Vaiman D, Mecier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, Lepingle A, Velmala R, Kaukinen J, Varvio SL, Martin P, Leveziel H, Guerin G. A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome. 1994;5:288–297. doi: 10.1007/BF00389543. [DOI] [PubMed] [Google Scholar]
- Yang QL, Kong JJ, Zhao SG, Liu LX, Wang DW, Li FS, Jiang TT, Feng Y. Polymorphism of SLA-DRA gene exon 2 and its correlation with piglet diarrhea. Acta Vet Zootech Sin. 2012;43:1020–1027. [in Chinese, English abstract] [Google Scholar]
- Yu H, Liu RH, Li H, Zuo QZ, Li Y, Wu ZF. Genetic diversity based on swine leukocyte antigen complex mi-crosatellites (SLA-MS) in five pig populations. Yi Chuan. 2012;34:1427–1433. doi: 10.3724/sp.j.1005.2012.01427. [DOI] [PubMed] [Google Scholar]
- Zhou H, Hickford JGH. Clonal polymerase chain reaction-single-strand conformational polymorphism analysis: An effective approach for identifying cloned sequences. Anal Biochem. 2008;378:111–112. doi: 10.1016/j.ab.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Zhu L, Ruan XD, Ge YF, Wan QH, Fang SG. Low major histocompatibility complex class II DQA diversity in the Giant Panda (Ailuropoda melanoleuca). BMC Genet. 2007;8:29. doi: 10.1186/1471-2156-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]



