Abstract
Lipolytic enzymes catalyze the hydrolysis of ester bonds in the presence of water. In media with low water content or in organic solvents, they can catalyze synthetic reactions such as esterification and transesterification. Lipases and esterases, in particular those from extremophilic origin, are robust enzymes, functional under the harsh conditions of industrial processes owing to their inherent thermostability and resistance towards organic solvents, which combined with their high chemo-, regio- and enantioselectivity make them very attractive biocatalysts for a variety of industrial applications. Likewise, enzymes from extremophile sources can provide additional features such as activity at extreme temperatures, extreme pH values or high salinity levels, which could be interesting for certain purposes. New lipases and esterases have traditionally been discovered by the isolation of microbial strains producing lipolytic activity. The Genome Projects Era allowed genome mining, exploiting homology with known lipases and esterases, to be used in the search for new enzymes. The Metagenomic Era meant a step forward in this field with the study of the metagenome, the pool of genomes in an environmental microbial community. Current molecular biology techniques make it possible to construct total environmental DNA libraries, including the genomes of unculturable organisms, opening a new window to a vast field of unknown enzymes with new and unique properties. Here, we review the latest advances and findings from research into new extremophilic lipases and esterases, using metagenomic approaches, and their potential industrial and biotechnological applications.
Keywords: Extremophiles, industrial biocatalysts, lipases and esterases, lipolytic enzymes classification, metagenomics, non-culturable microorganisms.
1. LIPASES AND ESTERASES
Lipolytic enzymes are members of the α/β hydrolase family and include two groups of enzymes, true lipases (EC 3.1.1.3 triacylglycerol lipases) and esterases (EC 3.1.1.1 carboxylesterases), which differ in their biochemical properties. Both types catalyze the hydrolytic cleavage of an ester bond between a carboxylic acid and an alcohol group in water. In organic solvents or non-aqueous media they can also catalyze the reverse reaction, the formation of an ester bond by transesterification or esterification. The above-mentioned reactions awaken great interest in diverse industrial fields.
These groups of enzymes share little primary sequence similarity but their tertiary structure is highly conserved. They present a typical α/β hydrolase fold with eight beta sheets, all parallel except the anti-parallel second, connected through alpha-helices. The active site, located in an alpha helix, contains a catalytic triad of amino acid residues always arranged in the same order along the sequence: serine (Ser), aspartate (Asp) or glutamate (Glu), and histidine (His), with the catalytic serine embedded in the consensus motif Gly-X-Ser-X-Gly [1].
The catalytic mechanism is common to every lipase and esterase. First, the catalytic serine binds the carbonyl carbon of the lipid ester bond. A tetrahedral intermediate is formed, stabilized by the catalytic residues His and Asp/Glu. The alcohol component of the ester bond is cleaved and esterification of the acid component to the catalytic serine forms the covalent intermediate. Second, a water molecule hydrolyzes the covalent intermediate by forming a new tetrahedral intermediate, and releases the acyl product [2]. In a (trans-) esterification reaction the water molecule is replaced by an alcohol (or an ester).
Lipases and esterases are ubiquitous in nature and can be found in animals, plants and microorganisms, but most industrial lipases are of microbial origin [3]. Bacterial lipolytic enzymes were classified by Arpigny and Jaeger into eight families, I to VIII, according to their amino acid sequences and biological properties [4]. This classification is still the reference most commonly used to assign a newly discovered enzyme to a family, though with certain modifications, because new families have been discovered through metagenomics since it was published [5-9]. The current state of this classification of bacterial lipolytic enzymes is summarized in Table 1.
Table 1.
Current bacterial lipolytic enzymes classification. Description of bacterial lipolytic families I-VIII in the Arpigny and Jaeger classification [4], and new families and subfamily discovered by functional metagenomics (*) [5-9].
| Family | Description |
|---|---|
| I |
|
| II |
|
| III |
|
| EstA* |
|
| IV |
|
| V |
|
| EstF* |
|
| VI |
|
| VII |
|
| VIII |
|
| LipG* |
|
| LipEH166* |
|
| EstY* |
|
There are several criteria for distinguishing between true lipases and esterases. They differ in substrate preference. Esterases hydrolyze only short-chain (< 12 carbon atoms) water soluble fatty acid esters, and lipases show preference for long-chain (≥ 12 carbon atoms) fatty acid esters, with low water solubility [2]. Also, there is a phenomenon called interfacial activation in most lipases and not in esterases. Whereas esterases show classical Michaelis–Menten kinetic behavior, most lipases possess a lid or loop covering the active site and its opening leads to sudden activation of the enzyme. This change from closed to open conformation, where the active site is accessible to the substrate so the enzyme becomes active, is driven by the lipidic interface of a substrate emulsion [10].
Nowadays, lipases and esterases represent a major portion with high growth potential in the World Industrial Enzymes Market. They have many applications in the food industry (modification of fats to develop organoleptic and nutritional qualities) and the paper industry (removal of pitch from paper pulp), as additives in detergents, synthesis of biopolymers, biodiesel production, synthesis of optically pure compounds and fine chemicals of interest in the pharmaceutical (antibiotics, anti-inflammatory drugs), cosmetic (flavor and fragrance compounds) and agrochemical (herbicides, insecticides) industries, and bioremediation and waste treatment [1, 11-13].
Lipases/esterases are inherently robust enzymes that can withstand the harsh conditions of industrial bioconversion such as broad pH range, presence of organic solvents and high temperatures [3]. Besides, they possess distinctive features of chemo-, stereo- and regioselectivity that are of special interest in certain applications such as the synthesis of optically pure compounds [12]. Their catalytic versatility, robustness and high specificity attract enormous attention as industrial biocatalysts.
As mentioned above, most industrial lipases are of microbial origin, a term that includes a great variety of sources. Extremophiles in particular are adapted to living in environments with extreme physicochemical conditions such as high temperatures (thermophiles), low temperatures (psychrophiles), high salt concentration (halophiles) or high pressure (barophiles). Their enzymes have evolved to be functional under such extreme conditions, which their mesophilic counterparts could not survive, providing additional features of high value for industrial process development [14].
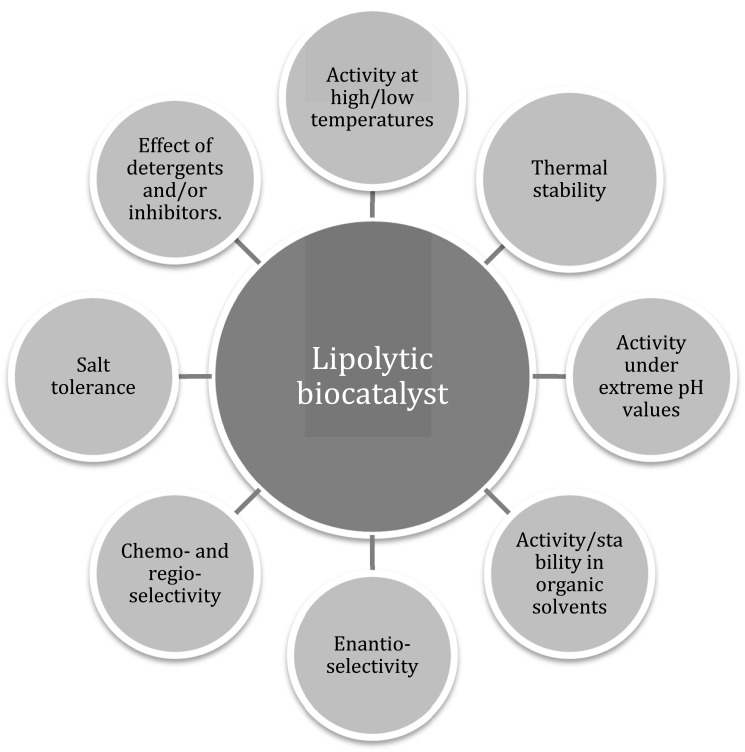
Known lipases and esterases have different combinations of substrate specificity, operative temperature and pH range, thermostability, tolerance to organic solvents and activity under the presence of diverse activators and inhibitors. The most valuable properties that determine their potential as industrial biocatalysts, either singly or in combination, are shown in (Fig. 1) and include:
Fig. (1).
Physicochemical properties of industrial interest of a lipolytic biocatalyst.
Activity at high temperatures and thermostability: most industrial processes in which lipases/esterases are used as biocatalysts are carried out at temperatures above 45ºC [12], so the enzymes need to be stable at this temperature and above. Besides, there are several advantages in performing industrial bioconversions at high temperatures, such as reducing the risk of contamination with mesophiles and higher reaction rates due to the reduction of viscosity and increased diffusion rate and substrate solubility [12, 15]. For instance, thermostable lipases can be used for processing lipids that are in the solid state at room temperature, when temperatures above 50ºC are necessary [16].
Activity at low temperatures: Low-temperature adapted enzymes show ten times more activity at low temperatures than their mesophilic counterparts, with optimal activity at 20-30ºC [17]. This activity profile allows industrial biotransformations to be performed at low temperature, maintaining high conversion rates with considerably lower energy costs. Low-temperature active lipases have potential applications as additives in detergents for cold washing, in the food industry to avoid changes in ingredients, in biocatalysis when thermolabile or volatile compounds are used, and in bioremediation of low-temperature soil or water [13, 18, 19].
Activity in alkaline pH: many industrial applications are performed under alkaline conditions. This feature is especially important in the laundry industry.
Activity/stability in the presence of organic solvents: When lipases/esterases are used in organic solvents, synthesis (esterification and transesterification) is thermodynamically favored over hydrolysis. Biocatalysis in the presence of organic solvents instead of water also favors the recovery of the product and higher global yield, increases the solubility of non-polar substrates and subsequently their conversion rate, avoids side reactions, and minimizes denaturation, deactivation and/or substrate/product inhibition [20]. Thus, lipases/esterases with activity in organic solvents are potentially useful in organic synthesis and other industrial bioconversions such as the production of pharmaceuticals [18]. Ionic liquids are environmentally friendly alternative media for lipase-catalyzed synthesis. Their advantageous properties include no measurable vapor pressure, non-flammability and a wide temperature range in the liquid phase (they are liquid at temperatures lower than 100ºC). Moreover, they have proven to have great potential in allowing generally better enantioselectivity and conversion rates to be achieved than those obtained with organic media, along with improved biocatalyst stability [21]. They can be designed to obtain the desired physical properties that fit a particular reaction with optimized enantioselectivity, by the selection of appropriate cations and anions [22].
Salt tolerance: enzymes from halophiles are currently underexploited, but they hold great potential. They can successfully compete for water and resist the denaturating effects of salts, a property that is related to resistance to organic solvents [23, 24].
High enantioselectivity: this feature make lipolytic enzymes extremely useful for the synthesis of optically pure compounds. One remarkable example is the kinetic resolution of tertiary alcohols, important building blocks in organic synthesis. The majority of lipases/esterases with activity towards tertiary alcohols possess the motif GGG(A)X in their oxyanon hole, which has been useful for identifying them among the enzymes revealed by metagenomic studies [25]. The production of biologically active enantiomers such as antibiotics or anti-inflammatory drugs also take advantage of this property of lipases/esterases.
Chemo- and regio-selectivity: Some industrial reactions are based on these properties, such as the release of ferulic acid from plant cell wall polysaccharides, which is used thereafter as substrate in the production of vanillin (a flavor compound) [1].
Activity in the presence of detergents and/or inhibitors: Resistance to denaturation under these conditions while maintaining high activity levels is of special interest in the laundry industry.
Given the importance of these industrial biocatalysts, the mining of new enzymes with unique feature combinations is crucial for industrial development. This is where metagenomics play a key role in the discovery of really novel enzymes.
2. METAGENOMICS IN THE DISCOVERY OF NEW LIPASES AND ESTERASES
Cultivation and isolation of microbial strains producing lipolytic enzymes have been traditionally used for discovering new lipases/esterases with potential applications. The task is facilitated by simple assays for detecting the presence of lipolytic enzymes, such as the formation of hydrolysis halos in solid medium plates containing emulsified substrates. Even today this strategy continues to be used.
Automation of sequencing methods and shotgun cloning have motivated the start-up of many Genome Projects that have provided vast amounts of genetic information. To date, 1078 bacterial genomes and 82 from Archaea are fully sequenced and available in the Genome Atlas Database [26]. Using sequenced data, many enzymes have been discovered through genome mining for novel genes by homology with known lipases and esterases, and they have subsequently been cloned, overexpressed and purified for biochemical characterization. In this way, several lipolytic enzymes from Thermus thermophilus HB27, whose genome is completely sequenced and publicly available [27], were cloned and expressed in mesophilic hosts. A recombinant esterase with interesting properties was obtained: extreme thermal stability, and high activity at mesophilic temperatures, remarkable given its thermophilic origin [28].
At that stage, the sources of new enzymes were technically limited to a minor fraction of total microbial diversity, the culturable microorganisms, which have been estimated as representing less than 1% of the real diversity in most environments [29]. The Metagenomics Era meant a step forward in this field with the study of the metagenome, the pool of genomes in an environmental microbial community [30]. Current molecular biology techniques make it possible to construct total environmental DNA libraries, including the genomes of unculturable organisms, opening a new window to a vast field of unknown enzymes with potentially new and unique properties. The growing number of enzymes of industrial interest discovered by metagenomic studies will probably very soon exceed the number of enzymes discovered by traditional techniques. It is remarkable that new biocatalysts search in metagenomic DNA libraries is mainly focused on the discovery of a small group of enzymes, where lipases and esterases are included [31].
With the development of next-generation sequencing technologies and new bioinformatic tools to carry out large-scale analysis and classification of metagenomic data, many metagenomic sequencing projects emerged, yielding a comprehensive view of the taxonomic and ecological diversity of microbial communities [32]. According to the Genomes Online Database (GOLD) there are 340 available sequenced metagenomes, 197 from natural environments (most of them of aquatic origin), 114 host-associated, and 29 from engineered environments [33].
In the sequence-based metagenomic approach, new enzymes are discovered by exploring these available metagenomic data for enzymes homologous to known lipases/esterases. Another common strategy is a PCR-based method with degenerate primers designed according to the conserved regions of already-known classes of lipolytic enzymes [34]. A disadvantage is that this approach tends to detect only enzymes related to previously reported families, and might overlook those with completely new sequences.
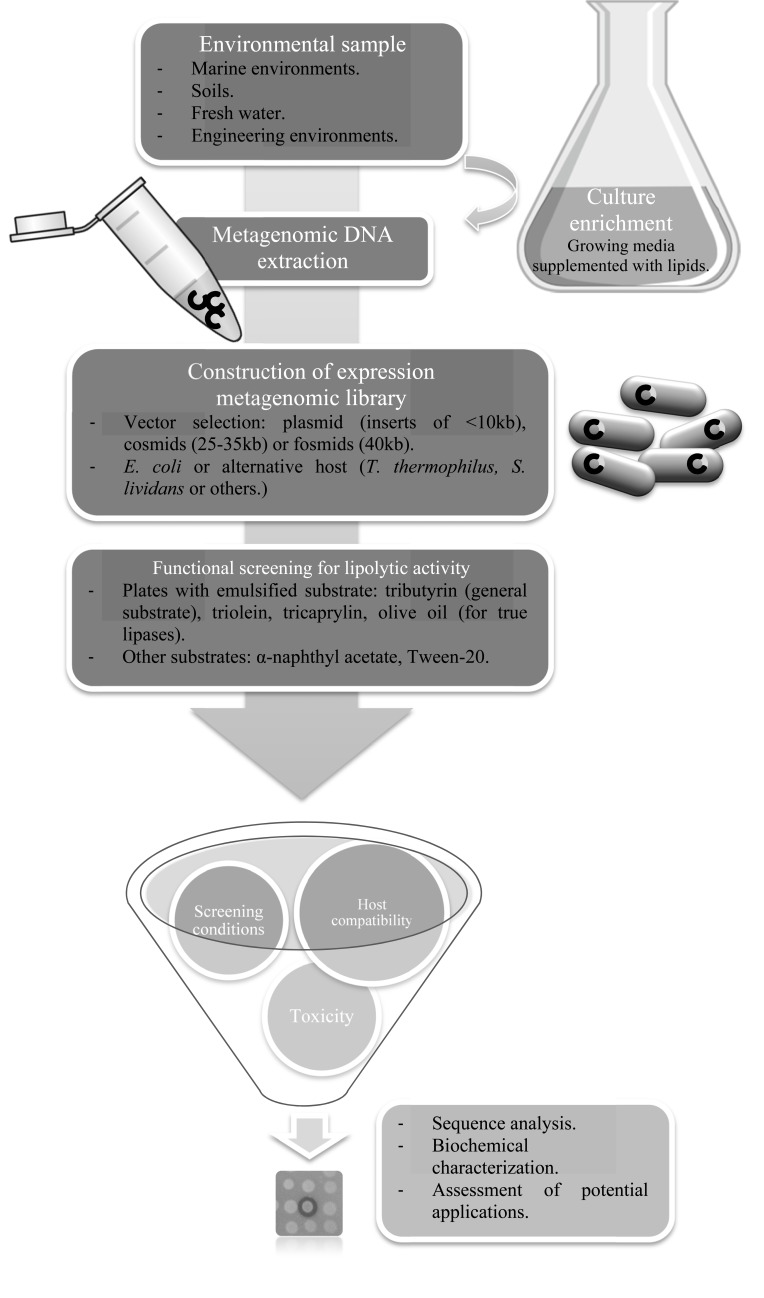
Alternatively, metagenomic libraries can be subjected to functional screening to detect clones that exhibit lipolytic activity (Fig. 2). The success of such screening relies on the compatibility of the cloned genes with the transcription and translation machinery of the heterologous host, usually Escherichia coli. Moreover, expression of lipases can be hampered by the requirement for specific chaperones for the correct folding of the enzyme or by its toxicity to the host cells. It has been reported that only a subset of enzymes with the desired activity present in a metagenomic library, about 40%, are recovered by functional screening when E. coli is used as host [35].
Fig. (2).
Steps for functional metagenomic mining of lipolytic enzymes.
The usefulness of Broad-Host Range vectors for overcoming the barrier of host compatibility has been assessed. One of the most recent studies proves its effectiveness using six different proteobacteria as hosts for the same metagenomic cosmid library, recovering different positive clones in each host [36]. More recently, Lussier and collaborators [37] developed a cosmid vector with two improvements: multi-host expression (E. coli and Streptomyces lividans) and transcription directed by T7 RNA polymerase, which has high activity, generates very long mRNAs and is very poorly terminated by unrelated transcription terminators, potentially enhancing the expression of foreign genes in large insert libraries, in this case lipase/esterase genes. A special fosmid vector has even been developed for the expression of metagenomic libraries from thermophiles, which allows the library to be constructed in E. coli and subsequently transferred to Thermus thermophilus for expression and screening [38].
The advantage of the function-driven approach is the potential for discovering entirely new classes of genes or enzymes, with no similarity to known lipases and esterases [31, 39].
Wang and collaborators [40] developed a variant of metagenomic screening named “truncated metagenomic gene-specific PCR” using previous information from function-driven screening to overcome the limitations of the typical sequence-based approach. The starting point of this method is a metagenome-derived lipolytic enzyme, which is used to design degenerate primers and explore the diversity of the sample through DNA shuffling. A diversified chimeric library of related lipolytic enzymes was obtained, with proven different specificities.
The most popular screening method for detecting positive clones exhibiting the desired lipolytic activity uses tributyrin agar plates, in which the appearance of clear halos around the colonies indicates hydrolysis of the substrate [41]. Screenings for specifically detecting true lipases have also been used with metagenomic libraries, using longer substrates that are not hydrolyzed by esterases (such as emulsified triolein, tricaprylin or olive oil) in the presence of the fluorescent dye rhodamine B. In this case, orange fluorescent halos appear around lipase-producing colonies when irradiated with UV at 350 nm [42].
Less frequently, metagenomic libraries are screened using alternative substrates such as α-naphthyl acetate in a soft agar overlay with fast blue R (detection of yellow halos) [43, 44] or agar plates with Tween-20 in the presence of CaCl2 (detection of halos formed by released fatty acid salts) [45].
The organic synthesis of optically pure compounds is one application of these enzymes that is attracting more attention. There is therefore a need to develop new methods to screen for enantioselectivity. In this context, Wang and collaborators have recently developed a sensitive, economic and versatile method based on the use of fluorescein sodium salt as indicator. This assay allows specific esters of different substrates to be identified in a 96-plate format, not only from chiral carboxylic acids but also from chiral alcohols [46].
Another important issue in metagenomic screening efficiency is the insert size. When plasmids are used, relatively short metagenomic sequences are cloned (<10kb), and more clones are needed to find a positive than in metagenomic libraries constructed with fosmid or cosmid vectors, where insert sizes are 40kb and 25-35kb respectively [31]. Besides, with short inserts, large clusters of genes and operons cannot be recovered. An alternative for reducing the plasmid library size needed to detect positive clones is to use plasmids that allow bidirectional transcription, with promoters on both sides of a multiple cloning site. This kind of plasmid has been used to search for lipolytic activity, yielding high frequencies of positive clones comparable to those obtained in cosmid libraries [47].
In functional metagenomics, the frequency of clones expressing the desired activity (harboring a lipase enzyme-coding gene that is correctly expressed) is usually less than 0.1%. This value can be raised by a culture-enrichment step prior to library construction, at the expense of a great decrease in microbial diversity. This strategy consists in favoring the growth of microorganisms harboring the target enzyme by the ability to use specific substrates or resistance under certain physicochemical pressures. For example, high culture temperatures favor the growth of thermophiles harboring thermophilic enzymes. In this way, seven novel alkaline and thermophilic lipases have been discovered by culture enrichment in a Sequence Fed-Batch Reactor at 50-70ºC during three months of culture [48, 49].
3. NOVEL EXTREME LIPASES AND ESTERASES MINED BY METAGENOMICS.
Enzymes usually show tailor-made properties according to the physiochemical conditions of the habitats in which their producing organisms grow. Therefore, the choice of samples for a metagenomic study should be established to a greater extent in agreement with the desired features in the recovered enzymes.
Lipases from extremophiles can provide special features that make them more suitable for specific applications where a lipolytic biocatalyst is required. In some cases this would be an advantage, e.g. enhanced stability that permits the recycling of the biocatalyst with no loss of activity, but in other cases it would be a requisite, e.g. high levels of catalytic activity at low temperatures for processing thermolabile compounds.
Here we present different extreme environments from which lipases/esterases have been isolated through metagenomics in recent years, and the new knowledge that these discoveries have brought to the field of lipolytic enzymes: bifunctional enzymes, several completely new families, protein engineering of metagenome-derived lipases and some very extreme features.
3.1. Hot Springs
Two types of protein thermostability are of interest for industrial purposes: thermodynamic stability (when a enzyme is used under denaturing conditions such as high temperature or presence of an organic solvent) and long term stability [50]. But enzymes from thermophiles, apart from their thermostability and activity at high temperatures, often show resistance towards organic solvents, detergents and extreme pH values [12], which make them potential biocatalysts for many industrial applications.
Two thermostable esterases have been isolated from metagenomic libraries from hot spring in recent years: EstE1 from a screening of four independent metagenomic libraries of thermal areas of Indonesia [51], and Est 1 from the Jae Sawn hot spring in Thailand [52]. These enzymes display typical thermophilic profiles: extremely stable at 80 and 70ºC in the absence of any stabilizer, with high optimal temperatures of 95ºC and 70ºC, respectively. The activity at lower temperatures is remarkably high in EstE1: 20% and 30% of its optimal activity is retained at 30ºC and 40ºC, respectively. The combination of high thermostability and activity at mesophilic temperatures might be valuable for reducing costs in industrial processes: prolonged useful life of the biocatalysts combined with energy saving.
More recently, a lipase was discovered using PCR with degenerated primers (designed using multiple sequence alignment of different lipases from thermophilic bacterial species) from metagenomic DNA from hot springs in Manikaran in India [50]. This lipase displayed unusual behavior given its thermophilic origin: loss of secondary structure at ambient temperature. A CD spectroscopy study showed complete distortion of the secondary structure above 35ºC. Notwithstanding the CD data, intrinsic fluorescence spectroscopy revealed that the enzyme retained its tertiary structure; indeed it showed maximal activity at 50ºC. This lipase is closely identical to lipases from Geobacillus with high thermostability but there are changes in certain regions at the amino acid level that might be responsible for its lower thermostability: an amino acid at position 310 and six other amino acids at the C-terminus. In any case, this lipase showed high activity in the presence of detergents and in 30% n-hexane, promising properties for industrial application. The modification of this enzyme by error-prone PCR yielded an improved mutant lipase with the mutation N355K in the mature polypeptide, close to the catalytic center of the enzyme, with 144-fold greater thermostability at 60ºC over the wild type enzyme, with an optimal temperature of 40ºC [53].
3.2. Compost
The temperature can rise to 80ºC during the thermogenic phase of composting, so compost could be a good source of thermostable enzymes. Various esterases have been isolated from compost through metagenomics in recent years, but not all of them were thermostable [47, 54]. One of the most interesting discoveries was a novel moderately thermostable esterase, EstCS2, which has an optimum temperature at 55ºC and remains stable up to 60ºC. With respect to stability in organic solvents, it showed remarkable stability in up to 50% (v/v) dimethyl sulfoxide (DMSO) or dimethylformamide (DMF). It can degrade polyurethane, which along with the ability to cleave sterically hindered esters of tertiary alcohols has potential use in industrial applications and bioremediation [55].
Despite the lack of thermostability, another esterase isolated from compost, named Est2K, showed alkaline tolerance, high tolerance to 30% methanol and enhanced activity (236.8% higher) in the presence of 5% methanol, properties really appreciated in organic synthesis [52].
3.3. Soils
Soil is known to be a great source of antibiotics, pharmaceuticals and biocatalysts. Soil metagenomics has yielded many diverse and novel enzymes for a broad range of applications.
The properties of two new lipases isolated from soils at different altitudes in Taishan (China) illustrate the influence of climate on adaptation of enzyme function: several screenings of the expression libraries at different temperatures revealed a cold-active lipase, pST1, in a sample from 1200 m altitude (optimal activity at 20ºC) and a moderately thermostable lipase, pTS2, in a sample from 400 m altitude (optimal activity at 40ºC) [56].
Other factors such as soil contamination can also favor the occurrence of lipases/esterases and the appearance of certain biochemical properties in the enzymes isolated from contaminated samples. A cold-active lipase, LipCE, was identified in the metagenome of oil-contaminated soil from the north part of Germany with maximum activity at 30ºC and remarkably high activity at low temperatures; 28% residual activity at 0ºC and 16% at −5ºC. This enzyme also exhibits high enantioselectivity and specificity for esters of primary alcohols, unique features that make LipCE a valuable biocatalyst in organic synthesis of pharmaceutical compounds [57]. More recently, a novel solvent-stable lipase named OSTL28 was identified by metagenomic study of top soil from an oil field in China. Its high stability in organic solvents and glycerol make it potentially useful for non-aqueous reactions such as biodiesel production [58].
Thermophilic esterases-lipases have long been studied, and during recent years there has been growing attention on their underexploited cold-adapted counterparts. A PCR-based metagenomic study was carried out on glacier soil in order to assess the lipolytic diversity of HSL lipases. Phylogenetic analysis of the released sequences revealed that even at low temperatures there is a great diversity of lipase genes, most of them with little similarity to known lipases [59]. Arctic and Antarctic soils have also been subjected to prospecting for new lipases/esterases [60] and two studies yielded particularly interesting examples. In the first of these, two esterases from Arctic sediment, EstM-N1 and EstM-N2, with optimal temperatures of 20 and 30ºC respectively, showed high homology with β-lactamases and unusually high β-lactamase activity for esterases of family VIII [61]. In the second, an extreme alkaliphilic esterase, CHA2, was isolated from dry valley soil from Antarctica. Its active temperature range is 5-40ºC, with an optimum at 20ºC, and the pH optimum is 11. Such extreme alkaliphilic esterases are rare, and there has been only one other report of a cold-active extremely alkaliphilic esterase [62]. This lipase cannot be classified clearly by sequence analysis [63]. The same metagenomic library had previously yielded a cold-active esterase, CHA3, with higher optimal temperature, 40ºC, and without such an extreme alkaliphilic tendency (pH optimum = 9) [64].
More extreme temperature-adapted lipases/esterases have been revealed by soil metagenomics: a moderately thermostable and thermally activated (80% after 1 h at 50ºC) lipase from Atlantic Rain Forest Soil [65] and a cold-active esterase from a mountain soil that retains 60% of its maximal activity at 1ºC [19].
Apart from the discovery of lipases/esterases with potential use in industrial applications, recent functional metagenomics studies on soils have yielded important findings that broaden our knowledge about these enzymes: (1) The discovery of a new family of lipases/esterases through the identification of the novel lipase Lip018ORF16 in Atlantic Rain Forest Soil, with no sequence similarity to known lipases/esterases, only to sequences annotated as hypothetical proteins in GeneBank [66]; (2) The study of the first moderately thermostable true lipase derived from a member of the phylum Acidobacteria revealed that it was probably acquired by the native host through horizontal transfer [65].
3.4. Marine Environments
There is a great diversity of microbial marine ecosystems ranging from coastal environments and microbial communities associated with other marine organisms to hydrothermal vents. Microbial inhabitants of these ecosystems are exposed to extreme environmental conditions such as high salinity, high or low temperatures and high pressure. They and their enzymes are adapted to survive under such conditions, so they have the potential to be well suited for many industrial processes [67].
3.4.1. Intertidal Flats
Microorganisms of intertidal flat sediments are exposed to periodically changing physicochemical conditions: salinity, water temperature and flood tides. Therefore, microbial diversity in this habitat is expected to be distinct from other marine environments. In fact, three new families and subfamilies of lipases/esterases have recently been reported in this habitat in three independent metagenomic studies, all of them, curiously, using samples from different points on Korean coasts [5, 7, 68].
The first of these studies revealed a new bacterial lipase named LipG, which along with six other putative lipases constitutes a new family of bacterial lipolytic enzymes. This family seems to be related to fungal lipases since its members exhibit an Arg-Gly oxyanon hole sequence, a signature sequence characteristic of filamentous fungal lipases [5].
In the second study, a novel cold-adapted alkaline lipase, LipEH166, was identified as a member of a novel lipolytic enzyme family, which also includes three putative open reading frames. This enzyme displays cold-adapted activity with optimum temperature at 30ºC, but it retains 47% of maximal activity at 5ºC; and it is functional over a broad pH range (5-11), maintaining 80% activity [7].
The last study yielded three esterases with high salt tolerance and optimum activity in the mesophilic range, 40ºC, and slightly alkaline pH. These novel enzymes belong to family IV of lipases/esterases but have a distinctive pentapeptide motif GT(S)SA(G)G harboring the catalytic serine [68].
3.4.2. Seawater and Coastal Environments
Surface seawater is a highly saline and cationic habitat that has been prospected for lipolytic enzymes with interesting findings in each metagenomic study. Two esterases with different properties valuable in industrial applications have been isolated from the South China Sea: EstA, highly stable in high concentrations of divalent ions and NaCl, and Est B, highly stable in 30% methanol, ethanol, dimethylformamide, and dimethyl sulfoxide. EstA also represents a novel family of bacterial lipolytic enzymes, related to family III but with a different active site motif [6]. Using another metagenomic library, REBr, the first enzyme with esterase and haloacid-dehalogenase activities in the same active site, was discovered in an area contaminated with crude oil. Four homologues of this enzyme were cloned and expressed but only REBr exhibited this unique activity [69].
Marine sediments are reported to harbor a diverse microbial community with a potentially high diversity of metabolic enzymes [18]. Several lipolytic enzymes have been isolated from marine sediments in coastal environments: one lipase from the Baltic Sea [18] and two esterases from the Arctic seashore [70]. One of these esterases, EstAT11, preferentially hydrolyzed (S)-racemic oflozacin butyl ester with an enantiomeric excess value of 70.3%, showing great potential for the chiral resolution of heat-labile substrates.
Deep-sea hypersaline anoxic basins of the Eastern Mediterranean represent extreme and largely unexplored habitats >3500 m below sea level, with very stable brines entrapped in sea floor basins and sharply stratified from the overlying water column. They are characterized by extremely high salinity, high hydrostatic pressure, absence of light and anoxia [44]. Functional metagenomics on the brine:seawater interface of the hypersaline Urania anoxic basin released five esterases with different combinations of tolerance at high levels of salinity. Only one of these, Oil.16, showed remarkable properties related to its environmental origin: 180x enhanced activity at 2-4M NaCl, and remaining active at 40 MPa. Oil.16 also exhibits unusually enhanced activity in the presence of 70% ethanol or n-propanol and remarkably high enantioselectivity in hydrolysis and transesterification of compounds important in the pharmaceutical, cosmetic, and food industries. This novel esterase, with no similarity to known enzymes, exhibits particular features: it exists in different active forms, generated by reversible transitions in its tertiary and quaternary structures, and it holds three putative active serines embedded in their respective consensus motifs [44].
3.4.3. Deep-Sea Sediments
Deep-sea marine sediments represent the most unexplored marine habitat and are considered an extreme environment, where inhabitants are continuously exposed to extremes in pressure, salinity, temperature and nutrient availability.
Many cold-adapted lipolytic enzymes have been isolated from deep-sea sediments: the lipase EML1 from Edison Seamount (South West Pacific) [71], which belongs to the family of lipolytic enzymes previously discovered by Lee and collaborators [5] in a metagenomic library from tidal flat sediments; six lipolytic enzymes with low homology to known lipases/esterases, yielded by a larger metagenomic library constructed later from the same sample [72]; and the esterase EM2L8, extremely alkaliphilic, optimum pH 10-11 with activity five times higher than at pH 8 [62].
Large-scale screening of a metagenomic library from South China Sea deep sediments yielded 15 new lipolytic enzyme genes, two of them possibly belonging to new families, most of them active in a mesophilic/moderately thermophilic range (40-50ºC), not in a cold-adapted range as expected on the basis of their environmental origin [73]. However, one esterase isolated from this metagenomic library exhibited very low-temperature activity, even at 0ºC, with an active range of 0-60ºC and optimum temperature of 50ºC. The reported characteristics and the enhancement of activity by the organic solvents DMSO ( >10%) and methanol (>20%) made them attractive for organic synthesis. Besides, this esterase and six related putative esterases from genome projects and metagenomes constitute a new family of bacterial enzymes, closely related to family VI but with unique conserved sequence motifs (a pentapeptide motif and two C-terminal motifs) [9].
3.4.4. Organism Associated
Marine sponges are particular environments with reported high microbial diversity [74], and several metagenomic studies have been carried out yielding lipolytic enzymes with potential uses in industry. In Okinawa (Japan), a new esterase was isolated from a bacterial metagenome associated with Hystios erecta. This esterase, named EstHE1, belongs to the family GDSL. Only a few esterases are included in this family, and none have the moderate thermostability and high salt tolerance exhibited by EstHE1, properties of interest in the industrial field [45]. In Ireland, a halotolerant lipase with optimal activity at 5 M NaCl and 40ºC was discovered, displaying activity over a broad range of pH (3-12) and temperature (4ºC-60ºC), and stability in organic solvents and thermostability [74]. Lipolytic enzymes with antibacterial activity (lipases hydrolyze lipids and release fatty acids with known antibacterial properties) have even been found by functional metagenomics in sponge- and alga-associated microbial communities [75].
3.5. Other Environments
Freshwater environments have not received as much attention as sea ecosystems, but they also harbor distinctive microbial diversity. The esterase EstY was detected in a metagenomic library from the water surface of the Yangtze River in China. This enzyme showed maximal activity at 50ºC, which does not correlate with the temperature of the river, 20ºC. Phylogenetic analysis, unique conserved motifs and conserved domains suggest that EstY and its relative lipolytic enzymes constitute a new bacterial lipolytic enzyme family [8].
In activated sludge, a new esterase with unique features, named Lipo1, was isolated from a metagenomic library. Lipo1 showed an optimum temperature of 10ºC and resistance in the presence of detergents, which could be useful in organic chemistry and the laundry industry [76].
Carnivorous plants of the genus Nepenthes have developed a trapping organ called the pitcher, which possesses specific bacteria that secrete digestive enzymes. Two lipases with optimal activity at acidic pH were isolated from the pitcher fluid. Acidophilic lipases could be useful to the dairy and food industries and in the treatment of waste oil, among other applications [77].
CONCLUSIONS AND PERSPECTIVES
The studies surveyed in this review prove that functional metagenomics is a powerful strategy for discovering new lipolytic enzymes with unique combinations of biochemical features and potential use in industrial applications. Among all the enzymes cited, those from marine sources are of outstanding relevance. The fact that several new families of bacterial lipases/esterases have been discovered with only a few metagenomic studies reveals that despite the growing number of functional metagenomic studies, the unexplored diversity is still enormous.
Further optimization of metagenomic DNA extraction protocols, vectors and expression strains for constructing metagenomic libraries, along with the improvement of screening methods, will contribute to a better understanding of this unexplored diversity by allowing more efficient functional metagenomic studies to be conducted. There seem to be no limits to the knowledge that could be gained using this methodology.
ACKNOWLEDGEMENTS
The work of Olalla López was supported by a Maria Barbeito research contract from Xunta de Galicia. This research was supported by project grant 10MDS373027PR (Xunta de Galicia) and from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 324439. General support to the laboratory was funded by Xunta de Galicia (Consolidación D.O.G. 10-10-2012. CN: 2012/118) co-financed by FEDER. The English style of the manuscript was improved by Biomedes.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002;26(1):73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts molecular biology three-dimensional structures and biotechnological applications of lipases. Annu. Rev. Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 3.Gupta R, Gupta N, Rathi P. Bacterial lipases an overview of production purification and biochemical properties. Appl Microbiol Biotechnol. 2004;64(6):763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- 4.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes classification and properties. Biochem J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MH, Lee CH, Oh TK, Song JK, Yoon JH. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments evidence for a new family of bacterial lipases. Appl Environ Microbiol. 2006;72(11):7406–7409. doi: 10.1128/AEM.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu X, He H, Guo C, Sun B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol. 2008;80(4):615–625. doi: 10.1007/s00253-008-1566-3. [DOI] [PubMed] [Google Scholar]
- 7.Kim EY, Oh KH, Lee MH, Kang CH, Oh TK, Yoon JH. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol. 2009;75(1):257–260. doi: 10.1128/AEM.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, Sun B. Identification of novel esterase from metagenomic library of Yangtze river. J. Microbiol. Biotechnol. 2009;19(2):187–193. doi: 10.4014/jmb.0804.292. [DOI] [PubMed] [Google Scholar]
- 9.Fu C, Hu Y, Xie F, Guo H, Ashforth EJ, Polyak SW, Zhu B, Zhang L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic li-brary. Appl. Microbiol. Biotechnol. 2011;90(3):961–970. doi: 10.1007/s00253-010-3079-0. [DOI] [PubMed] [Google Scholar]
- 10.Verger R. Interfacial activation’ of lipases: facts and artifacts. Tibtech. 1997;15:32–38. [Google Scholar]
- 11.Jaeger KE, Eggert T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002;13(4):390–397. doi: 10.1016/s0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- 12.Haki GD, Rakshit SK. Developments in industrially important thermostable enzymes: a review. Bioresour. Technol. 2003;89(1):17–34. doi: 10.1016/s0960-8524(03)00033-6. [DOI] [PubMed] [Google Scholar]
- 13.Joseph B, Ramteke PW, Thomas G, Shrivastava N. Standard review cold-active microbial lipases: a versatile tool for industrial applications. Biotechnol. Mol. Biol. Rev. 2007;2(2):39–48. [Google Scholar]
- 14.Demirjian DC, Morís-Varas F, Cassidy CS. Enzymes from extremophiles. Curr. Opin. Chem. Biol. 2001;5(2):144–151. doi: 10.1016/s1367-5931(00)00183-6. [DOI] [PubMed] [Google Scholar]
- 15.Niehaus F, Bertoldo C, Kähler M, Antranikian G. Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 1999;51:711–729. doi: 10.1007/s002530051456. [DOI] [PubMed] [Google Scholar]
- 16.Cho AR, Yoo SK, Kim EJ. Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 2000;186(2):235–238. doi: 10.1111/j.1574-6968.2000.tb09110.x. [DOI] [PubMed] [Google Scholar]
- 17.Feller G. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life. Sci. 2003;60(4):648–662. doi: 10.1007/s00018-003-2155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hårdeman F, Sjöling S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007;59(2):524–534. doi: 10.1111/j.1574-6941.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- 19.Ko KC, Rim SO, Han Y, Shin BS, Kim GJ, Choi JH, Song JJ. Identification and characterization of a novel cold-adapted esterase from a metagenomic library of moun-tain soil. J. Ind. Microbiol. Biotechnol. 2012;39(5):681–689. doi: 10.1007/s10295-011-1080-y. [DOI] [PubMed] [Google Scholar]
- 20.Ghanem A, Aboul-Enein HY. Application of lipases in kinetic resolution of racemates. Chirality. 2005;17(1):1–15. doi: 10.1002/chir.20089. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Pan W. Ionic liquids: Green solvents for nonaqueous biocatalysis. Enzyme Microb. Technol. 2005;37:19–28. [Google Scholar]
- 22.van Rantwijk F, Sheldon RA. Biocatalysis in ionic liquids. Chem. Rev. 2007;107(6):2757–2785. doi: 10.1021/cr050946x. [DOI] [PubMed] [Google Scholar]
- 23.Oren A. Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 2010;31(8-9):825–834. doi: 10.1080/09593330903370026. [DOI] [PubMed] [Google Scholar]
- 24.DasSarma S, DasSarma P. Halophiles. eLS Reference available from: http://onlinelibrary.wiley.com/doi/10.1002/9780470015902. 2012.
- 25.Kourist R, Domínguez de María P, Bornscheuer UT. Enzymatic synthesis of optically active tertiary alcohols: expanding the biocatalysis toolbox. Chembiochem. 2008;9(4):491–498. doi: 10.1002/cbic.200700688. [DOI] [PubMed] [Google Scholar]
- 26.CBS Genome Atlas Database: http://www.bs.dtu.dk/services/GenomeAtlas-3.0/. Accesed September 5. 2012 [Google Scholar]
- 27.Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ. The genome sequence of the extreme thermophile. Thermus thermophilus. Nat. Biotechnol. 2004;22(5):547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- 28.López-López O, Fuciños P, Pastrana L, Rúa ML, Cerdán ME, González-Siso MI. Heterologous expression of an esterase from Thermus thermophilus HB27 in Saccharomyces cerevisiae. J. Biotechnol. 2010;145(3):226–232. doi: 10.1016/j.jbiotec.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5(10):R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 31.Streit WR, Schmitz RA. Metagenomics: the key to the uncultured microbes. Curr. Opin. Microbiol. 2004;7(5):492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 2011;77(4):1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Genomes OnLine Database (GOLD). http://www.genomesonline.org/cgi-bin/GOLD/index.cgi. Accessed September 5. 2012 [Google Scholar]
- 34.Bell PJ, Sunna A, Gibbs MD, Curach NC, Nevalainen H, Bergquist PL. Prospecting for novel lipase genes using PCR. Microbiology. 2002;148(Pt 8):2283–2291. doi: 10.1099/00221287-148-8-2283. [DOI] [PubMed] [Google Scholar]
- 35.Gabor EM, Alkema WBL, Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004;6(9):879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 36.Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microbiol. 2010;76(5):1633–1641. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lussier FX, Chambenoit O, Côté A, Hupé JF, Denis F, Juteau P, Beaudet R, Shareck F. Construction and functional screening of a metagenomic library using a T7 RNA polymerase-based expression cosmid vector. J. Ind. Microbiol. Biotechnol. 2011;38(9):1321–1328. doi: 10.1007/s10295-010-0915-2. [DOI] [PubMed] [Google Scholar]
- 38.Angelov A, Mientus M, Liebl S, Liebl W. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 2009;32(3):177–185. doi: 10.1016/j.syapm.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68(4):669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Wu H, Wang A, Du P, Pei X, Li H, Yin X, Huang L, Xiong X. Prospecting metagenomic enzyme subfamily genes for DNA family shuffling by a novel PCR-based approach. J. Biol. Chem. 2010;285(53):41509–41516. doi: 10.1074/jbc.M110.139659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence RC, Fryer TF, Reiter B. Rapid method for the quantitative estimation of microbial lipases. Nature (London): 1967;213:1264–1265. [Google Scholar]
- 42.Kouker G, Jaeger KE. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 1987;53(1):211–213. doi: 10.1128/aem.53.1.211-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SY, Shin HJ, Kim GJ. Screening and identification of a novel esterase EstPE from a metagenomic DNA library. J. Microbiol. 2011;49(1):7–14. doi: 10.1007/s12275-011-0201-7. [DOI] [PubMed] [Google Scholar]
- 44.Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Martins Dos Santos VA, Yakimov MM, Timmis KN, Golyshin PN. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 2005;12(8):895–904. doi: 10.1016/j.chembiol.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Okamura Y, Kimura T, Yokouchi H, Meneses-Osorio M, Katoh M, Matsunaga T, Takeyama H. Isolation and characterization of a GDSL esterase from the metagenome of a marine sponge-associated bacteria. Mar. Biotechnol. (NY) 2010;12(4):395–402. doi: 10.1007/s10126-009-9226-x. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Tang X, Ren G, Liu J, Yu H. A new high-throughput screening method for determining active and enantioselective hydrolases. Biochem. Eng. J. 2009;46(3):345–349. [Google Scholar]
- 47.Lämmle K, Zipper H, Breuer M, Hauer B, Buta C, Brunner H, Rupp S. Identification of novel enzymes with different hydrolytic activities by metagenome expression cloning. J. Biotechnol. 2007;127(4):575–592. doi: 10.1016/j.jbiotec.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 48.Meilleur C, Hupé JF, Juteau P, Shareck F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J. Ind. Microbiol. Biotechnol. 2009;36(6):853–861. doi: 10.1007/s10295-009-0562-7. [DOI] [PubMed] [Google Scholar]
- 49.Côté A, Shareck F. Expression and characterization of a novel heterologous moderately thermostable lipase derived from metagenomics in Streptomyces lividans. J. Ind. Microbiol. Biotechnol. 2010;37(9):883–891. doi: 10.1007/s10295-010-0735-4. [DOI] [PubMed] [Google Scholar]
- 50.Sharma PK, Singh K, Singh R, Capalash N, Ali A, Mohammad O, Kaur J. Characterization of a thermostable lipase showing loss of secondary structure at ambient temperature. Mol. Biol. Rep. 2012;39(3):2795–2804. doi: 10.1007/s11033-011-1038-1. [DOI] [PubMed] [Google Scholar]
- 51.Rhee JK, Ahn DG, Kim YG, Oh JW. New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl. Environ. Microbiol. 2005;71(2):817–825. doi: 10.1128/AEM.71.2.817-825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirawongsaroj P, Sriprang R, Harnpichamchai P, Thongaram T, Champreda V, Tanapongpipat S, Pootanakit K, Eurwilaichitr L. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 2008;133:42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 53.Sharma PK, Kumar R, Kumar R, Mohammad O, Singh R, Kaur J. Engineering of a metagenome derived lipase toward thermal tolerance: effect of asparagine to lysine mutation on the protein surface. Gene. 2012;491(2):264–271. doi: 10.1016/j.gene.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Kim YH, Kwon EJ, Kim SK, Jeong YS, Kim J, Yun HD, Kim H. Molecular cloning and characterization of a novel family VIII alkaline esterase from a compost metagenomic library. Biochem. Biophys. Res. Commun. 2010;393(1):45–49. doi: 10.1016/j.bbrc.2010.01.070. [DOI] [PubMed] [Google Scholar]
- 55.Kang CH, Oh KH, Lee MH, Oh TK, Kim BH, Yoon J. A novel family VII esterase with industrial potential from compost metagenomic library. Microb. Cell. Fact. 2011;10:41. doi: 10.1186/1475-2859-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei P, Bai L, Song W, Hao G. Characterization of two soil metagenome-derived lipases with high specificity for p-nitrophenyl palmitate. Arch. Microbiol. 2009;191:233–240. doi: 10.1007/s00203-008-0448-5. [DOI] [PubMed] [Google Scholar]
- 57.Elend C, Schmeisser C, Hoebenreich H, Steele HL, Streit WR. Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J. Biotechnol. 2007;130(4):370–377. doi: 10.1016/j.jbiotec.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 58.Fan X, Liu X, Wang K, Wang S, Huang R, Liu Y. Highly soluble expression and molecular characterization of an organic solvent-stable and thermotolerant lipase originating from the metagenome. J. Mol. Catal. B: Enzym. 2011;72(3-4):319–326. [Google Scholar]
- 59.Yuhong Z, Shi P, Liu W, Meng K, Bai Y, Wang G, Zhan Z, Yao B. Lipase diversity in glacier soil based on analysis of metagenomic DNA fragments and cell culture. J. Microbiol. Biotechnol. 2009;19(9):888–897. doi: 10.4014/jmb.0812.695. [DOI] [PubMed] [Google Scholar]
- 60.Berlemont R, Pipers D, Delsaute M, Angiono F, Feller G, Galleni M, Power P. Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev. Argent. Microbiol. 2011;43(2):94–103. doi: 10.1590/S0325-75412011000200005. [DOI] [PubMed] [Google Scholar]
- 61.Yu EY, Kwon MA, Lee M, Oh JY, Choi JE, Lee JY, Song BK, Hahm DH, Song JK. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl. Microbiol. Biotechnol. 2011;90(2):573–581. doi: 10.1007/s00253-011-3132-7. [DOI] [PubMed] [Google Scholar]
- 62.Park HJ, Jeon JH, Kang SG, Lee JH, Lee SA, Kim HK. Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Protein Expr. Purif. 2007;52(2):340–347. doi: 10.1016/j.pep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Hu XP, Heath C, Taylor MP, Tuffin M, Cowan D. A novel, extremely alkaliphilic and cold-active esterase from Antarctic desert soil. Extremophiles. 2012;16(1):79–86. doi: 10.1007/s00792-011-0407-y. [DOI] [PubMed] [Google Scholar]
- 64.Heath C, Hu XP, Cary SC, Cowan D. Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from Antarctic desert soil. Appl. Environ. Microbiol. 2009;75(13):4657–4659. doi: 10.1128/AEM.02597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faoro H, Glogauer A, Couto GH, de Souza EM, Rigo LU, Cruz LM, Monteiro RA, de Oliveira Pedrosa F. Characterization of a new Acidobacteria-derived moderately thermostable lipase from a Brazilian Atlantic Forest soil metagenome. FEMS Microbiol. Ecol. 2012;81(2):386–394. doi: 10.1111/j.1574-6941.2012.01361.x. [DOI] [PubMed] [Google Scholar]
- 66.Faoro H, Glogauer A, Souza EM, Rigo LU, Cruz LM, Monteiro RA, Pedrosa FO. Identification of a new lipase family in the Brazilian Atlantic Forest soil metagenome. Envi-ron. Microbiol. Rep. 2011;3(6):750–755. doi: 10.1111/j.1758-2229.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 67.Kennedy J, O'Leary ND, Kiran GS, Morrissey JP, O'Gara F, Selvin J, Dobson AD. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 2011;111(4):787–99. doi: 10.1111/j.1365-2672.2011.05106.x. [DOI] [PubMed] [Google Scholar]
- 68.Jeon JH, Lee HS, Kim JT, Kim SJ, Choi SH, Kang SG, Lee JH. Identification of a new subfamily of salt-tolerant esterases from a metagenomic library of tidal flat sediment. Appl. Microbiol. Biotechnol. 2012;93(2):623–631. doi: 10.1007/s00253-011-3433-x. [DOI] [PubMed] [Google Scholar]
- 69.Beloqui A, Polaina J, Vieites JM, Reyes-Duarte D, Torres R, Golyshina OV, Chernikova TN, Waliczek A, Aharoni A, Yakimov MM, Timmis KN, Golyshin PN, Ferrer M. Novel hybrid esterase-haloacid dehalogenase enzyme. Chembiochem. 2010;11(14):1975–1978. doi: 10.1002/cbic.201000258. [DOI] [PubMed] [Google Scholar]
- 70.Jeon JH, Kim JT, Kang SG, Lee JH, Kim SJ. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar. Biotechnol. (NY): 2009;11(3):307–316. doi: 10.1007/s10126-008-9145-2. [DOI] [PubMed] [Google Scholar]
- 71.Jeon JH, Kim JT, Kim YJ, Kim HK, Lee HS, Kang SG, Kim SJ, Lee JH. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009;81(5):865–874. doi: 10.1007/s00253-008-1656-2. [DOI] [PubMed] [Google Scholar]
- 72.Jeon JH, Kim JTM, Lee HS, Kim SJ, Kang SG, Choi SH, Lee JH. Novel lipolytic enzymes identified from metagenomic library of deep-sea sediment Evid Based Complement Alternat. Med 2011. Article ID 271419 9 pages. Reference available from: http://www.hindawi.com/journals/ecam/ 2011/271419. . 2011 doi: 10.1155/2011/271419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu. Y. Fu C, Huang Y, Yin Y, Cheng G, Lei F, Lu N, Li J, Ashforth EJ, Zhang L, Zhu B. Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiol. Ecol. 2010;72(2):228–237. doi: 10.1111/j.1574-6941.2010.00851.x. [DOI] [PubMed] [Google Scholar]
- 74.Selvin J, Kennedy J, Lejon DP, Kiran S, Dobson AD. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb. Cell. Fact. 2012;11(1):72. doi: 10.1186/1475-2859-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yung PY, Burke C, Lewis Novel antibacterial proteins from the microbial communities associated with the sponge Cymbastela concentrica and the green alga Ulva australis. Appl. Environ. Microbiol . 2011; 77(4):1512–1515. doi: 10.1128/AEM.02038-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roh C, Villatte F. Isolation of a low-temperature adapted lipolytic enzyme from uncultivated micro-organism. J. Appl. Microbiol. 2008;105(1):116–123. doi: 10.1111/j.1365-2672.2007.03717.x. [DOI] [PubMed] [Google Scholar]
- 77.Morohoshi T, Oikawa M, Sato S, Kikuchi N, Kato N, Ikeda T. Isolation and characterization of novel lipases from a metagenomic library of the microbial community in the pitcher fluid of the carnivorous plant Nepenthes hybrida. J. Biosci. Bioeng. 2011;112(4):315–320. doi: 10.1016/j.jbiosc.2011.06.010. [DOI] [PubMed] [Google Scholar]