Abstract
In order to evaluate the genetic diversity and discrimination among five Korean native chicken lines, a total of 86 individuals were genotyped using 150 microsatellite (MS) markers, and 15 highly polymorphic MS markers were selected. Based on the highest value of the number of alleles, the expected heterozygosity (He) and polymorphic information content (PIC) for the selected markers ranged from 6 to 12, 0.466 to 0.852, 0.709 to 0.882 and 0.648 to 0.865, respectively. Using these markers, the calculated genetic distance (Fst), the heterozygote deficit among chicken lines (Fit) and the heterozygote deficit within chicken line (Fis) values ranged from 0.0309 to 0.2473, 0.0013 to 0.4513 and −0.1002 to 0.271, respectively. The expected probability of identity values in random individuals (PI), random half-sib (PIhalf-sibs) and random sibs (PIsibs) were estimated at 7.98×10−29, 2.88×10−20 and 1.25×10−08, respectively, indicating that these markers can be used for traceability systems in Korean native chickens. The unrooted phylogenetic neighbor-joining (NJ) tree was constructed using 15 MS markers that clearly differentiated among the five native chicken lines. Also, the structure was estimated by the individual clustering with the K value of 5. The selected 15 MS markers were found to be useful for the conservation, breeding plan, and traceability system in Korean native chickens.
Keywords: Discrimination, Diversity, Microsatellite, Korean Native Chicken, Traceability
INTRODUCTION
The Korean native chicken has been documented since approximately 2,000 years ago. Due to their poor commercial performance, Korean native chicken breeds almost became extinct. For this reason, Korean native chicken conservation strategies have been launched by the Korean government in 1994. Based on the 2 decades of the conservation project’s duration, five native chicken breeds with nine lines have been developed. In Korea, approximately 90% of chicken meat consumption is based on imported breeds. Recently, the poultry meat production has steadily increased, accounting for up to 20% of the total meat consumption in Korea (MIFAFF, 2009). Nowadays, many Korean consumers have a higher preference for native chicken meat than before, even though they may be 2 to 3 times as expensive as broilers.
Evaluation of genetic diversity for local breeds is becoming more challenging, and large efforts have been concentrated on maintaining minimum number of animals for each native species (FAO, 2007). There are extensive advantages of microsatellite (MS) markers because MS markers are abundant repeats of one to six bases, exhibit co-dominant inheritance, and are highly polymorphic and dispersed throughout the genome (Cheng and Crittenden, 1994; Kaya and Yildiz, 2008). In the Ark Database, the documented chicken MS markers are 2,483 markers, of which 435 are unmapped (Jacobsson et al., 2004). Until now, MS markers are the most widely used for the improvement of genetic selection management, parentage studies, evolutionary analysis, genetic traceability systems and QTL mapping (Blott et al., 1999; Dalvit et al., 2007; Almasy and Blangero, 2009). Previously, twenty two MS markers were used to assess chicken domestication in 52 populations. The results from identified alleles and the amount of genetic variation supported the hypothesis that the red jungle fowl was the ancient progenitor (Hillel et al., 2003). Therefore, MS markers are more suitable for the study of genetic diversity, correlation studies, and for identifying population structure among the chicken populations (Kong et al., 2006; Muchadeyi et al., 2007; Mwacharo et al., 2007; Tadano et al., 2007a; Tadano et al., 2007b; Berthouly et al., 2008; Bodzsar et al., 2009; Ding et al., 2010). On the other hand, the single nucleotide polymorphism (SNP) markers in MC1R gene were not sufficient for the discrimination of these Korean native chicken lines (data has not shown). Also, the phylogenetic relationships between Korean native chicken and other breeds have been investigated using D-loop sequence variations in mtDNA and attempts for discrimination of Korean native chicken lines were performed using mtDNA and LEI0258 marker (Hoque et al., 2009; Hoque et al., 2011).
In our studies, 15 markers have been selected from the 150 MS markers in the Ark database in order to investigate Korean native chicken lines to identify their genetic relationships. Also, these markers were used for the calculation of discrimination probabilities, which can be used for chicken traceability systems. Also, these results can be used in further breeding and conservation strategies for Korean native chicken.
MATERIALS AND METHODS
Sample collection and DNA extraction
Five Korean native chicken lines were collected from the National Institute of Animal Science (NIAS) in Korea. These lines were basically classified according to their feather colors, which were white (KNC_W), black (KNC_B), red-brown (KNC_R), yellow-brown (KNC_Y) and gray (KNC_G) lines. A total of 86 individuals were used for DNA extraction from blood samples collected from wing veins in tubes containing EDTA. Samples were stored at −20°C and genomic DNAs were extracted using a manual extraction method (Miller et al., 1988).
PCR amplification and genotyping
Initially, 150 MS markers were selected from the Ark Database (http://www.thearkdb.org/arkdb/) and were genotyped (Table 1). The primers used for genotyping were labeled with four fluorescence dyes (FAM, NED, VIC, PET) in forward primers. For the discrimination analysis of chicken lines, 15 highly polymorphic microsatellite markers were selected based on the number of alleles, expected heterozygosity (He) and polymorphic information content (PIC) values (Table 2). PCR was performed in an initial denaturation at 95°C for 10 min, followed by 35 cycles of 30 s at 95°C, 30 s at 60°C, 30 s at 72°C and a final extension at 72°C for 10 min using My-Genie96 Thermal Cycler (Bioneer, Korea). The PCR products were initially electrophoresed on 4% agarose gel with ethidium bromide, and DNA bands were visualized under ultraviolet light. For genotyping, the final genotyping reactions were based on 1 μl of 20X diluted PCR products, 10 μl of Hi-Di™ formamide (Applied Biosystems, USA) and 0.1 μl of GeneScan™-500LIZ™ size standard (Appilied Biosystems, USA) in a total volume of 11.1 μl. The microsatellite genotyping was performed using a Genetic Analyzer 3130xl (Applied Biosystems, USA) and the genotyping results were obtained using Genemapper (ver. 3.0, Applied Biosystems, USA).
Table 1.
The investigated 150 MS markers for Korean native chicken lines. The numbers of alleles are indicated, and the selected 15 MS markers are in bold
| Marker | Chr. | Map position (cM) | No. of allele | Allele Size (bp) | Marker | Chr. | Map position (cM) | No. of allele | Allele size(bp) | Marker | Chr. | Map position (cM) | No. of allele | Allele size (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||
| MCW0248 | 1 | 21 | 3 | 215–223 | MCW0016 | 3 | 247 | 5 | 134–146 | GCT0016 | 9 | 41 | 8 | 108–154 |
| ADL0160 | 1 | 33 | 2 | 113–133 | GCT0053 | 3 | 263 | 5 | 128–154 | ADL0191 | 9 | 44 | 6 | 128–150 |
| LEI0194 | 1 | 81 | 7 | 130–174 | ADL0237 | 3 | 275 | 8 | 133–153 | ADL0021 | 9 | 53 | 8 | 166–188 |
| MCW0111 | 1 | 118 | 5 | 98–106 | MCW0040 | 3 | 282 | 8 | 129–147 | ADL0259 | 9 | 122 | 9 | 106–146 |
| ADL0188 | 1 | 133 | 6 | 145–165 | LEI0166 | 3 | 300 | 3 | 346–356 | MCW0134 | 9 | 132 | 7 | 260–284 |
| MCW0297 | 1 | 162 | 7 | 288–304 | MCW0037 | 3 | 317 | 3 | 152–156 | MCW0228 | 10 | 0 | 6 | 221–239 |
| LEI0146 | 1 | 169 | 5 | 248–268 | ADL0143 | 4 | 0 | 4 | 152–166 | MCW0067 | 10 | 59 | 3 | 176–182 |
| MCW0106 | 1 | 94 | 3 | 125–131 | ADL0255 | 4 | 3 | 4 | 97–109 | ADL0158 | 10 | 101 | 4 | 188–204 |
| MCW0101 | 1 | 248 | 4 | 274–280 | ADL0317 | 4 | 12 | 9 | 174–216 | ADL0112 | 10 | 120 | 3 | 127–133 |
| ADL0268 | 1 | 288 | 5 | 105–117 | ADL0203 | 4 | 35 | 7 | 168–194 | MCW0097 | 11 | 18 | 3 | 267–273 |
| LEI0108 | 1 | 300 | 10 | 256–310 | MCW0295 | 4 | 75 | 5 | 88–100 | ADL0123 | 11 | 22 | 3 | 106–138 |
| LEI0169 | 1 | 400 | 5 | 232–248 | ADL0241 | 4 | 80 | 7 | 201–215 | MCW0332 | 12 | 90 | 2 | 196–200 |
| LEI0107 | 1 | 424 | 8 | 206–240 | ADL0246 | 4 | 112 | 9 | 146–164 | ADL0147 | 13 | 32 | 4 | 211–217 |
| MCW0145 | 1 | 455 | 7 | 182–210 | ADL0194 | 4 | 118 | 4 | 198–214 | LEI0251 | 13 | 47 | 12 | 98–132 |
| MCW0020 | 1 | 460 | 4 | 183–189 | ROS0024 | 4 | 148 | 6 | 312–328 | MCW0216 | 13 | 47 | 4 | 136–146 |
| LEI0134 | 1 | 527 | 6 | 291–311 | LEI0094 | 4 | 153 | 9 | 246–280 | ADL0310 | 13 | 51 | 10 | 132–158 |
| MCW0107 | 1 | 565 | 4 | 110–118 | MCW0098 | 4 | 217 | 2 | 260–262 | ROS0083 | 13 | 55 | 7 | 109–129 |
| LEI0234 | 2 | 50 | 10 | 217–315 | LEI0085 | 4 | 231 | 5 | 245–259 | MCW0322 | 13 | 67 | 3 | 252–268 |
| MCW0131 | 2 | 102 | 6 | 196–216 | MCW0263 | 5 | 28 | 4 | 227–249 | MCW0104 | 13 | 74 | 11 | 188–226 |
| MCW0206 | 2 | 104 | 5 | 226–240 | MCW0193 | 5 | 50 | 9 | 298–318 | ADL0200 | 14 | 16 | 7 | 112–138 |
| ADL0176 | 2 | 116 | 5 | 184–200 | ROS0013 | 5 | 79 | 8 | 220–236 | LEI0098 | 14 | 37 | 6 | 150–170 |
| MCW0063 | 2 | 119 | 8 | 132–150 | ADL0292 | 5 | 83 | 7 | 112–138 | MCW0123 | 14 | 45 | 5 | 80–90 |
| ADL0217 | 2 | 121 | 4 | 150–156 | MCW0214 | 5 | 88 | 10 | 268–302 | MCW0080 | 15 | 49 | 4 | 270–280 |
| MCW0065 | 2 | 142 | 6 | 98–122 | MCW0078 | 5 | 93 | 3 | 135–143 | ADL0293 | 17 | 26 | 7 | 105–119 |
| LEI0089 | 2 | 165 | 6 | 182–200 | LEI0145 | 5 | 98 | 11 | 303–333 | MCW0330 | 17 | 41 | 4 | 254–286 |
| MCW0039 | 2 | 202 | 5 | 128–142 | MCW0223 | 5 | 123 | 4 | 177–195 | MCW0151 | 17 | 57 | 6 | 250–266 |
| MCW0034 | 2 | 233 | 7 | 217–237 | MCW0029 | 5 | 128 | 11 | 137–187 | ADL0304 | 18 | 7 | 8 | 127–161 |
| LEI0096 | 2 | 233 | 6 | 216–240 | MCW0081 | 5 | 151 | 2 | 113–131 | MCW0219 | 18 | 47 | 4 | 224–240 |
| ADL0181 | 2 | 241 | 3 | 175–179 | ADL0166 | 5 | 162 | 9 | 124–162 | MCW0266 | 19 | 0 | 3 | 163–175 |
| MCW0173 | 2 | 243 | 12 | 230–272 | ADL0298 | 5 | 198 | 6 | 105–121 | MCW0119 | 20 | 0 | 7 | 102–142 |
| MCW0087 | 2 | 252 | 9 | 267–287 | MCW0014 | 6 | 50 | 4 | 173–187 | ADL0324 | 20 | 18 | 6 | 157–181 |
| MCW0009 | 2 | 261 | 2 | 162–172 | MCW0250 | 6 | 59 | 5 | 226–240 | ADL0034 | 20 | 26 | 6 | 111–121 |
| MCW0137 | 2 | 273 | 7 | 240–264 | ADL0230 | 6 | 63 | 6 | 105–115 | SLC2A1 | 21 | 71.04 | 2 | 293–295 |
| MCW0288 | 2 | 275 | 5 | 108–118 | ADL0159 | 6 | 67 | 10 | 78–126 | ADL0262 | 23 | 0 | 3 | 105–109 |
| LEI0070 | 2 | 379 | 11 | 177–213 | MCW0120 | 7 | 44 | 10 | 258–286 | MCW0165 | 23 | 1 | 3 | 114–118 |
| ROS0074 | 2 | 302 | 3 | 315–321 | MCW0201 | 7 | 79 | 4 | 299–309 | ADL0289 | 23 | 7 | 3 | 173–177 |
| MCW0264 | 2 | 320 | 6 | 224–240 | MCW0183 | 7 | 86 | 3 | 291–319 | MCW0301 | 24 | 48 | 6 | 264–292 |
| GCT0002 | 2 | 349 | 5 | 154–172 | ADL0279 | 7 | 92 | 8 | 87–115 | MCW0285 | 26 | 38 | 7 | 179–195 |
| MCW0282 | 2 | 378 | 5 | 286–310 | ROS0019 | 7 | 101 | 10 | 119–147 | MCW0069 | 26 | 47 | 7 | 155–173 |
| LEI0141 | 2 | 382 | 8 | 220–242 | MCW0236 | 7 | 109 | 6 | 306–328 | LEI0074 | 26 | 67 | 6 | 224–240 |
| MCW0157 | 2 | 474 | 6 | 285–297 | MCW0316 | 7 | 127 | 2 | 158–186 | MCW0300 | 27 | 11 | 3 | 122–130 |
| MCW0261 | 3 | 0 | 8 | 225–251 | ADL0315 | 7 | 140 | 2 | 245–247 | ROS0073 | 22 | 0 | 4 | 280–292 |
| MCW0083 | 3 | 51 | 5 | 78–86 | MCW0275 | 8 | 6 | 3 | 128–150 | LEI0135 | 28 | 0 | 6 | 132–142 |
| MCW0222 | 3 | 85 | 4 | 217–223 | ROS0026 | 8 | 14 | 6 | 109–119 | ROS0249 | 32 | 20 | 4 | 148–162 |
| MCW0212 | 3 | 154 | 3 | 192–206 | MCW0095 | 8 | 26 | 5 | 72–82 | ADL0273 | Z | 73 | 4 | 144–168 |
| ADL0248 | 3 | 164 | 7 | 122–158 | MCW0160 | 8 | 35 | 5 | 205–229 | ADL0201 | Z | 87 | 4 | 138–144 |
| MCW0127 | 3 | 167 | 8 | 227–247 | ADL0154 | 8 | 46 | 8 | 125–171 | MCW0154 | Z | 95 | 3 | 170–186 |
| MCW0103 | 3 | 201 | 2 | 267–271 | ADL0278 | 8 | 94 | 4 | 111–119 | LEI0144 | Z | 131 | 4 | 251–269 |
| MCW0224 | 3 | 218 | 4 | 292–300 | MCW0351 | 8 | 105 | 5 | 149–159 | LEI0121 | Z | 131 | 3 | 257–273 |
| MCW0126 | 3 | 231 | 3 | 112–132 | ROS0078 | 9 | 0 | 16 | 172–246 | LEI0075 | Z | 165 | 8 | 164–200 |
Table 2.
Primer information for 15 selected microsatellite markers
| marker | Chr | Dye | Forward (5′ - 3′) | Reverse (5′ - 3′) |
|---|---|---|---|---|
| LEI0107 | 1 | NED | GCTGCTCAGAAGCATCTGTGC | ATCATTGCTACACCATGGTTC |
| MCW0145 | 1 | FAM | ACTTTATTCTCCAAATTTGGCT | AAACACAATGGCAACGGAAAC |
| MCW0063 | 2 | FAM | GGCTCCAAAAGCTTGTTCTTAGCT | GAAAACCAGTAAAGCTTCTTAC |
| MCW0087 | 2 | NED | ATTTCTGCAGCCAACTTGGAG | CTCAGGCAGTTCTCAAGAACA |
| MCW0264 | 2 | FAM | CTTACTTTTCACGACAGAAGC | AGACTGAGTCACACTCGTAAG |
| MCW0261 | 3 | FAM | GAGCAGTTCATATGAAGTGCAG | GTAGTAGCAGCTACACCAGAG |
| ADL0292 | 5 | FAM | CCAAATCAGGCAAAACTTCT | AAATGGCCTAAGGATGAGGA |
| MCW0029 | 5 | VIC | GTGGACACCCATTTGTACCCTATG | CATGCAATTCAGGACCGTGCA |
| ADL0021 | 9 | PET | GCTCCTCGCTTTGCTCTGAA | GCTTAGCCTCATCTCTTGTA |
| ADL0259 | 9 | VIC | CTCATTGCAGAGGAAGTTCT | GTAATGGAGGATGCTCAGGT |
| GCT0016 | 9 | NED | TCCAAGGTTCTCCAGTTC | GGCATAAGGATAGCAACAG |
| LEI0251 | 13 | NED | GATCTAGAAATGGCTGACTGAC | GGGTTACTCTTATGTTTAATGATGTC |
| MCW0104 | 13 | FAM | TAGCACAACTCAAGCTGTGAG | AGACTTGCACAGCTGTGTACC |
| ADL0304 | 18 | FAM | GGGGAGGAACTCTGGAAATG | CCTCATGCTTCGTGCTTTTT |
| ADL0324 | 20 | NED | TTGCCTCGACGGACCACAAT | GCAGCCCCGCCAAGTAACTG |
Statistical analysis
The number of alleles, expected heterozygosity (He), observed heterozygosity (Ho) and polymorphic information content (PIC) and F-statistics were calculated for the selected 15 MS markers using the Cervus (ver 3.0) program (Marshall et al., 1998). Expected heterozygosity was derived from an unbiased formula (Nei, 1987) using allele frequencies assuming Hardy-Weinberg equilibrium which is a useful measure of informativeness of a locus. The polymorphism information content (PIC) is a closely related diversity measure which is estimated as:
Here, l denotes the lth locus and Plu and Plv are the sample frequencies of a series of alleles Au and Av at the lth locus (Botstein et al., 1980). F-statistics describe the amount of inbreeding-like effects within subpopulations (Fst), among subpopulations (Fis), and within the entire population (Fit) (Wright, 1965).
The expected probability of identity values among genotypes of random individuals (PI), random half sibs (PIhalf-sibs) and random sibs (PIsibs) were calculated using API-CALC (ver 1.0) (Ayres and Overall, 2004). This formula is only used for pairs of unrelated individuals such as relatives share genes, and consequently additional loci are likely to be required in order to adequately determine whether two profiles are from distinct individuals as follows:
Where, pi and pj are population allele proportions.
Also, genetic distance values were calculated among five native chicken lines using PowerMarker (Ver 3.25) (Liu and Muse, 2005). A phylogenetic tree was also constructed by Neighbor-Joining tree method (Nei, 1983) method embedded in PowerMarker software package. Finally, in Structure (Ver 2.3.3) program, we assumed five populations (i.e., K = 5) in these chicken lines to get the estimates of proportion of individual’s ancestry from those population (Pritchard et al., 2000).
RESULTS AND DISCUSSION
Survival genetic diversity and differentiation
The highest values in the number of alleles, expected heterozygosity (He), observed heterozygosity (Ho) and polymorphism information content (PIC) are the vital index for the selection of markers in chicken line discrimination. In this study, we selected 15 microsatellite markers out of the 150 MS markers for the discrimination of Korean native chicken lines. The heterozygosity (He and Ho) and polymorphic information content (PIC) value for the Korean native chicken lines are summarized in Table 3. Among these selected 15 MS markers, LEI0251 is contained highest value of the number of allele, He, Ho and PIC for 12, 0.882, 0.852 and 0.865, respectively. While, MCW0264 marker is showed lowest He and PIC value of 0.709 and 0.648, respectively, but the Ho value lowest in GCT0016 marker. The selection process of MS markers were evaluated for the genetic diversity as of the number of alleles, He, Ho and PIC values range of 6 to 12, 0.709 to 0.882, 0.466 to 0.852 and 0.648 to 0.865, respectively. In order to investigate genetic relationships and breed differentiation, highly polymorphic MS markers were selected. Estimation of genotypic diversity of heterozygosity and PIC value informativeness of MS markers were previously used for the determining the animal breed selection (Berthouly et al., 2008). For the animal traceability, PIC>0.5 and He>0.6 are the most reasonable informative locus for application in genetics (Botstein et al., 1980). In this study, selected 15 MS markers were highly informative among the five chicken lines and these MS markers are appropriate for discrimination as well.
Table 3.
The statistical analysis of heterozygosity (He and HO), polymorphism information content (PIC), and F-statistics value using selected 15 microsatellite markers among the native chicken lines
| Locus | Chr | No. of allele | He | Ho | PIC | Fst | Fit | Fis |
|---|---|---|---|---|---|---|---|---|
| LEI0107 | 1 | 8 | 0.77 | 0.716 | 0.739 | 0.1076 | 0.1001 | −0.0083 |
| MCW0145 | 1 | 7 | 0.791 | 0.727 | 0.759 | 0.1141 | 0.0774 | −0.0413 |
| MCW0063 | 2 | 8 | 0.712 | 0.648 | 0.665 | 0.0956 | 0.1142 | 0.0206 |
| MCW0087 | 2 | 9 | 0.83 | 0.761 | 0.806 | 0.1219 | 0.1074 | −0.0165 |
| MCW0264 | 2 | 6 | 0.709 | 0.713 | 0.648 | 0.0309 | 0.0013 | −0.0306 |
| MCW0261 | 3 | 8 | 0.842 | 0.83 | 0.817 | 0.1324 | 0.0455 | −0.1002 |
| ADL0292 | 5 | 7 | 0.821 | 0.716 | 0.791 | 0.1345 | 0.1610 | 0.0306 |
| MCW0029 | 5 | 11 | 0.789 | 0.739 | 0.77 | 0.1215 | 0.0761 | −0.0517 |
| ADL0021 | 9 | 8 | 0.849 | 0.795 | 0.825 | 0.1111 | 0.0892 | −0.0246 |
| ADL0259 | 9 | 9 | 0.846 | 0.773 | 0.826 | 0.0890 | 0.0923 | 0.0036 |
| GCT0016 | 9 | 8 | 0.804 | 0.466 | 0.773 | 0.2473 | 0.4513 | 0.2710 |
| LEI0251 | 13 | 12 | 0.882 | 0.852 | 0.865 | 0.0927 | 0.0538 | −0.0429 |
| MCW0104 | 13 | 11 | 0.846 | 0.659 | 0.823 | 0.1965 | 0.2349 | 0.0478 |
| ADL0304 | 18 | 8 | 0.768 | 0.678 | 0.735 | 0.1004 | 0.1449 | 0.0495 |
| ADL0324 | 20 | 6 | 0.767 | 0.568 | 0.723 | 0.2174 | 0.2853 | 0.0868 |
| Total/Mean | 8 | 126/8.4 | 0.802 | 0.709 | 0.771 | 0.1290 | 0.1370 | 0.0093 |
He = Expected heterozygosity, Ho = Observed heterozygosity, PIC = Polymorphism information content, Fit = Total inbreeding, Fst = Genetic distance, Fis = Within inbreeding.
F-statistics (Wright, 1965) were estimated in a fixation index as genetic differentiation (Fst), the global heterozygote deficit among five chicken lines (Fit) and the heterozygote deficit within line (Fis) among the 15 MS markers (Table 3). Among these markers, estimation of fixation index has been discovered for Fst, Fit and Fis values ranging from 0.0309 to 0.2473, 0.0013 to 0.4513 and −0.1002 to 0.271, respectively. The estimated mean value of the total inbreeding (Fit), within line inbreeding (Fis) and genetic distance were 0.137, 0.0093 and 0.129, respectively. The high F-statistics value was contained in GCT0016 marker of 0.2473, 0.4513 and 0.271 for Fst, Fit and Fis, respectively. While, the lowest value for genetic distance (Fst) and total inbreeding (Fit) was 0.0309 and 0.0013, respectively, and lowest within inbreeding value of −0.1002 in MCW0261 marker. In addition, pair wise co-ancestry matrix genetic distance was confirmed 0.083 to 0.171 among Korean native chicken lines (Table 4). The highest genetic distance was obtained between KNC_B and KNC_W (17.1%), while the lowest genetic distance was observed between KNC_B and KNC_Y (8.3%).
Table 4.
Pair-wise genetic distance among five chicken lines
| KNC_B | KNC_G | KNC_R | KNC_W | |
|---|---|---|---|---|
| KNC_G | 0.156 | - | - | - |
| KNC_R | 0.103 | 0.164 | - | - |
| KNC_W | 0.171 | 0.137 | 0.137 | - |
| KNC_Y | 0.083 | 0.107 | 0.111 | 0.107 |
The expected probability of identity values of 15 MS markers were calculated in random individuals (PI), random half-sib (PIhalf-sibs) and random sibs (PIsibs), which were estimated as 7.98×10−29, 2.88×10−20 and 1.25×10−08, respectively (Table 5). Also, acceptance of marker accuracy for discrimination power was evaluated (Figure 1). The expected probability of chicken lines identity among the genotypes of random individuals (PI), random half-sib (PIhalf-sibs) and random sibs (PIsibs) were suggested approximately 12 markers. Thus, the expected probability of identity values from 12 MS markers in random individuals (PI), random half-sib (PIhalf-sibs) and random sibs (PIsibs) were estimated as 1.01×10−20, 3.85×10−15 and 1.69×10−7, respectively. Overall, the total expected probability of identity values was 99.9% for the discrimination of Korean native chicken. Our study identified markers in Korean native chicken lines which are applicable to future breeding plans, as well as discrimination markers for these lines.
Table 5.
The expected probability values among genotypes of random individuals (PI), random half-sib (PIhalf-sibs), random sibs (PIsibs) and total expected probability (PE) for discrimination chicken lines using 12 and 15 markers
| No. of marker | PI | PIhalf-sibs | PIsibs | PE |
|---|---|---|---|---|
| 15 | 7.98×10−29 | 2.28×10−20 | 1.25×10−8 | 0.999858 |
| 12 | 1.01×10−20 | 3.85×10−15 | 1.69×10−7 | 0.999555 |
Figure 1.
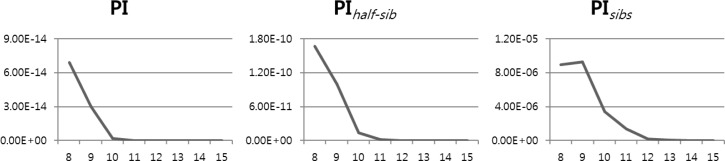
The expected probability of identity values among genotypes of random individuals (PI), random half-sib (PIhalf-sibs) and random sibs (PIsibs) were suggested markers for discrimination of chicken lines.
Phylogenetic and structure analysis
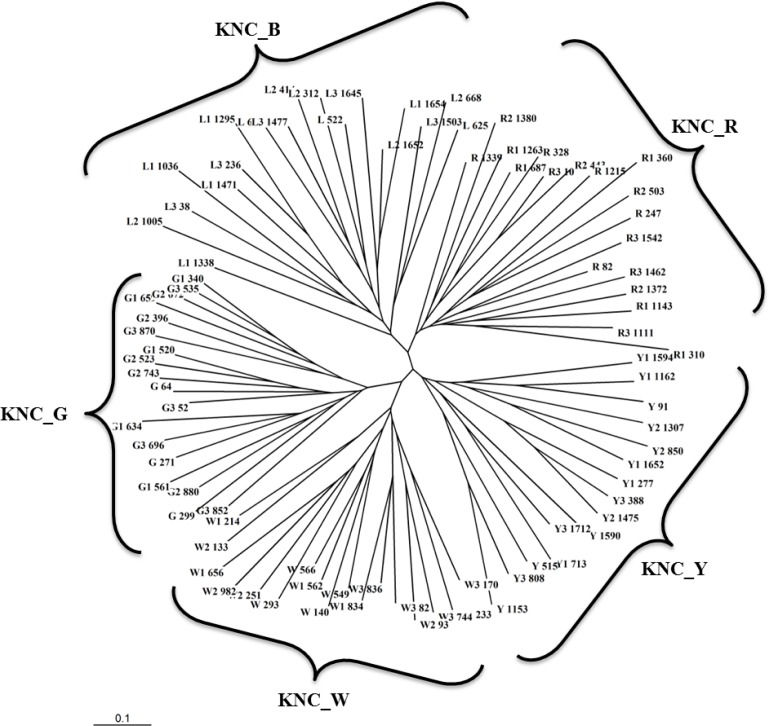
The unrooted phylogenetic neighbor-joining (NJ) tree was constructed using 15 MS markers that clearly differentiated among five native chicken lines (Figure 2). Based on the equation of Nei et al. (1983), a phylogenetic tree has been estimated by the distribution of allele sharing with genetic distance (Fst). In our analysis, the KNC_W line is different from the KNC_B and KNC_R lines. Also, the KNC_G line shows a long genetic distance from KNC_R. However, the KNC_Y line is very close to the KNC_W and KNC_R lines. The dendrogram drawn from the genetic distance matrix using 15 MS markers can also be used for the conservation of Korean native chicken lines. Also, five different lines in Korean native chickens were well discriminated by using these 15 MS markers.
Figure 2.
Construction of unrooted phylogenetic neighbor-joining (NJ) tree among the five chicken lines using 15 selected MS markers.
In 1994, the conservation policy for the development of Korean native chicken lines was launched. As a result, five breeds with nine chicken lines were documented. In order to investigate the genetic structure of the five Korean native chicken lines, a structured program of genetic analysis was applied (Figure 3). Based on the line specific clusters, chicken line structure was estimated with a K value of 5. The estimated average individual cluster value in the specific line was accurate to more than 95% (data was not shown). Our structure result for the five Korean native chicken lines indicates around 5% genetic admixture with other lines. In conclusion, our study shows the genetic diversity, genetic distance, and population structure among five Korean native chicken lines, using 15 selected MS markers. The maintaining of Korean native chickens with appropriate discrimination markers is the essential for conservation of this breed. Our results indicated that these MS markers will be used to aid the conservation, traceability and future improvement of Korean native chicken lines.
Figure 3.
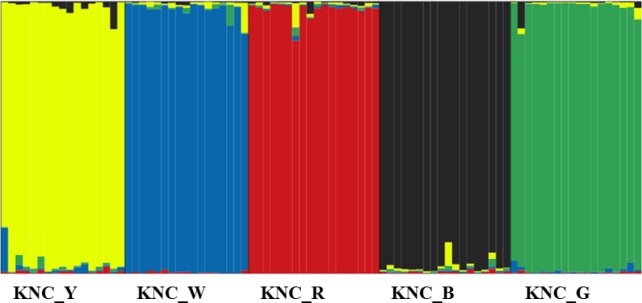
Construction of genetic structure using individual cluster (K value of 5) among the chicken lines.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008133), Rural Development Administration, Korea.
REFERENCES
- Almasy L, Blangero J. Human QTL linkage mapping. Genetica. 2009;136:333–340. doi: 10.1007/s10709-008-9305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthouly C, Bed'Hom B, Tixier-Boichard M, Chen CF, Lee YP, Laloe D, Legros H, Verrier E, Rognon X. Using molecular markers and multivariate methods to study the genetic diversity of local european and asian chicken breeds. Anim Genet. 2008;39:121–129. doi: 10.1111/j.1365-2052.2008.01703.x. [DOI] [PubMed] [Google Scholar]
- Blott SC, Williams JL, Haley CS. Discriminating among cattle breeds using genetic markers. Heredity. 1999;82:613–619. doi: 10.1046/j.1365-2540.1999.00521.x. [DOI] [PubMed] [Google Scholar]
- Bodzsar N, Eding H, Revay T, Hidas A, Weigend S. Genetic diversity of hungarian indigenous chicken breeds based on microsatellite markers. Anim Genet. 2009;40:516–523. doi: 10.1111/j.1365-2052.2009.01876.x. [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnik M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Crittenden LB. Microsatellite markers for genetic-mapping in the chicken. Poult Sci. 1994;73:539–546. doi: 10.3382/ps.0730539. [DOI] [PubMed] [Google Scholar]
- Dalvit C, DeMarchi M, Cassandro M. Genetic traceability of livestock products: A review. Meat Sci. 2007;77:437–449. doi: 10.1016/j.meatsci.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Ding FX, Zhang GX, Wang JY, Li Y, Zhang LJ, Wei Y, Wang HH, Zhang L, Hou QR. Genetic diversity of a Chinese native chicken breed, Bian chicken, based on twenty-nine microsatellite markers. Asian-Aust J Anim Sci. 2010;23:154–161. [Google Scholar]
- FAO . Status of animal genetic resources. In: Rischkowsky B, Pilling D, editors. The state of the world’s animal genetic resources for food and agriculture. Commission on Genetic Resources for Food and Agriculture; Rome, Italy: 2007. pp. 23–49. [Google Scholar]
- Hillel J, Groenen MA, Tixier-Boichard M, Korol AB, David L, Kirzhner VM, Burke T, Dirie AB, Crooijmans RPA, Elo K, Feldman MW, Freidlin PJ, Maki-Tanila A, Oortwijn M, Thomson P, Vignal A, Wimmers K, Weigend S. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet Sel Evol. 2003;35:533–557. doi: 10.1186/1297-9686-35-6-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MR, Jung KC, Park BK, Choi KD, Lee JH. Genetic variability of mtDNA D-loop region in Korean native chickens. Korean J Poult Sci. 2009;36:323–328. [Google Scholar]
- Hoque MR, Lee SH, Jung KC, Kang BS, Park MN, Lim HK, Choi KD, Lee JH. Discrimination of Korean native chicken populations using SNPs from mtDNA and MHC polymorphisms. Asian-Aust J Anim Sci. 2011;24:1637–1643. [Google Scholar]
- Jacobsson L, Park HB, Wahlberg P, Jiang S, Siegel PB, Andersson L. Assignment of fourteen microsatellite markers to the chicken linkage map. Poult Sci. 2004;83:1825–1831. doi: 10.1093/ps/83.11.1825. [DOI] [PubMed] [Google Scholar]
- Kaya M, Yildiz MA. Genetic diversity among turkish native chickens, denizli and gerze, estimated by microsatellite markers. Biochem Genet. 2008;46:480–491. doi: 10.1007/s10528-008-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HS, Oh JD, Lee JH, Jo KJ, Sang BD, Choi CH, Kim SD, Lee SJ, Yeon SH, Jeon GJ, Lee HK. Genetic variation and relationships of Korean native chickens and foreign breeds using 15 microsatellite markers. Asian-Aust J Anim Sci. 2006;19:1546–1550. [Google Scholar]
- Liu K, Muse SV. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Marshall TC, Slate J, Kruuk LE, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchadeyi FC, Eding H, Wollny CB, Groeneveld E, Makuza SM, Shamseldin R, Simianer H, Weigend S. Absence of population substructuring in zimbabwe chicken ecotypes inferred using microsatellite analysis. Anim Genet. 2007;38:332–339. doi: 10.1111/j.1365-2052.2007.01606.x. [DOI] [PubMed] [Google Scholar]
- MIFAFF . Primary statistics of Food. Agriculture, Forestry and Fisheries; Korea: 2009. [Google Scholar]
- Mwacharo JM, Nomura K, Hanada H, Jianlin H, Hanotte O, Amano T. Genetic relationships among kenyan and other east african indigenous chickens. Anim Genet. 2007;38:485–490. doi: 10.1111/j.1365-2052.2007.01641.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadano R, Nishibori M, Imamura Y, Matsuzaki M, Kinoshita K, Mizutani M, Namikawa T, Tsudzuki M. High genetic divergence in miniature breeds of japanese native chickens compared to red junglefowl, as revealed by microsatellite analysis. Anim Genet. 2008;39:71–78. doi: 10.1111/j.1365-2052.2007.01690.x. [DOI] [PubMed] [Google Scholar]
- Tadano R, Nishibori M, Nagasaka N, Tsudzuki M. Assessing genetic diversity and population structure for commercial chicken lines based on forty microsatellite analyses. Poult Sci. 2007a;86:2301–2308. doi: 10.3382/ps.2007-00233. [DOI] [PubMed] [Google Scholar]
- Tadano R, Sekino M, Nishibori M, Tsudzuki M. Microsatellite marker analysis for the genetic relationships among japanese long-tailed chicken breeds. Poult Sci. 2007b;86:460–469. doi: 10.1093/ps/86.3.460. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by f-statistics with special regard to systems of mating. Evol. 1965;19:395–420. [Google Scholar]