Abstract
AIM: To evaluate the association between genetic polymorphisms of the gene encoding AMP-activated protein kinase (PRKAA1) and the risk of gastric cancer.
METHODS: The study subjects consisted of 477 age- and sex-matched case-control pairs. Genotyping was performed for 5 tag single nucleotide polymorphisms (SNPs): rs13361707, rs154268, rs3805486, rs6882903, and rs10074991. Associations between gastric cancer and putative risk factors (including the SNPs) were analyzed with multivariate conditional logistic regression models, after adjusting for potential confounding factors. Multiple testing corrections were implemented following methodology for controlling the false discovery rate. Gene-based association tests were performed by using the versatile gene-based association study (VEGAS) method.
RESULTS: In the dominant model, SNPs rs13361707 [odds ratio (OR) = 1.51, 95%CI: 1.07-2.11)], rs154268 (OR = 1.65, 95%CI: 1.22-2.22), rs6882903 (OR = 1.48, 95%CI: 1.09-2.00), and rs10074991 (OR = 1.53, 95%CI: 1.09-2.16) were significantly associated with an increased risk of gastric cancer. In the recessive model, SNPs rs154268 (OR = 1.66, 95%CI: 1.22-2.26), rs3805486 (OR = 0.63, 95%CI: 0.46-0.85), and rs10074991 (OR = 1.47, 95%CI: 1.15-1.88) were significant risk or protective factors for gastric cancer. In the codominant model, the ORs of each of the 5 SNPs were statistically significant. All SNPs in the model showed a dose-response relationship between the minor allele frequency and the risk of gastric cancer. Most notably, subjects with a homozygous minor allele in SNP rs10074991 showed 2.15 times the risk of gastric cancer as subjects without a minor allele. The PRKAA1 gene showed a significant gene-based association with gastric cancer in the VEGAS test.
CONCLUSION: All 5 tested tag SNPs of the PRKAA1 gene (rs13361707, rs154268, rs3805486, rs6882903, and rs10074991) were significantly associated with gastric cancer.
Keywords: AMP-activated protein kinase, Gastric cancer, PRKAA1, Single nucleotide polymorphism, Case-control study
Core tip: There were a few studies to evaluate association between PRKAA1 gene and gastric cancer. However, in previous study, only one single nucleotide polymorphism (SNP) (rs13361707) of PRKAA1 gene was focused. The purpose of this study was to evaluate the association between 5 SNPs of the gene encoding AMP-activated protein kinase (PRKAA1) and the risk of gastric cancer. All SNPs in the model showed a dose-response relationship between the minor allele frequency and the risk of gastric cancer. The PRKAA1 gene showed a significant gene-based association with gastric cancer in the versatile gene-based association study test.
INTRODUCTION
Gastric cancer is the second most common cause of cancer-related mortality worldwide[1]. Since the 1980s, South Korea, Japan, China, and other Asian countries have had a particularly high incidence of this disease, despite general trends of decreasing incidence and mortality[2,3].
A model of gastric carcinogenesis in humans has been derived based on evidence from various epidemiological and pathological studies. According to this model, gastric cancer arises in a sequence of stages: chronic gastritis, atrophic gastritis, intestinal metaplasia, and dysplasia[4]. Helicobacter pylori, high salt intake, alcohol intake, smoking, diet, and genetic factors have been reported to be involved in gastric carcinogenesis[5-9].
AMP-activated protein kinase (AMPK) is an energy sensing/signaling intracellular protein, and a conserved serine/threonine kinase that regulates energy homeostasis and metabolic stress[10]. AMPK is activated by phosphorylation when the AMP/ATP ratio is high[11]. Activated AMPK switches on ATP-generating (catabolic) pathways and switches off ATP-consuming (anabolic) pathways[12,13]. AMPK activation is known to inhibit the accumulation of lipid in the body, decrease the biosynthesis of fatty acids and cholesterol, and increase the oxidation of fatty acids[12].
Considerable evidence indicates that AMPK activation suppresses cell proliferation in both tumor and non-malignant cells. These results of AMPK activation are mediated through various mechanisms, including G1 phase arrest in the cell cycle[14], and the inhibition of fatty acid and protein synthesis[13,15]. Regulation of the cell cycle by AMPK is mediated through activation of the p53-p21 axis pathway, activation of tumor suppressor LKB1, inhibition of the mammalian target of rapamycin pathway, and other similar mechanisms[16]. Based on this evidence, research on AMPK function has focused on its important role in development, and on its potential use as a therapeutic target for some cancers[16,17]. It is possible that AMPK plays an important role in gastric carcinogenesis and, therefore, polymorphic alleles of the encoding gene could modify individual susceptibility to gastric cancer. Recently, Song et al[18] reported that the rs13361707 single nucleotide polymorphism (SNP) of the protein kinase, AMP-activated alpha 1 catalytic subunit (PRKAA1) gene was associated with an increased risk of gastric cancer in the Korean population. However, because their study only examined the rs13361707 SNP of the PRKAA1 gene, it remains important to elucidate the associations between other SNPs of PRKAA1 and gastric cancer.
Accordingly, the aim of the present study was to evaluate the associations between 5 polymorphic alleles of PRKAA1, the gene that encodes AMPK, and gastric carcinogenesis in Koreans.
MATERIALS AND METHODS
Ethics
This work has been carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association. This study was approved by the institutional review boards of Chungbuk National University Hospital, South Korea (IRB No. 2011-09-071). All subjects had provided written informed consent.
Study subjects
The subjects included in this study consisted of 477 newly diagnosed gastric cancer patients and an equal number of age- (within 3 years) and sex-matched controls. The diagnoses of patients with gastric cancer were confirmed at Chungbuk National University Hospital and Eulji University Hospital, which are located in a geographically central region of the South Korea. Control subjects did not have a previous diagnosis of any type of cancer, and were selected from individuals who had received routine medical examinations at these hospitals. The case and control groups each included 301 men and 176 women. The mean ± SD age was 58.7 ± 9.9 years in the case group and 57.8 ± 10.2 years in the control group.
Trained interviewers interviewed all subjects by using a structured questionnaire, which included questions on demographic factors, smoking habits, alcohol consumption, and dietary habits. Peripheral blood and urine samples were collected from all the subjects.
Selection of PRKAA1 SNPs
We selected SNPs of PRKAA1 from several prominent online databases (GeneCards, HUGE navigator, NCBI; www.ncbi.nlm.nih.gov/SNP) because this gene may be related to diet risk factors for gastric carcinogenesis. To select tagging SNP, we identified functional elements from the Functional Elements SNPs Database, used the tagger pairwise method from the International HapMap Project, and finally selected SNPs with a minor allele frequency ≥ 0.05 in JPT (Japanese in Tokyo, Japan) and CHB (Han Chinese in Beijing, China) samples. SNPs that significantly deviated from the Hardy-Weinberg equilibrium were discarded.
Genomic DNA was extracted from whole blood by using a QuickGene-810 nucleic acid isolation system (Fujifilm, Tokyo, Japan) and QuickGene DNA Whole Blood Kit S (Kurabo, Osaka, Japan), in accordance with the manufacturer’s instructions. DNA was stored at 4 °C until use. SNP genotyping was performed by using a GoldenGate Genotyping Assay with VeraCode technology (Illumina, San Diego, CA, United States). A custom GoldenGate assay was designed for the analysis of the selected SNPs in the PRKAA1 gene. Those SNPs were then assessed for suitability for the GoldenGate genotyping platform, and the analysis was carried out on the validated SNPs. The average call rate was 99.40%. Genotyping was carried out by Macrogen (Seoul, South Korea).
Statistical analysis
Testing for deviation from the Hardy-Weinberg equilibrium was performed for each SNP in both cases and controls by using Pearson’s χ2 test. D values were measured by using Lewontin’s method for all combinations of biallelic loci, and linkage disequilibrium blocks were structured by using Haploview version 4.2 (Daly Lab at the Broad Institute Cambridge, MA, United States). Haplotype blocks were constructed and statistically compared between cases and controls by using SNP Analyzer version 2.0 (ISTEC, Goyang, South Korea). Haplotype blocks which frequency over 5% were selected for analysis.
Student’s t-test was used to compare the values of continuous variables in the patient and control groups. Associations between gastric cancer and putative risk factors (including the SNPs) were estimated by using odds ratios (ORs) and their corresponding 95%CI, as derived from multivariate conditional logistic regression models, after adjusting for potential confounding factors such as age, sex, smoking history, alcohol intake amount, total calorie intake, and education level. Homozygous reference genotypes, heterozygous alleles, and homozygous risk alleles in each SNP were coded as 0, 1, and 2 in the codominant model; 0, 1, and 1 in the dominant model; and 0, 0, and 1 in the recessive model, respectively. Benjaminin and Hochberg’s methods for control of the false discovery rate (FDR) were used for multiple testing corrections[19]. Two-sided adjusted P values < 0.05 were considered to be statistically significant. FDR Q values were calculated separately for the SNPs and haplotypes based on those numbers.
Gene-based association tests were performed by using the versatile gene-based association study (VEGAS) method[20].
The study power calculations were performed by using the “case - control for discrete traits” mode in the Genetic Power Calculator created by Shaun Purcell (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html). The following parameters were applied: a risk allele frequency of 0.4, an alpha error of 0.01, and a disease prevalence of 0.1%. The power of a codominant model was 0.7768 when the heterozygous odds ratio was set to 1.5. The power of a dominant model was 0.8821 when the odds ratio for a genotype with 1 or 2 risk allele(s) was taken to be 2. The power of a recessive model was 0.8182 when a value of 2 was input for the odds ratio for a genotype with 2 risk allele(s). SAS version 9.2 (SAS Institute, Cary, NC) was used for all statistical analyses.
RESULTS
We explored the associations between 5 DNA polymorphisms in the PRKAA1 gene, which encodes AMPK, and gastric cancer risk. A total of 954 subjects were included in the analysis, comprising 477 gastric cancer cases and equal number of matched controls. There were no significant differences between the two groups in terms of age, gender, smoking status, or alcohol intake.
Table 1 presents the frequencies of the 5 selected SNPs in the study subjects. The genotype distributions at all 5 SNPs were in Hardy-Weinberg equilibrium, with non-significant χ2 values.
Table 1.
Frequencies of PRKAA1 polymorphisms in cases and controls
| SNP | Position | Genotype |
Case |
Control |
|||||
| case/control | Freq1 | HWE2 | Freq1 | HWE2 | |||||
| rs6882903 | 40801619 | CC | CA | AA | N | 0.183 | 0.034 | 0.148 | 0.371 |
| 311/342 | 156/125 | 9/8 | 476/475 | ||||||
| rs10074991 | 40826308 | AA | AG | GG | N | 0.541 | 0.573 | 0.456 | 0.412 |
| 97/136 | 242/244 | 136/94 | 475/474 | ||||||
| rs13361707 | 40827641 | TT | TC | CC | N | 0.542 | 0.632 | 0.459 | 0.510 |
| 97/135 | 241/242 | 137/96 | 475/473 | ||||||
| rs154268 | 40831625 | TT | TC | CC | N | 0.239 | 0.020 | 0.188 | 0.576 |
| 267/311 | 192/149 | 18/15 | 477/475 | ||||||
| rs3805486 | 40831802 | TT | TC | CC | N | 0.224 | 0.474 | 0.296 | 0.786 |
| 283/233 | 170/199 | 21/40 | 474/472 | ||||||
Freq: Allele frequency;
P value deviation from Hardy-Weinberg Equilibrium (HWE). SNP: Single nucleotide polymorphism.
Haplotype linkage disequilibrium block and haplotype frequencies for PRKAA1 are presented in Figure 1. Common haplotypes (frequency > 5%) of the block were found in 93.6% of cases, and 95.7% of controls.
Figure 1.
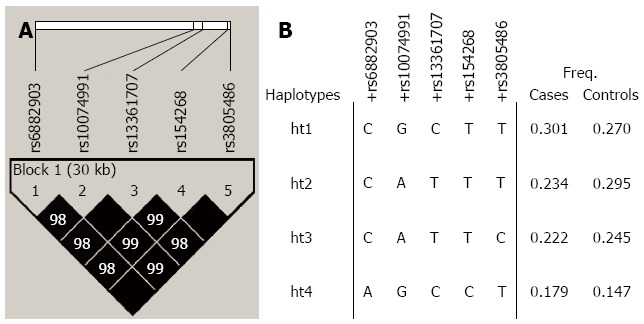
Haplotype linkage disequilibrium blocks and haplotype frequencies for PRKAA1. A: Linkage disequilibrium blocks and correlation coefficients among PRKAA1 polymorphisms. Black squares indicate statistically significant allelic association between the pair of single nucleotide polymorphisms (SNPs), as measured by using the D statistic; darker gray indicate higher values of D, up to a maximum of 1; B: Haplotype frequencies of PRKAA1 polymorphisms in the case and control groups. Freq: Allele frequency.
The frequencies and distribution of genotypes are presented in Tables 2 and 3, which also report the odds ratios for the associations between each polymorphism and gastric cancer. We performed the PRKAA1 gene association analysis with dominant, recessive, and codominant models by using conditional logistic regression. In the dominant model, 4 of 5 SNPs we observed were significantly associated with an increased risk of gastric cancer. SNPs rs13361707 (“C” allele, OR = 1.51, 95%CI: 1.07-2.11, P = 0.018, FDR Q = 0.023), rs154268 (“C” allele, OR = 1.65, 95%CI: 1.22-2.22, P = 0.001, FDR Q = 0.006), rs6882903 (“A” allele, OR = 1.48, 95%CI: 1.09-2.00, P = 0.012, FDR Q = 0.023), and rs10074991 (“G” allele, OR = 1.53, 95%CI: 1.09-2.16, P = 0.014, FDR Q = 0.023) were significantly associated with an increased risk of gastric cancer. In the recessive model, SNP rs154268 (OR = 1.66, 95%CI: 1.22-2.26, P = 0.001, FDR Q = 0.005), rs3805486 (OR = 0.63, 95%CI: 0.46-0.85, P = 0.003, FDR Q = 0.005) and rs10074991 (OR = 1.47, 95%CI: 1.15-1.88, P = 0.002, FDR Q = 0.005) were significant risk factors for gastric cancer (Table 2). In the codominant model, the odds ratios were statistically significant for all 5 SNPs. All SNPs in the model showed dose-response relationships between minor allele frequency and the risk of gastric cancer. Most notably, subjects with a homozygous minor allele in SNP rs10074991 showed 2.15 times of risk for gastric cancer, compared with subjects who did not have a minor allele (Table 3).
Table 2.
Association between PRKAA1 polymorphisms and gastric cancer
| SNP |
Dominant model |
Recessive model |
||||
| OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | |
| rs6882903 | 1.48 (1.09-2.00) | 0.0121 | 0.0230 | 1.21 (0.60-2.46) | 0.5906 | 0.7383 |
| rs10074991 | 1.53 (1.09-2.16) | 0.0139 | 0.0230 | 1.47 (1.15-1.88) | 0.0021 | 0.0045 |
| rs13361707 | 1.51 (1.07-2.11) | 0.0184 | 0.0230 | 1.13 (0.43-2.92) | 0.8085 | 0.8085 |
| rs154268 | 1.65 (1.22-2.22) | 0.0012 | 0.0060 | 1.66 (1.22-2.26) | 0.0012 | 0.0045 |
| rs3805486 | 0.61 (0.34-1.09) | 0.0916 | 0.0916 | 0.63 (0.46-0.85) | 0.0027 | 0.0045 |
| VEGAS statistics (P) | 30.0356 (0.0054) | 56.0515 (0.0001) | ||||
Reference alleles of each single nucleotide polymorphism (SNP) in logistic analysis are as follows; CC for rs6882903, AA for rs10074991, and TT for rs13361707, rs154268 and rs3805486.
P values for logistic analysis of two alternative models (dominant and recessive) adjusted with calorie intake, smoking history, alcohol intake and educational level; 2False discovery rate Q value.
Table 3.
Codominant model odds ratios of the PRKAA1 polymorphisms for gastric cancer
| SNPs | Adjusted OR (95%CI) | P value1 | Q2 | |
| rs6882903 | CC | 1.00 | 0.0123 | 0.0123 |
| CA | 1.46 (1.07-2.00) | |||
| AA | 1.75 (0.60-5.08) | |||
| rs10074991 | AA | 1.00 | 0.0005 | 0.0023 |
| AG | 1.30 (0.90-1.87) | |||
| GG | 2.15 (1.40-3.30) | |||
| rs13361707 | TT | 1.00 | 0.0009 | 0.0023 |
| TC | 1.29 (0.90-1.85) | |||
| CC | 2.05 (1.35-3.14) | |||
| rs154268 | TT | 1.00 | 0.0017 | 0.0028 |
| TC | 1.63 (1.20-2.22) | |||
| CC | 1.77 (0.82-3.83) | |||
| rs3805486 | TT | 1.00 | 0.0099 | 0.0123 |
| TC | 0.72 (0.40-1.32) | |||
| CC | 0.54 (0.30-0.98) | |||
| VEGAS statistics (P) | 46.0927 (0.0004) | |||
P values for logistic analysis of codominant model adjusted with age, sex, calorie intake, smoking history, alcohol intake and educational level; 2False discovery rate Q value. SNP: Single nucleotide polymorphism; VEGAS: Versatile gene-based association study.
To evaluate the association between gastric cancer and all SNPs within the PRKAA1 gene (rather than each SNP individually) we performed a gene-based analysis following the VEGAS method, the results of which indicated that SNPs in PRKAA1 had a statistically significant association with gastric cancer in all 3 models (P = 0.0054, 0.0001, and 0.0004 for the dominant, recessive, and codominant models, respectively).
The haplotype block was also evaluated for an association with the risk of gastric cancer (Table 4), but none of the results were significant in each of the 3 models.
Table 4.
Association between PRKAA1 haplotypes and gastric cancer
| Haplotypes |
Codominant |
Dominant |
Recessive |
|||||||
| OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | ||
| PRKAA1 | CGCTT | 1.57 (0.93-2.65) | 0.205 | 0.980 | 1.18 (0.92-1.53) | 0.195 | 0.980 | 1.48 (0.89-2.47) | 0.126 | 1.000 |
| haplotype | CATTT | 0.79 (0.46-1.36) | 0.574 | 0.980 | 1.02 (0.79-1.32) | 0.895 | 0.980 | 0.77 (0.45-1.32) | 0.339 | 1.000 |
| block 1 | CATTC | 0.44 (0.25-0.77) | 0.002 | 0.062 | 0.67 (0.52-0.87) | 0.002 | 0.063 | 0.50 (0.29-0.87) | 0.012 | 0.346 |
| CGCCT | 1.44 (0.53-3.90) | 0.032 | 0.456 | 1.45 (1.10-1.92) | 0.009 | 0.124 | 1.29 (0.48-3.50) | 0.614 | 1.000 | |
P values for logistic analysis of three alternative models (codominant, dominant and recessive), the P values for haplotype association were calculated by the single nucleotide polymorphism Analyzer™ 2.0 software; 2False discovery rate Q value.
DISCUSSION
In this study, we hypothesized that genetic polymorphisms in PRKAA1 might contribute to gastric cancer development by affecting the regulation of energy metabolism. Activated AMPK inactivates a number of metabolic enzymes involved in ATP-consuming cellular events (such as fatty acid, cholesterol, and protein synthesis) and also activates ATP-generating processes (such as the uptake and oxidation of glucose and fatty acids)[12,13]. Besides energy metabolism, AMPK also functions as a suppressor of cell proliferation[14]. Consequently, some research on AMPK has focused on its potential role as a therapeutic target for cancer.
A recent genome-wide association study identified a new SNP (rs13361707) in the PRKAA1 gene that is significantly associated with increased susceptibility to gastric cancer[21,22]. Additionally, Song et al[18] reported that the rs13361707 SNP was associated with an increased risk of gastric cancer in the Korean population. In their replication study, rs13361707 TC and CC genotypes were associated with a significantly increased risk of gastric cancer (OR = 1.29 for TC vs TT, OR = 1.68 for CC vs TT). In agreement with these findings, our result also showed that the rs13361707 SNP is associated with gastric cancer (OR = 1.29 for TC vs TT, OR = 2.05 for CC vs TT). Together, these results suggest that rs13361707 SNP might play an important role in the development of gastric cancer. However, since rs13361707 is not the only SNPs found in this gene, it remained important to examine associations between other SNPs of PRKAA1 and gastric cancer development.
In the present study, we evaluated the associations of 5 SNPs of PRKAA1 gene with gastric cancer. Interestingly, we found that all 5 of the tested SNPs of PRKAA1 we tested were associated with a significantly increased risk of gastric cancer. Most notably, subjects with a homozygous minor allele in SNP rs10074991 were at 2.15 times the risk of gastric cancer, compared with subjects who did not have a minor allele. After controlling the FDR, the associations of these SNPs remained statistically significant in the codominant model. In a gene-based association test, the PRKAA1 gene was found to be significantly associated with gastric cancer. These results suggest that genetic polymorphism of PRKAA1 might play an important role in gastric carcinogenesis.
Although the biological mechanism underlying the association between PRKAA1 and gastric cancer has not been clarified, these significant associations could potentially be explained by the ability of activated AMPK phosphorylates p53 to induce G1/S arrest. Further, the AMPK-p53 connection may represent a cell cycle checkpoint[23]. Therefore, individuals with mutant PRKAA1 alleles, which encode inactive AMPK, may be vulnerable to gastric cancer. Anti-inflammatory action by AMPK could provide another explanation of the association between SNPs of PRKAA1 and gastric cancer. A recent study has reported that activated AMPK can counter-regulate macrophage inflammatory function[24] and activate some anti-inflammatory agents[25]. Loss of anti-inflammatory action by AMPK in the body of individuals with mutant PRKAA1 alleles results in more severe injury of the epithelium[26]. Bone marrow-derived cells are recruited at these sites of epithelial damage, and these cells can be a potential source of malignancy[27]. To our knowledge, there is no study addressed association between PRKAA1 gene and gastric cancer except in Chinese and Korean population. Since SNPs frequency are different according to the population, it is need to further study in different races other than Asian.
The present study has several limitations. First, a relatively small number of patients and controls were enrolled in this study. Second, we could not obtain detailed data on the histological tumor types for the cases of gastric cancer. Finally, because the data of environmental factor for gastric cancer such as H. pylori infection and diet was not available in this study, we could not evaluate the gene-environmental interaction. It is needed further study about it.
In summary, the PRKAA1 gene and 5 of its SNPs (rs13361707, rs154268, rs3805486, rs6882903, and rs10074991) were associated with an increased risk of gastric cancer in Koreans.
COMMENTS
Background
Gastric cancer is the second most common cause of cancer-related mortality worldwide. AMP-activated protein kinase (AMPK) is an energy sensing/signaling intracellular protein, and a conserved serine/threonine kinase that regulates energy homeostasis and metabolic stress. It is known that AMPK activation suppresses cell proliferation in both tumor and non-malignant cells. Therefore, it is possible that AMPK plays an important role in gastric carcinogenesis and polymorphic alleles of the encoding gene could modify individual susceptibility to gastric cancer. The aim of the present study was to evaluate the associations between 5 polymorphic alleles of PRKAA1, the gene that encodes AMPK, and gastric carcinogenesis in Koreans.
Research frontiers
In the present study, the authors evaluated the associations of 5 single nucleotide polymorphisms (SNPs) of PRKAA1 gene with gastric cancer. To our knowledge, this is the first replication study to indicate an association between rs154268, rs3805486, rs6882903, and rs10074991 and gastric cancer development. Interestingly, all 5 SNPs of PRKAA1 tested SNP were associated with a significantly increased risk of gastric cancer. Most notably, subjects with a homozygous minor allele in SNP rs10074991 were at 2.15 times the risk of gastric cancer, compared with subjects who did not have a minor allele.
Innovations and breakthroughs
There were a few studies to evaluate association between PRKAA1 gene and gastric cancer. However, in previous study, only one SNP (rs13361707) of PRKAA1 gene was focused. In this study, authors evaluated 5 SNPs of PRKAA1 gene including rs13361707 in associated with gastric cancer risk.
Applications
The result of this study showed that PRKAA1 gene and its 5 SNPs (rs13361707, rs154268, rs3805486, rs6882903, and rs10074991) were associated with an increased risk of gastric cancer in Koreans. Further studies are needed to determine the mechanism which by PRKAA1 and other environmental factors interact and influence the development of gastric cancer.
Peer review
This is a nice paper with a good summary of the issue and a well described methodology for studying 5 polymorphic alleles of PRKAA1, the gene that encodes AMPK, and gastric cancer in Koreans. In this study, the authors test the association between 5 SNPs of PRKAA1 gene and gastric cancer in a Korean population. The PRKAA1 gene has been implicated in carcinogenesis at several levels and the authors provide a reasonable rationale for this selection. The SNPs show nominally significant association to gastric cancer.
Footnotes
Supported by A grant from the National R & D Program for Cancer Control, Ministry of Health and Welfare, South Korea, No. 1120330
P- Reviewers: Bader EL Din NGBG, Garcia-Elorriaga G, Park WS S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, Bray F. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet. 2012;380:1840–1850. doi: 10.1016/S0140-6736(12)60919-2. [DOI] [PubMed] [Google Scholar]
- 4.Kim HY. [What is the most important factor for gastric carcinogenesis in Koreans: Helicobacter pylori, host factor or environmental factor?] Korean J Gastroenterol. 2007;49:60–71. [PubMed] [Google Scholar]
- 5.Mayne ST, Navarro SA. Diet, obesity and reflux in the etiology of adenocarcinomas of the esophagus and gastric cardia in humans. J Nutr. 2002;132:3467S–3470S. doi: 10.1093/jn/132.11.3467S. [DOI] [PubMed] [Google Scholar]
- 6.Galanis DJ, Lee J, Kolonel LN. The influence of cigarette smoking, alcohol, and green tea consumption on the risk of carcinoma of the cardia and distal stomach in Shanghai, China. Cancer. 1997;79:1840–1841. doi: 10.1002/(sici)1097-0142(19970501)79:9<1840::aid-cncr29>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 7.Gammon MD, Schoenberg JB, Ahsan H, Risch HA, Vaughan TL, Chow WH, Rotterdam H, West AB, Dubrow R, Stanford JL, et al. Tobacco, alcohol, and socioeconomic status and adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst. 1997;89:1277–1284. doi: 10.1093/jnci/89.17.1277. [DOI] [PubMed] [Google Scholar]
- 8.Ji BT, Chow WH, Yang G, McLaughlin JK, Gao RN, Zheng W, Shu XO, Jin F, Fraumeni JF, Gao YT. Body mass index and the risk of cancers of the gastric cardia and distal stomach in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 1997;6:481–485. [PubMed] [Google Scholar]
- 9.Pelucchi C, Tramacere I, Bertuccio P, Tavani A, Negri E, La Vecchia C. Dietary intake of selected micronutrients and gastric cancer risk: an Italian case-control study. Ann Oncol. 2009;20:160–165. doi: 10.1093/annonc/mdn536. [DOI] [PubMed] [Google Scholar]
- 10.Hardie DG, Corton J, Ching YP, Davies SP, Hawley S. Regulation of lipid metabolism by the AMP-activated protein kinase. Biochem Soc Trans. 1997;25:1229–1231. doi: 10.1042/bst0251229. [DOI] [PubMed] [Google Scholar]
- 11.Kemp BE, Stapleton D, Campbell DJ, Chen ZP, Murthy S, Walter M, Gupta A, Adams JJ, Katsis F, van Denderen B, et al. AMP-activated protein kinase, super metabolic regulator. Biochem Soc Trans. 2003;31:162–168. doi: 10.1042/bst0310162. [DOI] [PubMed] [Google Scholar]
- 12.Hardie DG. New roles for the LKB1--> AMPK pathway. Curr Opin Cell Biol. 2005;17:167–173. doi: 10.1016/j.ceb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Luo Z, Saha AK, Xiang X, Ruderman NB. AMPK, the metabolic syndrome and cancer. Trends Pharmacol Sci. 2005;26:69–76. doi: 10.1016/j.tips.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Igata M, Motoshima H, Tsuruzoe K, Kojima K, Matsumura T, Kondo T, Taguchi T, Nakamaru K, Yano M, Kukidome D, et al. Adenosine monophosphate-activated protein kinase suppresses vascular smooth muscle cell proliferation through the inhibition of cell cycle progression. Circ Res. 2005;97:837–844. doi: 10.1161/01.RES.0000185823.73556.06. [DOI] [PubMed] [Google Scholar]
- 15.Kuhajda FP, Pizer ES, Li JN, Mani NS, Frehywot GL, Townsend CA. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc Natl Acad Sci USA. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motoshima H, Goldstein BJ, Igata M, Araki E. AMPK and cell proliferation--AMPK as a therapeutic target for atherosclerosis and cancer. J Physiol. 2006;574:63–71. doi: 10.1113/jphysiol.2006.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinnett-Smith J, Kisfalvi K, Kui R, Rozengurt E. Metformin inhibition of mTORC1 activation, DNA synthesis and proliferation in pancreatic cancer cells: dependence on glucose concentration and role of AMPK. Biochem Biophys Res Commun. 2013;430:352–357. doi: 10.1016/j.bbrc.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song HR, Kim HN, Kweon SS, Choi JS, Shim HJ, Cho SH, Chung IJ, Park YK, Kim SH, Choi YD, et al. Genetic variations in the PRKAA1 and ZBTB20 genes and gastric cancer susceptibility in a Korean population. Mol Carcinog. 2013;52 Suppl 1:E155–E160. doi: 10.1002/mc.22063. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful pproach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 20.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, Li JM, Kong GQ, Qi H, Cui J, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759–763. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 23.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Sag D, Carling D, Stout RD, Suttles J. Adenosine 5’-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J Immunol. 2008;181:8633–8641. doi: 10.4049/jimmunol.181.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald TT, Horton MA, Choy MY, Richman PI. Increased expression of laminin/collagen receptor (VLA-1) on epithelium of inflamed human intestine. J Clin Pathol. 1990;43:313–315. doi: 10.1136/jcp.43.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]


