Abstract
The aim of this study was to investigate allele frequencies at the CSN1S2 locus in two Chinese dairy goat breeds and the effects of its variation on dairy goat economic traits. Seven hundred and eight goats from Xinong Saanen (XS, n = 268) and Guanzhong (GZ, N = 440) breeds were selected. The milk samples of 268 XS goats were collected during the middle of lactation, body size parameters (708 goats) and daily milk yield (202 goats) were registered. The RFLP (restriction fragment length polymorphism) and SSCP (single strand conformation polymorphism) were used to detect the polymorphisms in CSN1S2. The Hardy-Weinberg (HW) equilibrium and the associations between body size, milk yield and composition and the genotypes were calculated. The results revealed that only A and F CSN1S2 alleles were found in the two Chinese dairy goat breeds. Allelic frequencies of A and F were 0.795, 0.205 and 0.739, 0.261 in Xinong Saanen and Guanzhong population respectively. Xinong Saanen breed was in Hardy-Weinberg equilibrium, while Guanzhong breed deviated from Hardy-Weinberg equilibrium (p<0.05). The association of polymorphism with economic traits indicated that the goats with FF genotype have higher milk fat and total solid concentration than those with AA and AF genotypes (p<0.05).
Keywords: CSN1S2, SNP, Dairy Goats, Milk, A and F Alleles
INTRODUCTION
Casein genes are organized as a cluster, including in order αS1-casein (CSN1S1), αS2-casein (CSN1S2), β-casein (CSN2) and κ-casein (CSN3) (Ferretti et al., 1990; Threadgill and Womack, 1990). In goats, the entire casein cluster region spans about 250 kb on chromosome 6 (Hayes et al., 1993). In recent years, the genetic polymorphism of goat casein has raised considerable research interest because goat casein polymorphisms are related to milk quality, milk composition and technological properties (Martin et al., 2002).
The CSN1S2 gene is 18.5 kb long and consists of 18 exons which vary from 21 to 266 bp (Groene et al., 1993). Seven alleles, showing three different synthesis levels, have been identified so far. The A, B, C, E and F alleles are characterized by point mutations related to single amino acid substitutions and associated with a normal amount of ααS2-casein (2.5 g/L). The CSN1S2 B allele, compared to the A allele, shows a G10A transition at exon 9, which causes a Glu64→Lys substitution in the mature protein, while the mutational events characterizing the C and E variants are substitution A5T (Lys167→Ile) and C83G (Pro193→Arg) at exon 16, respectively (Bouniol et al., 1994). The CSN1S2 F allele is characterized by a G13C transition occurring at exon 3, which results in an amino acid substitution Val7→Ile (Bouniol et al., 1994; Veltri et al., 2000 Ramunno et al., 2001a). CSN1S2 D is a rare defective allele characterized by a deletion, involving part of the exon 11 and of the following intron. The null allele CSN1S2 O associated with a non-detectable amount of this casein fraction shows a G→A transition at the 80th nucleotide in exon 11, creating a premature stop codon at position 110 (Ramunno et al., 2001a).
China, with nearly four million goats, is one of largest producers of dairy goats in the world. Among which, the Xinong Saanen (SN) and Guanzhong (GZ) breeds are the best-known, not only due to their tolerance of crushed feed and the local harsh weather, but also to their high milk yield (Xu et al., 2003; Deng et al., 2010). While as for Chinese dairy goats, we found few reports about studying DNA polymorphism and their association with economic traits. The aim of the present study was to investigate the genetic structure of CSN1S2 locus in these two important Chinese dairy goat breeds. Therefore, the genetic variations analysis of the CSN1S2 gene in 708 Chinese dairy goats may provide useful information related to the understanding of their genetic characteristics. The genetic associations related to goat milk yield and composition in Chinese dairy goat breeds will possibly contribute to improving the quality of dairy goat breeds in China.
MATERIALS AND METHODS
Samples collection and DNA extraction
Blood samples of 708 goats were collected from two Chinese dairy goat breeds of Xinong Saanen (XS) and Guanzhong breed (GZ) as follows: Qianyang Dairy Goat Breeding Farm (XS breed, n = 202), Northwest A&F University Goat Farm (XS breed, n = 66) and Shaanxi Dairy Goat Breeding Centre (GZ breed, n = 440) in Sanyuan county, Shaanxi province of China. The genomic DNA was extracted from blood by a standard phenol-chloroform method (Chomczynski and Sacchi, 1987).
Milk samples and data collection
The milk yield of second lactation of the XS breed (n = 202) in Qianyang Dairy Goat Breeding Farm was recorded. In addition, a 50 ml sample of morning milk obtained from goats of the XS breed in Northwest A&F University Goat Farm and Qianyang Dairy Goat Breeding Farm was collected (n = 268). The milk samples were stored at 4°C until the next day when they could be analyzed by MilkoScan FT120 (FOSS Corporation, Denmark) for concentrations of milk fat, protein, lactose, total solids (TS), milk solids-not-fat (SNF) and density of milk. Body size parameters of all goats are documented, including Withers height, Body length and Chest circumference.
Genotyping of CSN1S2 locus
CSN1S2 F, O and D Alleles were detected by means of RFLP-PCR (restricted fragment length polymorphism-PCR) according to Ramunno et al. (2001a). Furthermore, based on the nucleotide sequence characterization of CSN1S2 B, C and E alleles, the PCR-SSCP procedure was developed for genotyping them by analyzing polymorphisms of exon 9 and exon 16 of CSN1S2 gene (Table 1). The PCR-SSCP and RFLP-PCR procedures were carried out based on Yue et al. (2011) and Ramunno et al. (2001a) respectively.
Table 1.
The primer pair sequences and their information of CSN1S2 gene in goat
| Loci | Sequences of primers | Annealing temperature | Amplified region/size bp | Allele |
|---|---|---|---|---|
| L1 | R: 5′-TCTCTTGCCATCAAAACA-3′ | 50.4 | Exon3 and flanking | F |
| F: 5′-TGGTCTTTATTCCTCTCT-3′ | region/310 bp | |||
| L2 | R:5′-GGACTCTAAATATACTTAATGAATT-3′ | 54.8 | Exon9 and flanking | B |
| F: 5′-GCTTATCGTCCACAGTAATCTT-3′ | region/261 bp | |||
| L3 | R: 5′-GACACATAGAGAAGATTC-3′ | 50.6 | Exon11 and partial | O, D |
| F: 5′-CGTTGGGACATTTTATCT-3′ | intron11/301 bp | |||
| L4 | R: 5′-GGTTAGGTCTAGGTGTTCTGA-3′ | 50.4 | Exon16 and flank | C, E |
| F: 5′-TTTTTATTACAAAAGACAACT-3′ | region/226 bp |
Statistical analysis
Genotypic and allelic frequencies, as well as Hardy–Weinberg equilibriums, were directly calculated. Mixed model analyses for milk yield, milk composite and body size parameters were performed using the general linear models procedure of SSPS 17.0 (SPSS Inc., Chicago, IL, USA). The model for dairy goats included marker genotype, year, kidding season (spring versus fall) and lactation number as fixed effects, the linear and quadratic effects of milk yield, milk composite and body size parameters included as covariables and doe as a random effect. The factor, number of kids, had not been included in the statistical model for following reason. The ewes in this study predominantly had one kid, but does that produced twins were also part of the dataset. Data were presented as least squares means with associated standard error.
RESULTS AND DISUSSION
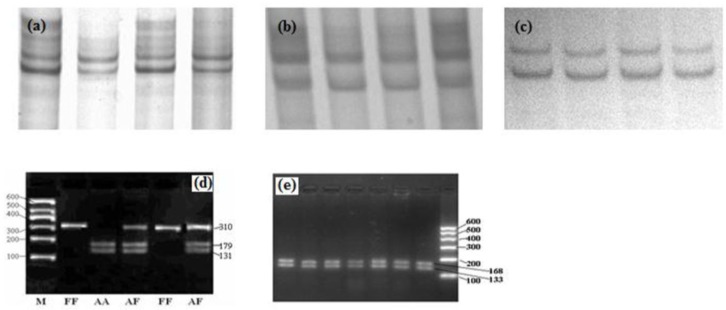
The CSN1S2 locus is characterized by the presence of seven alleles, of which, A, B, C, E and F are associated with normal αs2-casein level (2.5 g/L). D allele is associated with reduced αs2-casein content, and O allele is associated with a non-detectable amount of this casein in milk (Ramunno et al., 2001a, b). Interestingly, in present study only CSN1S2 F and A alleles were detected (Figure 1), while B, C, E, D and O alleles were absent. Overall, three different genotypes, AA, AF and FF were identified in the two Chinese dairy goat breeds. Allele and genotype frequencies are shown in the Table 2. Frequencies of A and F alleles in XS and GZ breeds are 0.795/0.205 and 0.739/0.261, respectively. XS breed was found to follow the Hardy-Weinberg equilibrium, whereas GZ breed was deviated from Hardy-Weinberg equilibrium (p<0.05). Associations of second lactation milk yield, body size and milk composition and genotype were calculated by SPSS 17.0 (Table 3). Statistical analysis indicated that the individuals with FF genotype have higher milk fat and total solid than those of AA and AF genotypes. However, this mutation has no influence on milk yield and body size parameters based on the current study (Table 3). Previous study revealed that dairy goats with CSN1S2 FF genotype had a significantly lower milk yield than other individuals, whereas this study did not show that goats carrying FF genotype had a significantly lower milk yield (Lan et al., 2005). This can be explained by the sample size of the previous study being too small (69 individuals).
Figure 1.
PCR products of five loci analyzed by PCR-SSCP and PCR-RFLP. Figure 1-a, b, c are the PAGE-electrophoresis patterns of PCR-SSCP for L1 (a), L3 (b), L5 (c) locus of CSN1S2 gene, respectively. They all show monopolymorphy. Figure 1-d is the 2.5% agarose gel electrophoresis for Alw26 I digesting PCR products of CSN1S2 L2 locus. Figure 1-e is the 2.5% agarose gel electrophoresis for Nco I digesting PCR products of CSN1S2 L4 locus.
Table 2.
Genotype and allelic frequencies of CSN1S2 gene in 2 dairy goat breeds
| Breeds | Genotypes
|
Samples
|
Genotype frequencies
|
Allele frequencies
|
x2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AF | FF | N | P AA | P AF | P FF | A | F | ||
| XS | 170 | 86 | 12 | 268 | 0.634 | 0.321 | 0.045 | 0.795 | 0.205 | 0.071 |
| GZ | 250 | 150 | 40 | 440 | 0.568 | 0.341 | 0.091 | 0.739 | 0.261 | 6.029* |
Value with * * and * differ significantly at p<0.01 and p<0.05 respectively. 2. x20.05 (df = 1) = 3.84, x20.01(df = 1) = 6.63.
Table 3.
Associations of polymorphism at CSN1S2 gene L2 locus with economic traits in dairy goats
| Traits | Genotypes
|
||
|---|---|---|---|
| AA | AF | FF | |
| Fat (%) | 2.71±0.12a | 2.69±0.19 a | 3.86±0.42 b |
| Protein (%) | 3.25±0.05 | 3.20±0.08 | 3.43±0.18 |
| TS (%) | 11.29±0.15 a | 11.18±0.23 a | 12.57±0.52 b |
| SNF (%) | 8.48±0.06 | 8.38±0.23 | 8.61±0.20 |
| Lactose (%) | 4.12±0.03 | 4.06±0.47 | 4.04±0.10 |
| Density (g/L) | 1,030.84±0.23 | 1,030.26±0.38 | 1029.69±0.78 |
| Milk yield (kg) | 860.90±33.20 | 825.54±13.43 | 822.79±8.48 |
| Withers height (cm) | 70.99±0.35 | 71.27±0.41 | 71.42±0.79 |
| Body length (cm) | 80.03±0.44 | 80.18±0.69 | 79.66±1.18 |
| Chest circumference (cm) | 91.98±2.62 | 94.05±2.98 | 93.68±5.17 |
Values in the same row labeled with the different letter (a, b) are significantly different.
The number of goats carrying AA, AF and FF genotypes with body size parameters (Withers height, Body length and Chest circumference) represent 420, 236 and 52 individuals, respectively.
In milk composite traits (Fat, Protein, TS, SNF, Lactose, and Density), AA, AF and FF genotypes represent 170, 86 and 12 individuals, respectively.
In the milk yield, AA, AF and FF genotypes represent 139, 54 and 9 individuals, respectively.
It is interesting that only two CSN1S2 alleles (A and F allele) were detected in two Chinese dairy goat breeds in the present study, which is greatly different from the CSN1S2 allelic distribution in European dairy goat breeds. The frequency of CSN1S2 A, B and C alleles were estimated to be 0.85, 0.04 and 0.11 in French Alpine and Saanen breeds before discovering the E, D, F and O alleles (Bouniol et al., 1994). While C and F alleles are predominant in local Italian goat breeds (Sacchi et al., 2005). The relatively high incidence of O allele (0.146) appeared in Hungarian Milking Goats (Kusza et al., 2007). There needs to be a larger number of Chinese dairy goats and more goat breeds sampled to confirm whether CSN1S2 alleles in addition to A and F exist in Chinese goat breeds.
A previous study (Ramunno et al., 2001a) reported that CSN1S2 homozygous genotypes were associated with good quality of milk proteins. We also detected this tendency, but this difference was not significant (p>0.05), maybe due to the small sample size. This study indicated that F allele was associated with good quality of milk fat. According to this finding, the selection of goats with higher milk fat should aim to increase the homozygous CSN1S2 FF genotype.
The defective D and O alleles were not identified in this study. Both alleles showed a very low or null frequency in previous studies (Sacchi et al., 2005; Marletta et al., 2005; Kusza et al., 2007). The results of the trial allow us to presume that the CSN1S2 genetic polymorphism in the two Chinese breed does not affect the as2-casein quantity. CSN1S2 locus is closely linked to CSN1S1, CSN2 and CSN3 loci and alleles of these loci are inherited together as allele groups called haplotype (Hayes et al., 1993; Rijnkels, 2002). It means that polymorphisms occurring at a casein locus have to be considered in the context of the casein gene cluster, using information deriving from the entire casein haplotype (Hayes et al., 2006; Finocchiaro et al., 2008). The goats analyzed in this study showed the occurrence of defective alleles at the CSN1S1 locus, coding for αs1-casein, such as the F, 01 and N alleles in our previous studies (Yue et al., 2011). Therefore further studies are required to determine the relationship between the absence of CSN1S2 D and CSN1S2 O alleles and the distribution of alleles at CSN1S1, CSN2 and CSN3 loci in the populations studied.
Acknowledgments
This work was supported by the National “863” Program of China (No.2008AA10Z138), National Natural Science Foundation of China (No.31172184), and Natural Science Foundation of Shaanxi province of China (No.2011JQ3009).
REFERENCES
- Bouniol C, Brignon G, Mahé MF, Printz C. Biochemical and genetic analysis of variant C of caprine αs2-casein (Capra hircus) Anim Genet. 1994;25:173–177. doi: 10.1111/j.1365-2052.1994.tb00106.x. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Deng CJ, Ma RN, Yue XP, Lan XY, Chen H, Lei CZ. Association of IGF-I gene polymorphisms with milk yield and body size in Chinese dairy goats. Genet Mol Biol. 2010;33:266–270. doi: 10.1590/S1415-47572010005000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L, Leone P, Sgaramella V. Long range restriction analysis of the bovine casein genes. Nucleic Acids Res. 1990;18:6829–6833. doi: 10.1093/nar/18.23.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finocchiaro R, Hayes BJ, Siwek M, Spelman RJ, van Kaam JBCHM, Ådnøy T, Portolano B. Comparison of casein haplotypes between two geographically distant European dairy goat breeds. J Anim Breed Genet. 2008;125:68–72. doi: 10.1111/j.1439-0388.2007.00687.x. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Dijkhof RJM, Verstege AJM, Van der Poel JJ. The complete sequence of the gene encoding bovine αs2-casein. Gene. 1993;123:187–193. doi: 10.1016/0378-1119(93)90123-k. [DOI] [PubMed] [Google Scholar]
- Hayes B, Hagesæther N, Ådnøy T, Pellerud G, Berg PR, Lien S. Effects on production traits of haplotypes among casein genes in Norwegian goats and evidence for a site of preferential recombination. Genetics. 2006;174:455–464. doi: 10.1534/genetics.106.058966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes H, Petit E, Bouniol C, Popescu P. Localization of the alpha S2-casein gene (CASAS2) to the homologous cattle, sheep and goat chromosomes 4 by in situ hybridization. Cytogenet Cell Genet. 1993;64:281–285. doi: 10.1159/000133593. [DOI] [PubMed] [Google Scholar]
- Kusza S, Veress G, Kukovics S, Jávor A, Sanchez A, Angiolillo A, Bősze A. Genetic polymorphism of αs1and αs2-caseins in Hungarian milking goats. Small Rumin Res. 2007;68:329–332. [Google Scholar]
- Lan XY, Chen H, Zhang RF, Tian Y, Zhang YD, Fang XT, Sun WB, Lei CZ, Hu SR. Association of polymorphisms of CSN1S2 gene with average milk yield and body sizes indexes in Xinong Saanen dairy goat. Acta Veterinaria Et Zootechnica Sinica. 2005;36:318–322. (in Chinese). [Google Scholar]
- Marletta D, Bordonaro S, Guastella AM, Criscione A, D’Urso G. Genetic polymorphism of the calcium sensitive casein in Sicilian Girgentana and Argentata dell’Etna goat breeds. Small Rumin Res. 2005;57:133–139. [Google Scholar]
- Martin P, Szymanowska M, Zwierzchowski L, Leroux C. The impact of genetic polymorphisms on the protein composition of ruminant’s milks. Reprod Nutr Dev. 2002;42:433–459. doi: 10.1051/rnd:2002036. [DOI] [PubMed] [Google Scholar]
- Ramunno L, Cosenza G, Pappalardo M, Longobardi E, Gallo D, Pastore N, Di Gregorio P, Rando A. Characterization of two new alleles at the goat CSN1S2 locus. Anim Genet. 2001a;32:264–268. doi: 10.1046/j.1365-2052.2001.00786.x. [DOI] [PubMed] [Google Scholar]
- Ramunno L, Longobardi E, Pappalardo M, Rando A, Di Gregorio P, Cosenza G, Mariani P, Pastore N, Masina P. An allele associated with a non-detectable amount of αs2-casein in goat milk. Anim Genet. 2001b;32:19–26. doi: 10.1046/j.1365-2052.2001.00710.x. [DOI] [PubMed] [Google Scholar]
- Rijnkels M. Multispecies comparison of the casein gene loci and evolution of casein gene family. J. Mammary Gland Biol Neoplasia. 2002;7:327–345. doi: 10.1023/a:1022808918013. [DOI] [PubMed] [Google Scholar]
- Sacchi P, Chessa S, Budelli E, Bolla P, Ceriotti G, Soglia D, Rasero R, Cauvin E, Caroli A. Casein haplotype structure in five Italian goat breeds. J Dairy Sci. 2005;88:1561–1568. doi: 10.3168/jds.S0022-0302(05)72825-3. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Womack JE. Genomic analysis of the major bovine milk proteins genes. Nucleic Acids Res. 1990;18:6935–6942. doi: 10.1093/nar/18.23.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri C, Lagonigro R, Pietrolà E, D’Andrea M, Pilla F, Chianese L. Molecular characterization of the goat αs2-casein E allele and its detection in goat breeds of Italy. Proceedings 7th International Conference on Goats; Tour, France. 15–18 May; 2000. p. 727. [Google Scholar]
- Xu GY, Pu JB, Chen TY. On development status and countermeasures of goat industry in western China. China Herbivores Suppl. 2003;1:31–32. [Google Scholar]
- Yue XP, Zhang XM, Wang W, Ma RN, Deng CJ, Lan XY, Chen H, Li F, Xu XR, Ma Y, Lei CZ. The CSN1S1 N and F alleles identified by PCR-SSCP and their associations with milk yield and composition in Chinese dairy goats. Mol Biol Rep. 2011;38:2821–2825. doi: 10.1007/s11033-010-0428-0. [DOI] [PubMed] [Google Scholar]