Abstract
Members of the family Iridoviridae are animal viruses that infect only invertebrates and poikilothermic vertebrates. Invertebrate iridescent virus 22 (IIV-22) was originally isolated from the larva of a blackfly (Simulium sp., order Diptera) found in the Ystwyth river, near Aberystwyth, Wales, UK. IIV-22 virions are icosahedral, with a diameter of about 130 nm and contain a dsDNA genome that is 197.7 kb in length, has a G+C content of 28.05 mol% and contains 167 coding sequences. Here, we describe the complete genome sequence of this virus and its annotation. This is the fourth genome sequence of an invertebrate iridovirus to be reported.
The family Iridoviridae consists of large dsDNA viruses that infect species of both poikilothermic vertebrates (fishes, amphibians and reptiles) and invertebrates (arachnids, cephalopods, crustaceans, insects, molluscs, nematodes and polychaetes; Williams, 2008). Iridoviruses are members of the nucleocytoplasmic large DNA viruses (NLCDVs; Iyer et al., 2001). The dsDNA genomes of iridoviruses are circularly permuted with terminal redundancy; the map of their genomes is represented as a circular molecule. Only one linear molecule is encapsidated in each virion, with the ends of individual encapsidated genomes being located at different positions on the map of different virions (Bigot et al., 2000). The genome of vertebrate iridoviruses is highly methylated, whereas little to no methylation occurs in the genomes of the invertebrate iridoviruses. Replication of the iridoviral genome includes distinct nuclear and cytoplasmic phases (Jancovich et al., 2011). The genomes are encapsidated within an icosahedral shell ranging between 120 and 180 nm in diameter. Capsids comprise predominantly a 50 kDa major capsid protein (MCP). The invertebrate iridoviruses studied by cryo-electron microscopy have 2 nm diameter surface fibrils (Yan et al., 2000).
The family Iridoviridae is currently organized into five genera: Chloriridovirus, Iridovirus, Lymphocystivirus, Megalocytivirus and Ranavirus. Members of the first two genera have a host range restricted to invertebrate species, whereas members of the three others infect only poikilothermic vertebrates. The type species for the genus Chloriridovirus is Invertebrate iridescent virus 3 (Delhon et al., 2006; Jancovich et al., 2011), the only species reported in this genus. The type species for the genus Iridovirus is Invertebrate iridescent virus 6. Invertebrate iridescent virus 1 is also recognized by the International Committee for Virus Taxonomy (ICTV) as a species in this genus. Ten other related viruses that may be members of the genus Iridovirus await biological and genomic data to determine whether they are valid species of this genus or variants of existing species. Interestingly, phylogenetic analyses of proteins encoded by invertebrate iridescent virus 3 (IIV-3), IIV-6 (Jakob et al., 2001) and IIV-9 (Wong et al., 2011) have revealed that IIV-9, one of the proposed members of the genus Iridovirus, is more closely related to IIV-3 than to IIV-6. This indicates that some species of the genus Iridovirus may be closer to members of the genus Chloriridovirus than to their Iridovirus relatives. Hence, it is possible that the genus Iridovirus contains several diverse species or species complexes. A division of the genus Iridovirus into three species complexes [Polyiridovirus (proposed type species Invertebrate iridescent virus 9), Oligoiridovirus (type species Invertebrate iridescent virus 6) and Crustaceoiridovirus (proposed type species Invertebrate iridescent virus 31)] has been proposed (Williams, 1994; Williams & Cory, 1994). This taxonomy has not yet been accepted by the ICTV because it implies that members of the Polyiridovirus complex would share a common iridovirus ancestor with those of the genus Chloriridovirus. Nevertheless, in agreement with this view, members of the Oligoiridovirus complex have been shown to be the closest iridovirus relatives of the family Ascoviridae (Bigot et al., 2011), and to share a common invertebrate iridovirus ancestor (Stasiak et al., 2003; Bigot et al., 2009). Moreover, ascoviruses and invertebrate iridoviruses are known to be closer to each other than to vertebrate iridoviruses. This was first confirmed by phylogenetic analyses, and is strongly supported by data demonstrating that they share 26 core genes, of which only 19 are found in vertebrate iridoviruses (Eaton et al., 2007; Bigot et al., 2009). A precise definition of genera among the invertebrate iridoviruses remains unresolved at present. Further investigations are required to determine whether the proposed species complexes should be elevated to genus rank.
To date, three genomes of invertebrate iridoviruses have been sequenced: IIV-3 (also known as mosquito iridescent virus (MIV); Delhon et al., 2006), IIV-6 (Chilo iridescent virus (CIV); Jakob et al., 2001) and IIV-9 (Wiseana iridovirus (WIV); Wong et al., 2011). Here, we present a summary classification and a set of features for IIV-22, the fourth invertebrate iridovirus to be sequenced, together with a description of the sequencing and annotation of its genome. Current data reveal that IIV-22 is a tentative member of the genus Iridovirus and is related to the Polyiridovirus complex (Williams, 1994; Williams & Cory, 1994; Jancovich et al., 2011).
IIV-22 (Cameron, 1990) was kindly supplied by Professor Trevor Williams (Instituto de Ecologia AC, Xalapa, Mexico) and Professor Primitivo Caballero (Universidad Publica de Navarra, Pamplona, Spain). IIV-22 was originally isolated in 1980 from a blackfly larva (Simulium sp., order Diptera) found in the Ystwyth river near Aberystwyth in Wales, UK (Williams, 1994). It was subsequently propagated in Aedes cells in culture, and the virions were purified and plaque assayed using Spodoptera frugiperda cells (Cameron, 1990). Large quantities of virions can also be produced using a secondary host, L3 larvae of Galleria melonella (Lepidoptera; Cameron, 1990). Here, IIV-22 was amplified by infecting third-instar larvae of Spodoptera frugiperda (order Lepidoptera, family Noctuidae). Seven days after infection, larvae were frozen at −80 °C. IIV-22 virions and their genomic DNA were purified as described previously (Bigot et al., 2000).
In 2009, the scientific committee of GENOSCOPE selected the IIV-22 genome for sequencing. The complete genome sequence and annotation are now available in GenBank (accession no. HF920633). A summary of the results of this project is shown in Table 1. The genome of IIV-22 was sequenced using the 454 FLX pyrosequencing platform (Roche). Library construction and sequencing were performed as described previously (Henn et al., 2010). Assembly metrics are described in Table 1. The assembled contig representing the entire IIV-22 genome sequence was confirmed by comparing five predicted restriction fragment profiles from the genome, for BamHI, EcoRI, HindIII, PstI and SalI, with the matching fragment profiles produced by actual restriction digestions of the IIV-22 genome (Williams, 1994).
Table 1. Genome sequencing project information and genome statistics of IIV-22.
The percentage of total is based on either the size of the genome (in bp) or the total number of protein-coding genes in the annotated genome, as applicable.
| MIGS ID | Property | Term |
| MIGS-31 | Finishing quality | Finished (>99 %) |
| Number of contigs | 1 | |
| Assembly size | 197 693 bp | |
| Assembly coverage | 81× | |
| Total number of reads used | 42 828 | |
| MIGS-29 | Sequencing platform | 454 |
| MIGS-30 | Assemblers | Newbler version 2.3 Post-release 11.19.2009 |
| Gene calling method | Annotation protocol (Bigot et al., 2009) | |
| EMBL ID | HF920633 | |
| Attribute | Value | % of total |
| Size (bp) | 197 693 | 100 |
| G+C content (mol%) | 55 529 | 28.05 |
| Coding region (bp) | 170 799 | 86.4 |
| Pseudogenes | 1 | 100 |
| Total genes (putatively functional) | 167 | 100 |
| Protein coding genes with function prediction | 67 | 40.1 |
| Protein coding genes with orthologues in databases | 166 | 99.4 |
| Family of gene paralogues | 3 | – |
| Genes in families of paralogous genes | 22 | 12.6 |
| Non-coding regions over 200 bp | 11 801 (16 segments) | 6 |
Genes were identified using the Broad Institute Automated Phage Annotation Protocol as described previously (Ashburner et al., 2000; Henn et al., 2010). Briefly, evidence-based and ab initio gene-prediction algorithms were used to identify putative genes, followed by construction of a consensus gene model using a rule-based evidence approach. Gene models were checked manually for errors such as in-frame stops, very short peptides, splits and merges. Additional gene-prediction analysis and functional annotation were performed as described previously (Bigot et al., 2009).
General features of the IIV-22 genome sequence (Table 1, Fig. 1) included a G+C content of 28.05 mol% and a total of 167 putatively functional genes. No tRNA genes were found. Of the 167 coding DNA sequences (CDSs), 109 occurred in forward orientation, 58 in reverse orientation and two overlapped (119L and 120R). Sixty-seven CDSs (40.1 %) were annotated with functional product predictions. A ‘damaged’ gene between ORFs 122L and 123L was found coding for a protein close to those of CIV 261R, MIV 091L and WIV 068L. The status of this gene was confirmed by PCR and sequencing; we annotated this as a pseudogene (i.e. a molecular fossil corresponding to a relative of a functional gene that has lost its protein-coding ability due to the accumulation of one or several mutations that interrupt or disrupt its coding frame).
Fig. 1.
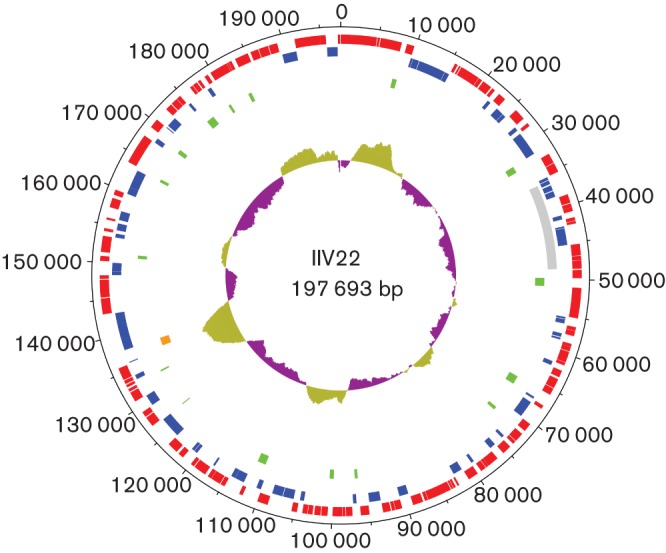
Circular map of the 197 693 bp IIV-22 genome. The outer scale is numbered clockwise in nt. Circles 1 and 2 (numbered from outside to inside) denote CDSs (forward strands in red; reverse strands in blue). The grey box in circle 3 represents the region that is found in inverse orientation in IIV-9. Green boxes in circle 4 represent ORF-free regions with a size over 200 bp. The orange box in circle 5 represents a pseudogene. Circle 6 represents local variations in G+C content along the genome sequence.
Pair alignment using blastn of nucleotide sequences of the IIV-22 and IIV-9 genomes revealed that they were 74–83 % identical over 60 % of their length. This also revealed that the region spanning nt 36 000–48 500 in the IIV-22 genome was inverted in IIV-9 between nt 165 500 and 180 000. No nucleotide sequence identity was found between IIV-22 (the isolate from the blackfly larva) and either isolate IIV-3 (from a mosquito larva) or isolate IIV-6 (from the lepidopteran Chilo suppressalis).
The annotations for the 167 genes are shown in Table S1 (available at JGV Online). This revealed that 166 of the 167 protein-coding genes had a related gene in databases with e-values below 10−3. Only seven IIV-22 genes, 012R, 036L, 089R, 104R, 120R, 126R and 167R, had no orthologues in the IIV-9 genome. Sequence analysis of the IIV-22 genome revealed that it is closely related to IIV-9. Thus, the functions of the 67 IIV-22 protein-coding genes are presumably the same as those described previously (Wong et al., 2011). The cluster of five collinear genes in IIV-9 (097R–101R) and IIV-3 (028R–032R; Wong et al., 2011) was also found to be conserved in IIV-22 (148R–152R).
With regard to repeats, three families of gene paralogues occurred in the IIV-22 genome. The first contained 13 members that were related to the 12 CIV genes: 006L, 019R, 029R, 146R, 148R, 211L, 212L, 238R, 313L, 388R, 420R and 468L. The second contained seven members related to CIV261R, 396L and 443R. The third contained only two members that belonged to the family bro-like genes, a widespread family of repeated genes in NLCDVs (Bideshi et al., 2003).
The presence of certain mobile genetic elements that occur in some NLCDVs belonging to the families Phycodnaviridae and Mimiviridae was investigated in the IIV-22 genome (Desnues et al., 2012). No transpovirons or group I introns were found. However, two inteins were found to be specifically inserted in-frame in 001R and 085L, as reported previously (Bigot et al., 2013).
Our reanalysis of the IIV-9 genome allowed us to detect a miniature transposon (MITE) between nt 179 089 and 180 010. This MITE of 921 bp was named WIV-MITE (Fig. 2a). We surmised that this was a derivative of a class II transposon (Wicker et al., 2007). Indeed, both of its extremities contained inverted terminal repeats (ITRs) about 280 bp in length that were juxtaposed at their outer ends by a TA dinucleotide that corresponded to a target site duplication (TSD), which apparently occurred at the time of the MITE insertion. In IIV-9, this MITE overlapped three small CDSs (161R, 162R and 163L), which had no orthologue in other sequenced iridoviruses. This strongly suggested that 161R, 162R and 163L are not true CDSs.
Fig. 2.
Nucleotide sequence of WIV-MITE (a) and IIV-22-MITE (b). ITRs at both ends are highlighted in black, and the duplicated TA or TTAA at the insertion site is in bold. In the central region of IIV-22-MITE, the (CT)15 stretch is in italics.
No WIV-MITE sequences were present in the IIV-22 genome, nor in the genomes of IIV-3 and IIV-6. Similar searches, however, enabled us to detect another MITE between nt 150 863 and 151 282. This MITE was named IIV-22-MITE (Fig. 2b). It was 420 bp in length and had ITRs of about 160 bp juxtaposed at their outer ends by a TTAA TSD. Such a TSD indicates that IIV-22-MITE is probably a member of the piggyBac transposon family (Wang et al., 2010) known from insects. The IIV-22-MITE was located in a non-coding sequence of 512 bp (Table S1). Interestingly, it contained a (CT)15 stretch in its central region. No IIV-22-MITE was detected in IIV-9 or in the genomes of IIV-3 and IIV-6.
IIV-22 is the fourth genome of an invertebrate iridovirus to be sequenced and reported to date. The genome reveals only one new putative protein. Many of the CDSs identified display high conservation with their counterparts in other IIVs, and insect and bacterial genomes. Further sequencing of related strains will increase our understanding of the genetic and functional diversity of these viruses.
The presence of eukaryotic class II DNA transposons in the genomes of IIV-9 and IIV-22 is of interest. This is the first report of the presence of MITEs in iridoviruses, suggesting that these viruses could act as vectors for horizontal transfer of transposable elements between host species.
Acknowledgements
This research was supported by the C.N.R.S. and GENOSCOPE, and funded by the Ministère de l’Education Nationale, de la Recherche et de la Technologie and the Groupement de Recherche CNRS 3546.
Footnotes
One supplementary table is available with the online version of this paper.
References
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S. & other authors (2000). Gene ontology: tool for the unification of biology. Nat Genet 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideshi D. K., Renault S., Stasiak K., Federici B. A., Bigot Y. (2003). Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J Gen Virol 84, 2531–2544 10.1099/vir.0.19256-0 [DOI] [PubMed] [Google Scholar]
- Bigot Y., Stasiak K., Rouleux-Bonnin F., Federici B. A. (2000). Characterization of repetitive DNA regions and methylated DNA in ascovirus genomes. J Gen Virol 81, 3073–3082 [DOI] [PubMed] [Google Scholar]
- Bigot Y., Renault S., Nicolas J., Moundras C., Demattei M. V., Samain S., Bideshi D. K., Federici B. A. (2009). Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS ONE 4, e6397. 10.1371/journal.pone.0006397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot Y., Asgari S., Bideshi D. K., Cheng X., Federici B. A., Renault S. (2011). Family Ascoviridae. In Viral Taxonomy, Ninth Report of the International Committee on the Taxonomy of Viruses, 3rd edn, pp. 147–150 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. London: Elsevier/Academic Press [Google Scholar]
- Bigot Y., Piégu B., Casteret S., Gavory F., Bideshi D. K., Federici B. A. (2013). Characteristics of inteins in invertebrate iridoviruses and factors controlling insertion in their viral hosts. Mol Phylogenet Evol 67, 246–254 10.1016/j.ympev.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Cameron I. R. (1990). Identification and characterization of the gene encoding the major structural protein of insect iridescent virus type 22. Virology 178, 35–42 10.1016/0042-6822(90)90376-3 [DOI] [PubMed] [Google Scholar]
- Delhon G., Tulman E. R., Afonso C. L., Lu Z., Becnel J. J., Moser B. A., Kutish G. F., Rock D. L. (2006). Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). J Virol 80, 8439–8449 10.1128/JVI.00464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues C., La Scola B., Yutin N., Fournous G., Robert C., Azza S., Jardot P., Monteil S., Campocasso A. & other authors (2012). Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A 109, 18078–18083 10.1073/pnas.1208835109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton H. E., Metcalf J., Penny E., Tcherepanov V., Upton C., Brunetti C. R. (2007). Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol J 4, 11. 10.1186/1743-422X-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn M. R., Sullivan M. B., Stange-Thomann N., Osburne M. S., Berlin A. M., Kelly L., Yandava C., Kodira C., Zeng Q. D. & other authors (2010). Analysis of high-throughput sequencing and annotation strategies for phage genomes. PLoS ONE 5, e9083. 10.1371/journal.pone.0009083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L. M., Aravind L., Koonin E. V. (2001). Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75, 11720–11734 10.1128/JVI.75.23.11720-11734.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob N. J., Müller K., Bahr U., Darai G. (2001). Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286, 182–196 10.1006/viro.2001.0963 [DOI] [PubMed] [Google Scholar]
- Jancovich J. K., Chinchar V. G., Hyatt A., Miyazaki T., Williams T., Zhang Q. Y. (2011). Family Iridoviridae. In Viral Taxonomy, Ninth Report of the International Committee on the Taxonomy of Viruses, 3rd edn, pp. 193–210 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. London: Elsevier/Academic Press [Google Scholar]
- Stasiak K., Renault S., Demattei M. V., Bigot Y., Federici B. A. (2003). Evidence for the evolution of ascoviruses from iridoviruses. J Gen Virol 84, 2999–3009 10.1099/vir.0.19290-0 [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang L., Meyer E., Matz M. V. (2010). Characterization of a group of MITEs with unusual features from two coral genomes. PLoS ONE 5, e10700. 10.1371/journal.pone.0010700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A., Bennetzen J. L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M. & other authors (2007). A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8, 973–982 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Williams T. (1994). Comparative studies of iridoviruses: further support for a new classification. Virus Res 33, 99–121 10.1016/0168-1702(94)90048-5 [DOI] [PubMed] [Google Scholar]
- Williams T. (2008). Natural invertebrate hosts of iridoviruses (Iridoviridae). Neotrop Entomol 37, 615–632 10.1590/S1519-566X2008000600001 [DOI] [PubMed] [Google Scholar]
- Williams T., Cory J. S. (1994). Proposals for a new classification of iridescent viruses. J Gen Virol 75, 1291–1301 10.1099/0022-1317-75-6-1291 [DOI] [PubMed] [Google Scholar]
- Wong C. K., Young V. L., Kleffmann T., Ward V. K. (2011). Genomic and proteomic analysis of invertebrate iridovirus type 9. J Virol 85, 7900–7911 10.1128/JVI.00645-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Olson N. H., Van Etten J. L., Bergoin M., Rossmann M. G., Baker T. S. (2000). Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol 7, 101–103 10.1038/72360 [DOI] [PMC free article] [PubMed] [Google Scholar]



