Abstract
Viral load measurements may predict whether human papillomavirus (HPV) type 16 infections may become persistent and eventually lead to cervical lesions. Today, multiple PCR methods exist to estimate viral load. We tested three protocols to investigate viral load as a predictor of HPV clearance. We measured viral load in 418 HPV16-positive cervical smears from 224 women participating in the Ludwig–McGill Cohort Study by low-stringency PCR (LS-PCR) using consensus L1 primers targeting over 40 known HPV types, and quantitative real-time PCR (qRT-PCR) targeting the HPV16 E6 and L1 genes. HPV16 clearance was determined by MY09/11 and PGMY PCR testing on repeated smears collected over 5 years. Correlation between viral load measurements by qRT-PCR (E6 versus L1) was excellent (Spearman’s rank correlation, ρ = 0.88), but decreased for L1 qRT-PCR versus LS-PCR (ρ = 0.61). Viral load by LS-PCR was higher for HPV16 and related types independently of other concurrent HPV infections. Median duration of infection was longer for smears with high copy number by all three PCR protocols (log rank P<0.05). Viral load is inversely related to HPV16 clearance independently of concurrent HPV infections and PCR protocol.
Introduction
Cervical cancer is the second most frequent malignancy among women worldwide. Persistent human papillomavirus (HPV) infection with oncogenic HPV types is now well established as the biological intermediate for the development of cervical cancer (Ylitalo et al., 2000; Schlecht et al., 2001b; Wentzensen et al., 2009). Previous studies have demonstrated increased HPV viral load to be a surrogate indicator for persistence and a significant predictor of risk of squamous intraepithelial lesions (SIL) (Schlecht et al., 2001a, 2003; zur Hausen 2002; Gravitt et al., 2007). Moreover, HPV type 16 (HPV16) infections have been shown to last longer than other oncogenic HPV types (Trottier et al., 2008).
There are several PCR assays for measuring HPV viral load (Bavin et al., 1993; Farthing et al., 1994; Caballero et al., 1995; Jacobs et al., 1999; Ylitalo et al., 2000; Iftner & Villa, 2003). We previously published results using a (GP5/6) consensus low-stringency PCR (LS-PCR) method for HPV viral load quantification (Caballero et al., 1995) that amplifies over 40 known HPV types including HPV16 (Schlecht et al., 2003). Recent studies employing methods based on quantitative real-time PCR (qRT-PCR) that target HPV16 E6 and L1 genes, however, have shown mixed results with respect to the association between mean viral burden and grade of SIL of the cervix (Gravitt et al., 2007; Fontaine et al., 2005).
This study sought to conduct a direct comparison for viral load quantification by LS-PCR (Caballero et al., 1995) and qRT-PCR protocols for HPV16 targeting HPV16 E6 (Gravitt et al., 2003) and L1 genes, and the resulting associations with duration of cervical HPV infection in a cohort of Brazilian women. Cervical smears were collected at repeated intervals over 5 years from women enrolled in a large natural history study of HPV infection and cervical neoplasia.
Results
The present analysis includes 418 cervical specimens positive for HPV16 DNA from 224 women. We retested all cervical specimens originally positive by (MY09/11) PCR with quantitative LS-PCR and qRT-PCR protocols to measure viral burden.
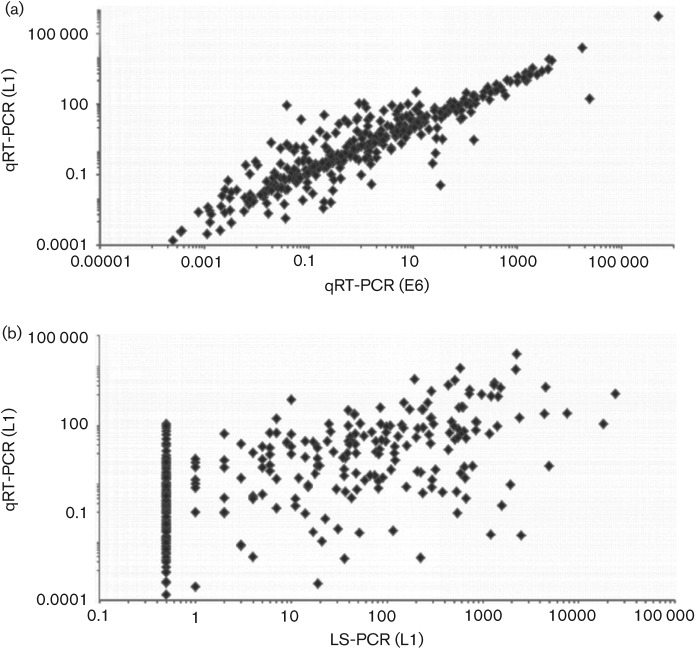
There was a strong correlation between viral load assessed by qRT-PCR targeting two different genes, E6 and L1, within the HPV genome (Spearman’s rank correlation, ρ = 0.88), although measurements based on L1 tended to be higher than those based on E6 (Wilcoxon signed-rank test, z = 5.224; P<0.0001), particularly in specimens with low copy number (Fig. 1a). In contrast, the correlation between viral load measurements by qRT-PCR and LS-PCR, with both targeting L1 was considerably less strong (ρ = 0.61; Fig. 1b). L1-based viral load was consistently higher with qRT-PCR than with LS-PCR, whether based on samples with HPV16 only (P<0.0001, n = 286), HPV16 plus other related (alpha 9) HPV types (P = 0.003, n = 35) and HPV16 plus other unrelated types (P = 0.0004, n = 97).
Fig. 1.
Correlation between HPV16 viral loads measured by two different protocols (axes show copies per cell on log10-transformed scale). (a) qRT-PCR targeting E6 (x-axis) and L1 genes (y-axis); n = 418; Spearman’s rank correlation ρ = 0.88; P = 0.0001. Thirty-one observations were excluded from this graph due to measurement values below the threshold of detection by either assay. (b) LS-PCR (x-axis) and qRT-PCR (y-axis) targeting L1 gene; n = 418; ρ = 0.61; P = 0.0001. Forty-eight observations were excluded from the graph due to measurement values below the threshold of detection by either assay. The vertical line is a result of all the values being below the detectable threshold (0.5 copies per cell).
In order to assess the relative contribution of known HPV types to viral burden measured by our consensus PCR method (LS-PCR), we estimated the mean change (in copies per cell) whenever a given HPV type was present in the specimen. Separate models were derived for all HPV-positive specimens (n = 3409) and for HPV16-positive specimens (n = 552), age and adjusting for all other concurrent types (Table 1). By grouping HPV types by phylogenetic relatedness, we found mean counts to be consistently higher whenever HPV16 and related types from alpha species 9 (i.e. HPV 31, 33, 35, 52, 58 and 67) were present. Higher relative viral burdens were also seen for oncogenic HPV types 18 and 56 and non-oncogenic types 6/11, 40, 54, 66, 72 and 81. Among HPV16-positive subjects, a relative increase in viral load was observed for specimens co-infected with related HPV types (HPV33, -35, -52 and -58), although this was only significant for HPV58.
Table 1. Mean change in viral load (copies per cell) measured by LS-PCR as a function of the presence of HPV types detected in the cervical specimen.
Change in mean viral load [with 95 % confidence intervals (CI) shown in parentheses] is presented based on loge-transformed data. Models were mutually adjusted for all concurrent HPV types detected (see Methods).
| Alpha papillomavirus species | Infection with HPV type | Mean change in viral load relative to | |||
| All HPV-positive specimens (n = 3409) | HPV16-positive specimens (n = 552) | ||||
| 9 | HPV16 | 2.7 | (2.0, 3.5) | – | (referent) |
| HPV31 | 3.1 | (2.1, 4.7) | −1.2 | (−5.9, 3.9) | |
| HPV33 | 3.2 | (1.8, 5.8) | 1.6 | (−3.7, 9.1) | |
| HPV35 | 7.5 | (4.7, 11.9) | 3.2 | (−1.4, 14.7) | |
| HPV52 | 1.9 | (1.3, 2.9) | 1.1 | (−10.0, 12.8) | |
| HPV58 | 3.7 | (2.5, 5.4) | 3.9 | (1.1, 13.6) | |
| 7 | HPV18 | 1.7 | (1.0, 2.6) | 1.6 | (−3.3, 7.8) |
| 3 | HPV72 | 2.7 | (1.1, 6.6) | na* | – |
| HPV81 | 4.4 | (2.3, 8.6) | 1.67 | (−213.4, 598.3) | |
| HPV83 | 1.4 | (−1.3, 2.5) | 43.5 | (5.1, 369.1) | |
| 5 | HPV51 | 1.1 | (−1.3, 1.5) | 4.8 | (1.5, 15.9) |
| 6 | HPV53 | −1.0 | (−1.4, 1.4) | 4.2 | (1.4, 12.3) |
| HPV56 | 4.2 | (2.5, 7.1) | 3.4 | (−2.6, 30.0) | |
| HPV66 | 2.2 | (1.3, 3.8) | 3.1 | (−1.6, 15.3) | |
| 8 | HPV40 | 7.2 | (3.6, 14.4) | −182.2 | (−158850, 4.8)† |
| <1?h -8p?>10 | HPV6/11 | 1.8 | (1.2, 2.8) | 1.7 | (−2.8, 7.8) |
| HPV44 | 1.4 | (−1.1, 2.2) | 16.8 | (1.9, 148.5) | |
| 13 | HPV54 | 1.8 | (1.1, 2.9) | −1.1 | (−5.8, 4.8) |
HPV67, -70, -32, -62, -72, -89, -57, -69 and -34 were dropped from the model due to collinearity.
Although adjusted for in the model, HPV types with viral count estimates that included the null or that fell beyond the predictable range for LS-PCR in both models are not shown. The complete information for all HPV types is shown in Table S1, available in JGV Online.
Kaplan–Meier analyses were used to assess time to clearance of incident and prevalent HPV16 infections by viral load (Fig. 2). Stratifying on the tertile cut-off in viral load, we found that women with a higher viral burden (third tertile) took significantly longer to clear their infections than women with a lower viral load (first tertile) for all three viral load assays. Corresponding tertile cut-off points were ≤0.50, 0.51–21 and >21 copies per cell for LS-PCR (Fig. 2a), <8, 8–825 and >825 for L1 qRT-PCR (Fig. 2b), and <7, 7–675 and >675 for E6 qRT-PCR (Fig. 2c).
Fig. 2.
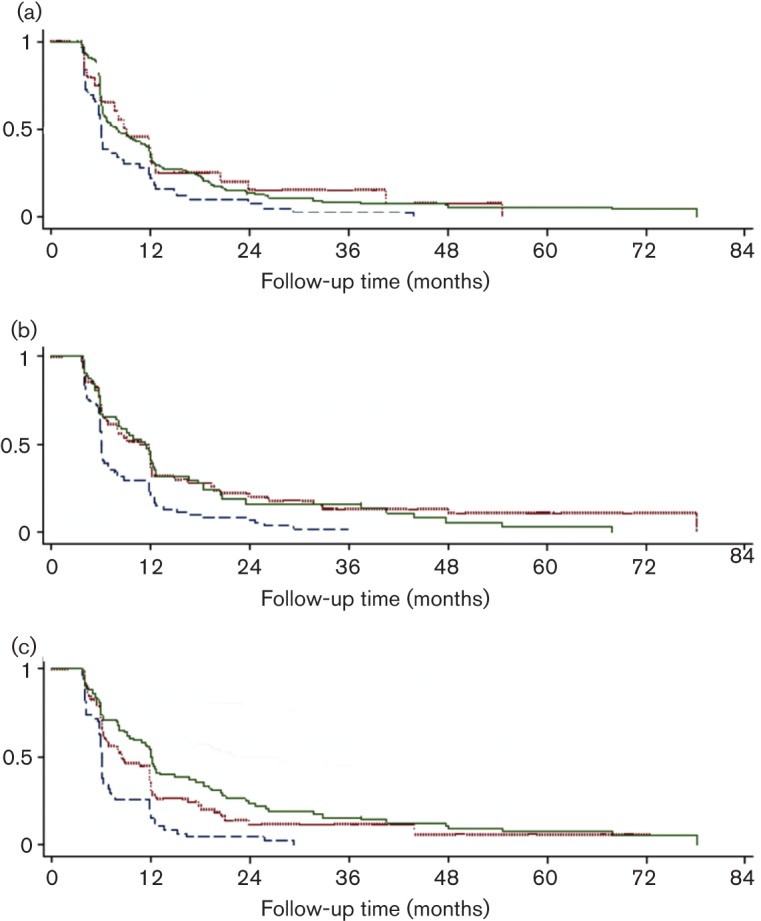
Clearance of HPV 16 positivity by viral load tertile (copies per cell) based on three different assays. (a) LS-PCR: first tertile ≤0.50; second tertile 0.51–21; and third tertile >21 (P = 0.0146) (P trend = 0.0083). (b) qRT-PCR L1: first tertile <8; second tertile 8–825; and third tertile >825 (P = 0.0009) (P trend = 0.0053). (c) qRT-PCR E6: first tertile <7; second tertile 7–675; and third tertile >675 (P = 0.0000) (P trend = 0.0000). The lower (first) tertile group is shown in blue (dashed), middle (second) tertile in red (dotted) and upper (third) tertile in green (solid).
To further investigate clearance rates by viral load in more absolute terms, we compared the clearance rates of HPV16 infections in specimens with multiple copies per cell to those with less than one copy (Table 2). Median durations of infection measured by all three protocols ranged from 8 to 12 months for infections with 1–100 and >100 copies per cell, while infections with <1 copy per cell cleared within 7 months. A monotonic increase in median duration of HPV DNA detection by viral level was only observed with the two qRT-PCR assays.
Table 2. Mean and median duration and clearance rate of HPV16 infection according to viral load in copies per cell measured by different assays.
| Assay | Viral load category (n) | Clearance rate (per 1000 women-months) (95 % CI) | Duration in months | |
| Mean (95 %CI) | Median (95 % CI) | |||
| qRT-PCR E6 | <1 (n = 97) | 102.2 (74.0–142.2) | 9.4 (6.9–12.0) | 6.0 (5.7–7.7) |
| 1–100 (n = 91) | 58.3 (45.4–76.1) | 17.1 (12.0–22.2) | 8.4 (6.2–11.9) | |
| >100 (n = 17) | 61.5 (49.2–76.2) | 16.3 (12.5–20.0) | 11.3 (8.1–12.2) | |
| qRT-PCR L1 | <1 (n = 108) | 104.1 (78.3–138.1) | 9.43 (7.3–11.3) | 6.1 (5.7–7.9) |
| 1–100 (n = 84) | 55.2 (40.4–75.4) | 18.8 (11.9–25.7) | 8.4 (5.9–11.9) | |
| >100 (n = 13) | 58.1 (47.1–72.5) | 16.7 (13.2–20.1) | 11.8 (8.2–12.2) | |
| LS-PCR L1 | <1 (n = 88) | 78.3 (64.4–93.0) | 13.1 (12.3–15.9) | 6.7 (6.1–8.7) |
| 1–100 (n = 41) | 78.4 (61.4–99.4) | 12.7 (9.7–15.7) | 8.2 (6.1–9.9) | |
| >100 (n = 91) | 60.5 (44.2–81.6) | 16.5 (10.9–22.2) | 7.4 (6.1–13.1) | |
Discussion
A number of studies have shown viral load measurements for oncogenic HPV types (including HPV16) in cervical specimens to be a suitable indicator of persistent infection and risk of SIL incidence (Schlecht et al., 2001b, 2003; zur Hausen, 2002). However, introduction of methods based on qRT-PCR that target HPV16 E6 and L1 genes have shown a discrepancy in results for viral burden and grade of SIL (Fontaine et al., 2005; Gravitt et al., 2007). To address this, we tested three established PCR-based protocols to investigate whether viral load is a predictor of HPV16 clearance.
In this study, we found women with high viral burden took significantly longer to clear their infections than women with low viral load for all three assays we used. Although infections with <1 copy per cell cleared within 7 months, the median duration of more productive infections ranged from 8 to 12 months. This was in line with our previously published data that showed the mean duration of single HPV16 infection to be about 11 months (Trottier et al., 2008). These findings are also consistent with results from a large population-based cervical cancer screening study in The Netherlands (Bulkmans et al., 2007).
We previously observed that viral load measured by LS-PCR increased with cytologic grade (from normal to low-grade SIL with koilocytosis) but then decreases in high-grade SIL (HSIL) (Schlecht et al., 2001a). Consistent findings were observed by Gravitt et al. (2003) analysing HPV18 in a population-based study conducted in Guanacaste, Costa Rica.
While our study is one of the largest to evaluate viral load in cervical specimens by multiple methods (Bavin et al., 1993; Farthing et al., 1994; Jacobs et al., 1999; Ylitalo et al., 2000; Wentzensen et al., 2012) we recognize that its design has some limitations.
Misclassification in HPV detection is one such limitation since our results were based, in part, on MY09/11 PCR. We opted for selective retesting by the more sensitive PGMY protocol where possible to reduce the potential for false negatives.
Although the LS-PCR is labour intensive, both protocols (LS-PCR and qRT-PCR), allow for direct measurement of viral DNA against the quantity of host DNA permitting the calculation of true viral load in terms of copies per cell, thus eliminating the confounding effect of variation in cell content among specimens. However, it is possible that abnormal cells found in high-grade lesions could have been sampled more readily than normal cells as these express fewer intercellular adhesion molecules than normal cells (Klein et al., 1994). Given the higher likelihood for HPV16 and other oncogenic types to develop into HSIL, our viral load association therefore may reflect underlying cytologic changes in the sample. We attempted to reduce this by excluding all prevalent cytologically high-grade specimens.
Quantification (in copies per cell) for our LS-PCR results was performed using standards prepared with pre-selected amounts of a reference plasmid containing HPV16. While this allowed for more stable viral load measurements and comparison with other HPV16-based assays, it may also explain in part why we observed higher viral counts for specimens infected with related types from the alpha 9 species (HPV33, -35 and -58), either alone or concurrently with HPV16. However, this was not specific only to alpha 9 types as higher viral levels were also observed for unrelated types (HPV6/11, -44, -18, -51, -56, -66 and -81). Despite the non-specific nature of the LS-PCR assay (which is based on broad detection of all mucosotropic HPV types), the relative associations between HPV16 viral load and clearance were consistent irrespective of assay platform. Nonetheless, qRT-PCR estimates appear to be more precise and show a stronger dose-dependent association with clearance, particularly with the HPV16 E6 gene.
The choice of region used for PCR has implications for HPV screening strategies in the clinical diagnosis and management of cervical cancer (Morris, 2005). The advantage of targeting the L1 region is that this has the potential to detect several HPV types. On the other hand, PCR primers that target the L1 region can be unreliable and may miss more advanced disease when compared with those directed at the E6 region, which encode an oncogenic product. The L1 region has the disadvantage of being lost during integration of viral DNA into host genomic DNA, a process that can represent an integral component of progression from infection to tumorigenesis. It is rare for this to happen with the E6 region. Furthermore, the E6 nucleotide sequence exhibits less nucleotide variation (Morris, 2005).
The difference between both reactions (L1- and E6-based qRT-PCR) can be statistically demonstrated. The slope of the standard curve generated in a qRT-PCR can be translated into an efficiency value by the formula: E = 10 (−1/slope) − 1. According to the equipment manufacturer, the PCR efficiency should be between 90 and 100 % (−3.6 ≥ slope ≥−3.3). If it is 100 %, the slope is −3.32. If the slope is below −3.6, then the PCR has poor efficiency. This can partially explain the discrepancies observed for viral load measured by L1- and E6-based qRT-PCR. In general, the slope of the β-globin reaction and the E6-based qRT-PCR assay ranged from −3.6 to −3.2. The L1-based qRT-PCR was shown to be an unreliable protocol due to a greater slope variation between different tests. Sometimes the L1-based assay was more efficient than the β-globin assay. Therefore, we have a clear example showing that even small differences in efficiency can lead to a dramatic effect on the final yield after several cycles (Morris, 2005).
We recognize that other methods for viral copy quantification exist that may have produced different results from those tested in this study. Such methods include assays based on end-point PCR (Bavin et al., 1993), quantitative competitive PCR (Jacobs et al., 1999) and hybrid capture (HC) (Farthing et al., 1994). Compared with qRT-PCR, these methods are laborious and require controlled sample conditions (Gravitt et al., 2003). Some, like HC, a widely used method for clinical detection of oncogenic HPV types, do not provide direct quantitative measures. We therefore opted to test the most common qRT-PCR method currently used in epidemiological studies to compare it with our previously demonstrated method based on LS-PCR.
Testing under a wide range of conditions has shown that the PCR-based techniques used to measure viral load in this study were in general reproducible and adaptable to large-scale testing in epidemiological studies. The viral load estimates by PCR methods, especially the E6-based qRT-PCR may be used as an important tool to identify women at increased risk of developing persistent infections and cervical lesions, particularly due to HPV16.
Methods
Study sample.
There are 2462 women enrolled in the Ludwig–McGill Cohort Study and half of them were diagnosed with HPV infection at least once during the follow-up. Out of 575 HPV16-positive samples from 284 women, 418 cervical specimens were assayed for qRT-PCR viral load (224 women) and all of the samples were also assayed for LS-PCR viral load. Subjects were followed up over an average of 5 years at scheduled returns every 4 months in the first year and once every 6 months thereafter. Subjects completed an interviewer-administered structured questionnaire and provided cervical smears for HPV DNA testing and Pap cytology at each visit. Additional details about the study design and methods of the Ludwig–McGill Cohort Study have been described elsewhere (Franco et al., 1999). The ethical review boards of all participating institutions approved this investigation.
Cervical cell specimens.
An Accelon biosampler (Medscand) was used to collect a sample of ectocervical and endocervical cells at each visit. After preparation of the Pap smear, the sampler with remaining exfoliated cells was immersed in Tris/EDTA buffer at pH 7.4, stored at 4 °C at the clinic for at most 5 days, and then frozen until testing. Cervical smears were blindly graded for HPV results using the Bethesda system.
HPV detection and typing.
Cervical specimen DNA was extracted and purified following standard techniques. In brief, samples collected prior to 1999 were digested with 100 µg proteinase K ml−1 for 3–18 h at 55 °C and the DNA purified by spin-column chromatography. After 1999, DNA isolation was performed using a GFX Genomic Blood DNA Purification kit (Amersham Pharmacia Biotech). Specimens were tested for the presence of HPV DNA by a previously described PCR protocol amplifying a highly conserved 450 bp segment of the L1 viral gene flanked by MY09/11 or PGMY09/11 primers (Manos et al., 1989; Gravitt et al., 2000). Typing of the amplified products was performed by hybridization with individual oligonucleotide probes specific for 27 HPV genital types, including HPV16 (Bauer et al., 1991; Hildesheim et al., 1994). Amplified products hybridizing to the generic probe but not to any of the type-specific probes were further tested by RFLP analysis (Bernard et al., 1994) extending the range of identifiable HPV to over 40 genital types. Testing for host DNA was performed using GH20 and PCO4 primers, which amplify a 268 bp region of the human β-globin gene (Saiki et al., 1988).
Viral load by LS-PCR.
Briefly, a consensus primer pair targeting the L1 gene regions of several HPV types (van den Brule et al., 1990) was employed under low-stringency conditions so that, in addition to the specific HPV DNA product, a series of host DNA fragments were also amplified. We selected a 192 bp DNA product homologous to a small region of the human chromosome X to serve as internal control for the reaction (Caballero et al., 1995). The ratio of the (140 bp) HPV band to that of the host genome internal band was then measured by densitometry and quantified (in copies per cell) by linear interpolation using a standard curve (Trottier et al., 2008). This standard was prepared with pre-selected amounts of a reference plasmid containing the entire HPV16 genome (kindly provided by E. M. de Villiers, Deutsches Krebsforschungszentrum, Heidelberg, Germany) against a constant background of human DNA extracted from human breast tissue (van den Brule et al., 1990; Caballero et al., 1995). All samples, controls and standards were analysed in duplicate and viral load derived from the mean values.
Viral load by qRT-PCR.
We quantified the HPV DNA in cervical specimens by real-time PCR using a 5′-exonuclease fluorescence (TaqMan) assay ABI Prism 7700 Sequence Detector System (Applied Biosystems) with primers and hybridization probes specific to the HPV16 L1 (Bryan et al., 2006; Iftner et al., 2009) and E6 genes and the human β-globin gene (van den Brule et al., 1990). Reactions for HPV16 L1 and human β-globin were performed using the 2× Universal Master Mix (Applied Biosystems) with 0.06 µM each primer and 0.09 µM probe for human β-globin and with 0.1 µM each primer and 0.05 µM probe for HPV16 L1 in a final volume of 50 µl. The thermal cycler conditions for the HPV16 L1 reaction were 50 °C/2 min and 95 °C/10 min, followed by 55 cycles of 94 °C/15 s and 60 °C/1 min. We used similar thermal conditions and fewer cycles for the globin reaction: 45 cycles of 94 °C/15 s and 60 °C/1 min. The thermal cycler conditions for the HPV16 E6 are described elsewhere (Gravitt et al., 2003). The sequence of primers and probes and respective protocols for L1 and human β-globin genes were kindly provided by F. Taddeo (Merck). Equal input amounts (2 µl) of each DNA control and amplified sample were added to 2 µl of MilliQ water corresponding to the DNA standards volume below. Viral load quantification (in copies µl−1) was achieved by comparing the threshold cycle (Ct) number for the cervical sample with the Ct numbers for samples with known starting viral copy numbers. Transformation into copies per cell was then obtained by linear interpolation using a standard curve constructed with the results of the DNA mixtures containing known amounts of HPV DNA.
DNA standards were prepared with pre-selected amounts of a reference plasmid containing the entire HPV16 genome (105 copies µl−1 for HPV16 L1 and E6 reactions) added to a constant background of Escherichia coli strain B DNA (Sigma-Aldrich) (2.5 µg ml−1) to final concentrations of 20 000 and 6000 copies of HPV16 µl−1. Solutions were then serially diluted tenfold to prepare standards containing 20 000, 6000, 2000, 600, 200, 60, 20, six and two copies HPV16 µl−1. Standards for the globin reaction were prepared using the same strategy above with pre-selected amounts of a reference plasmid, kindly provided by F. Taddeo (Merck), containing the 254 bp of the human β-globin gene (106 copies µl−1) to generate standards containing 250 000, 25 000, 2500, 250, 25 and 2.5 copies µl−1. The input amount of each standard amplified in all experiments was 4 µl. The conversion from copies per microlitre to copies per cell was done by calculating the ratio of HPV copy number to half the globin gene copy number in each sample.
Cervical carcinoma cell lines.
DNA extracted from HPV16-positive cervical carcinoma cell lines, SiHa (one to two copies per cell) and CaSki (400–600 copies per cell) (Yee et al., 1985; Mincheva et al., 1987) were used as controls to assess the precision and reproducibility of the standard curves to estimate HPV viral load in cervical smears. Geometric means were estimated for each control sample. DNA extracted from the human cervical carcinoma cell line C33A was used as a negative control for viral load determination (Yee et al., 1985). DNA extraction was performed as described elsewhere (Wright & Manos 1990) with minor modifications.
Mean viral load based on the E6 was 2.95 copies per cell (95 % CI: 2.94–2.96) in SiHa cells (expected: one to two copies) and 395 copies per cell (95 % CI: 350–447) in CaSki (expected: 400–600 copies), whereas the L1 load in SiHa and CaSki was 15 (95 % CI: 12–19) and 7318 (95 % CI: 5801–9235) copies per cell, respectively.
Statistical analyses.
To assess the correlation between viral load protocols, we computed Spearman’s rank (ρ) correlation coefficients between pairs of viral load data measured at HPV16-positive visits using log-transformed viral counts. Comparison of viral load measurements between assays on the same specimen was done by Wilcoxon signed-rank test. To estimate the contribution of co-infections with additional HPV types, we modelled the mean change in viral load measured using LS-PCR, first using all HPV-positive specimens and then restricting the analysis to specimens that were HPV16 positive, either alone or concurrently with other HPV types. We used multivariate, random-effects, generalized least-squares regression for repeated data. Observations were restricted to cases with no evidence of high-grade disease to avoid potential confounding effects. Kaplan–Meier analyses and log-rank tests were used to evaluate HPV16 clearance rates by viral load level, defined by tertile-based and predetermined cut-offs (<1, 1–100 and 100+ copies per cell), at the first HPV16 positivity for each protocol. Duration of infection was determined from date of first incident or prevalent (at baseline) HPV16 DNA detection episode to date of first negative result by PCR (i.e. clearance).
Acknowledgements
Collaborators in the Ludwig–McGill Study Group include: Alex Ferenczy, Helen Trottier, Salaheddin Mahmud (McGill University, Montreal, Canada), Thomas Rohan (Albert Einstein College of Medicine, Bronx, USA), Otávia L. S. D. Caballero, João Candeias, Paula Rahal, Paulo C. Maciag, Patricia Thomann, Laura S. Gonzales, Patricia Savio, Maria Luiza Baggio, Lenice Galan, Maria C. Costa, Romulo Myamura, Silvaneide Ferreira, Lara Termini, João S. Sobrinho, José C. M. Prado, Maria Antonieta Andreoli (Ludwig Institute for Cancer Research, São Paulo, Brazil). This work was supported by Ludwig Institute for Cancer Research; the US National Cancer Institute (CA70269); the Canadian Institutes of Health Research (MOP-49396 and CRN-83320); and Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (99/10790-0 to A. T.). Luisa Lina Villa has been a consultant of Merck, Sharp & Dohme for the quadrivalent HPV vaccine; of Qiagen and Roche Molecular Diagnostics for HPV DNA tests. Eduardo L. Franco has served as a consultant to Merck and GSK, on HPV vaccine matters, to BD, Roche and Gen-Probe, on issues related to HPV diagnostics. Nicolas F. Schlecht has served in the past as an advisory board member for Merck and GSK on HPV vaccine matters. The other authors disclose no conflict of interest.
Footnotes
One supplementary table is available with the online version of this paper.
References
- Bauer H. M., Ting Y., Greer C. E., Chambers J. C., Tashiro C. J., Chimera J., Reingold A., Manos M. M. (1991). Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA 265, 472–477 10.1001/jama.1991.03460040048027 [DOI] [PubMed] [Google Scholar]
- Bavin P. J., Giles J. A., Deery A., Crow J., Griffiths P. D., Emery V. C., Walker P. G. (1993). Use of semi-quantitative PCR for human papillomavirus DNA type 16 to identify women with high grade cervical disease in a population presenting with a mildly dyskaryotic smear report. Br J Cancer 67, 602–605 10.1038/bjc.1993.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H. U., Chan S. Y., Manos M. M., Ong C. K., Villa L. L., Delius H., Peyton C. L., Bauer H. M., Wheeler C. M. (1994). Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis 170, 1077–1085 10.1093/infdis/170.5.1077 [DOI] [PubMed] [Google Scholar]
- Bryan J. T., Taddeo F., Skulsky D., Jansen K. U., Frain B. M., Qadadri B., Brown D. R. (2006). Detection of specific human papillomavirus types in paraffin-embedded sections of cervical carcinomas. J Med Virol 78, 117–124 10.1002/jmv.20512 [DOI] [PubMed] [Google Scholar]
- Bulkmans N. W., Berkhof J., Bulk S., Bleeker M. C., van Kemenade F. J., Rozendaal L., Snijders P. J., Meijer C. J., POBASCAM Study Group (2007). High-risk HPV type-specific clearance rates in cervical screening. Br J Cancer 96, 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero O. L., Villa L. L., Simpson A. J. G. (1995). Low stringency-PCR (LS-PCR) allows entirely internally standardized DNA quantitation. Nucleic Acids Res 23, 192–193 10.1093/nar/23.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing A., Masterson P., Mason W. P., Vousden K. H. (1994). Human papillomavirus detection by hybrid capture and its possible clinical use. J Clin Pathol 47, 649–652 10.1136/jcp.47.7.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine J., Gravitt P., Duh L. M., Lefevre J., Pourreaux K., Hankins C., Coutlée F. (2005). High level of correlation of human papillomavirus-16 DNA viral load estimates generated by three real-time PCR assays applied on genital specimens. Cancer Epidemiol Biomarkers Prev 14, 2200–2207 10.1158/1055-9965.EPI-05-0055 [DOI] [PubMed] [Google Scholar]
- Franco E., Villa L., Rohan T., Ferenczy A., Petzl-Erler M., Matlashewski G., Ludwig-McGill Study Group (1999). Design and methods of the Ludwig-McGill longitudinal study of the natural history of human papillomavirus infection and cervical neoplasia in Brazil. Rev Panam Salud Publica 6, 223–233 10.1590/S1020-49891999000900001 [DOI] [PubMed] [Google Scholar]
- Gravitt P. E., Peyton C. L., Alessi T. Q., Wheeler C. M., Coutlée F., Hildesheim A., Schiffman M. H., Scott D. R., Apple R. J. (2000). Improved amplification of genital human papillomaviruses. J Clin Microbiol 38, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravitt P. E., Burk R. D., Lorincz A., Herrero R., Hildesheim A., Sherman M. E., Bratti M. C., Rodriguez A. C., Helzlsouer K. J., Schiffman M. (2003). A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol Biomarkers Prev 12, 477–484 [PubMed] [Google Scholar]
- Gravitt P. E., Kovacic M. B., Herrero R., Schiffman M., Bratti C., Hildesheim A., Morales J., Alfaro M., Sherman M. E. & other authors (2007). High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer 121, 2787–2793 10.1002/ijc.23012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildesheim A., Schiffman M. H., Gravitt P. E., Glass A. G., Greer C. E., Zhang T., Scott D. R., Rush B. B., Lawler P. & other authors (1994). Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis 169, 235–240 10.1093/infdis/169.2.235 [DOI] [PubMed] [Google Scholar]
- Iftner T., Villa L. L. (2003). Chapter 12: Human papillomavirus technologies. J Natl Cancer Inst Monogr 31, 80–88 10.1093/oxfordjournals.jncimonographs.a003487 [DOI] [PubMed] [Google Scholar]
- Iftner T., Germ L., Swoyer R., Kjaer S. K., Breugelmans J. G., Munk C., Stubenrauch F., Antonello J., Bryan J. T., Taddeo F. J. (2009). Study comparing human papillomavirus (HPV) real-time multiplex PCR and Hybrid Capture II INNO-LiPA v2 HPV genotyping PCR assays. J Clin Microbiol 47, 2106–2113 10.1128/JCM.01907-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. V., Walboomers J. M., van Beek J., Voorhorst F. J., Verheijen R. H., Meijer C. J., van den Brule A. J., Helmerhorst T. J., Snijders P. J. (1999). A quantitative polymerase chain reaction-enzyme immunoassay for accurate measurements of human papillomavirus type 16 DNA levels in cervical scrapings. Br J Cancer 81, 114–121 10.1038/sj.bjc.6690659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. S., Ho G. Y., Vermund S. H., Fleming I., Burk R. D. (1994). Risk factors for squamous intraepithelial lesions on Pap smear in women at risk for human immunodeficiency virus infection. J Infect Dis 170, 1404–1409 10.1093/infdis/170.6.1404 [DOI] [PubMed] [Google Scholar]
- Manos M. M., Ting Y., Wrigth D. K., Lewis A. J., Broker T. R., Wolinsky S. M. (1989). The use of polymerase chain reaction amplification for the detection of genital human papillomaviroses. In Molecular Diagnostic of Human Cancer, Cancer Cells, pp. 209–214 Edited by Furth M., Greaves M. New York, NY: Cold Spring Harbor Press [Google Scholar]
- Mincheva A., Gissmann L., zur Hausen H. (1987). Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med Microbiol Immunol (Berl) 176, 245–256 10.1007/BF00190531 [DOI] [PubMed] [Google Scholar]
- Morris B. J. (2005). Cervical human papillomavirus screening by PCR: advantages of targeting the E6/E7 region. Clin Chem Lab Med 43, 1171–1177 10.1515/CCLM.2005.203 [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491 10.1126/science.2448875 [DOI] [PubMed] [Google Scholar]
- Schlecht, N., Trevisan, A., Rohan, T., Ferenczy, A., Villa, L. L. & Franco, E. L. (2001a).Viral load as a predictor of lesion grade severity in cervical intraepithelial neoplasia. In Program and Oral Presentations for 19th International Papillomavirus Conference, Florianópolis, Santa Catarina, Brazil, p. 138, O-93. [Google Scholar]
- Schlecht N. F., Kulaga S., Robitaille J., Ferreira S., Santos M., Miyamura R. A., Duarte-Franco E., Rohan T. E., Ferenczy A. & other authors (2001b). Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. JAMA 286, 3106–3114 10.1001/jama.286.24.3106 [DOI] [PubMed] [Google Scholar]
- Schlecht N. F., Trevisan A., Duarte-Franco E., Rohan T. E., Ferenczy A., Villa L. L., Franco E. L. (2003). Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int J Cancer 103, 519–524 10.1002/ijc.10846 [DOI] [PubMed] [Google Scholar]
- Trottier H., Mahmud S., Prado J. C., Sobrinho J. S., Costa M. C., Rohan T. E., Villa L. L., Franco E. L. (2008). Type-specific duration of human papillomavirus infection: implications for human papillomavirus screening and vaccination. J Infect Dis 197, 1436–1447 10.1086/587698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule A. J. C., Snijders P. J. F., Gordijn R. L., Bleker O. P., Meijer C. J. L. M., Walboomers J. M. M. (1990). General primer-mediated polymerase chain reaction permits the detection of sequenced and still unsequenced human papillomavirus genotypes in cervical scrapes and carcinomas. Int J Cancer 45, 644–649 10.1002/ijc.2910450412 [DOI] [PubMed] [Google Scholar]
- Wentzensen N., Schiffman M., Dunn T., Zuna R. E., Gold M. A., Allen R. A., Zhang R., Sherman M. E., Wacholder S. & other authors (2009). Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer 125, 2151–2158 10.1002/ijc.24528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzensen N., Gravitt P. E., Long R., Schiffman M., Dunn S. T., Carreon J. D., Allen R. A., Gunja M., Zuna R. E. & other authors (2012). Human papillomavirus load measured by Linear Array correlates with quantitative PCR in cervical cytology specimens. J Clin Microbiol 50, 1564–1570 10.1128/JCM.06240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. K., Manos M. M. (1990). Sample preparation from paraffin-embedded tissues. In PCR Protocols: a Guide to Methods and Applications, pp. 153–158 Edited by Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. San Diego: Academic Press [Google Scholar]
- Yee C., Krishnan-Hewlett I., Baker C. C., Schlegel R., Howley P. M. (1985). Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol 119, 361–366 [PMC free article] [PubMed] [Google Scholar]
- Ylitalo N., Sørensen P., Josefsson A. M., Magnusson P. K., Andersen P. K., Pontén J., Adami H. O., Gyllensten U. B., Melbye M. (2000). Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet 355, 2194–2198 10.1016/S0140-6736(00)02402-8 [DOI] [PubMed] [Google Scholar]
- zur Hausen H. (2002). Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2, 342–350 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]