Abstract
Cost-effective, high-throughput epigenomic technologies have begun to emerge, rapidly replacing the candidate gene approach to molecular epidemiology and offering a comprehensive strategy for the study of epigenetics in human subjects. Epigenome-wide association studies (EWAS) provide new opportunities for advancing our understanding of epigenetic changes associated with complex disease states. However, such analyses are complicated by the dynamic nature of DNA methylation. In contrast to genomic studies, where genotype is essentially constant across somatic cells, EWAS present a new set of challenges, largely due to differential DNA methylation across distinct cell types, particularly for studies involving heterogeneous tissue sources, and changes in the epigenetic profile that occur over time. This review describes potential applications of EWAS from the viewpoint of the molecular epidemiologist, along with special considerations and pitfalls involved in the design of such studies.
Keywords: EWAS, DNA methylation, molecular epidemiology
Introduction
The term epigenetics was originally applied in the early half of the 20th century to describe “causal mechanisms” of embryogenesis in the field of developmental biology [Waddington 1942]. Since that time it has burgeoned into a flourishing discipline with a much broader context, particularly over the course of the last few decades. During this transformation, the definition of epigenetics has expanded beyond developmental biology and evolved to encompass any mitotically stable modification that influences (or at least has the potential to influence) gene expression without altering the underlying DNA sequence. Epigenetic modifications include DNA methylation, histone modifications (e.g. acetylation, methylation, ubiquitylation or sumoylation, etc.), and the now large field encompassing the many effects of non-coding RNAs. Disturbances or variations in these mechanisms have been associated with a myriad of human diseases and thus have garnered the attention of molecular epidemiologists as a major target for research.
DNA methylation is credited as the first epigenetic modification to be described on a molecular basis [Holliday and Pugh 1975] and has been the most widely studied to date, particularly in the context of molecular epidemiology, and thus will be the focus of the remainder of this review. Much of this attention can be attributed to the general stability of DNA methylation and its amenability to measurement using polymerase chain reaction (PCR) based laboratory techniques. DNA methylation involves the covalent attachment of a methyl group to cytosine at the 5-carbon position of its pyrimidine ring, resulting in 5-methylcytosine, in a reaction catalyzed by DNA methyltransferase (DNMT). This occurs primarily in the context of CpG dinucleotides, where a cytosine base is positioned upstream and adjacent to a guanine base in the DNA sequence [Tost 2010]. CpGs are overrepresented in enriched regions referred to as CpG islands (CGI). These CGIs are situated throughout the genome and can be found within the 5′ promoter region of approximately 60% of transcribed genes. Methylation of CGIs in promoter regions (commonly referred to as simply “promoter methylation”) has been generally associated with transcriptional repression (Figure 1), which experimental evidence suggests operates through recruitment of repressors that signal changes in chromatin conformation through histone modification and via interference with the binding of transcriptional activators [Choudhuri et al. 2010]. Promoter methylation may arise as part of pathologic states or during normal genomic processes including X-inactivation, imprinting [Esteller 2008] or tissue differentiation [Eckhardt et al. 2006; Schilling and Rehli 2007; Illingworth et al. 2008; Rakyan et al. 2008]. An estimated 8-12% of gene promoters are methylated under normal conditions [Meaburn and Schulz 2012]. Recent evidence suggests that methylation of DNA sequences located outside but within 2kb distance of a CpG island (dubbed “shores”) is also associated with transcriptional repression and has key involvement in cellular differentiation [Doi et al. 2009; Irizarry et al. 2009]. Clearly, genomic context plays a very important (and complex) role in the phenotypic expression of any effects associated with DNA methylation, specifically including transcriptional repression.
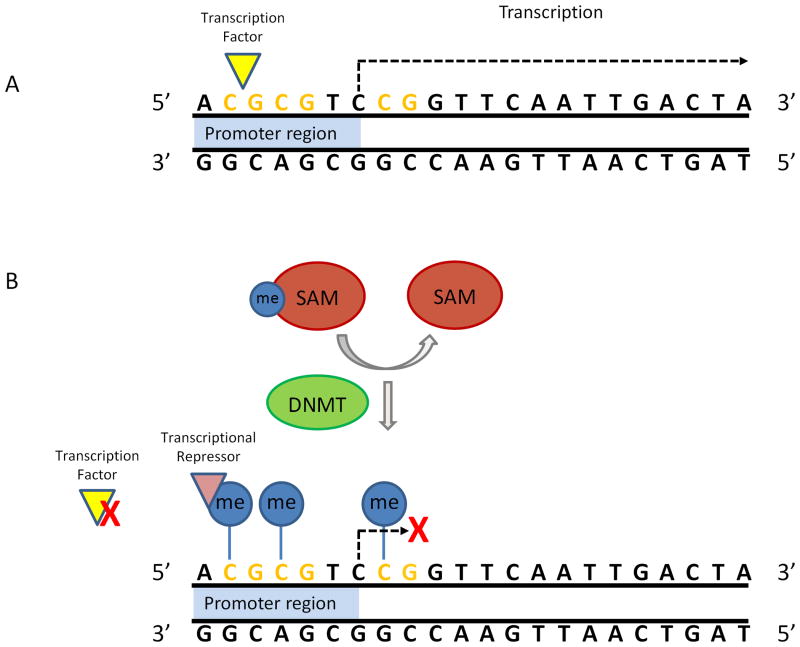
Figure 1.
Promoter methylation and transcriptional regulation. (A) The gene is transcriptionally competent when the promoter region is unmethylated. (B) S-adenosylmethionine (SAM) donates a methyl group, which is covalently attached to the 5-carbon of a CpG (represented by the blue lollipops) in a reaction catalyzed by DNA methyltransferase (DNMT). Methylation of the promoter region can inhibit the binding of transcription factors and/or recruit repressors and is typically associated with transcriptional inactivation.
At the same time, transcriptional repression is only part of the story, as it is not the only way in which DNA methylation can influence the genome. The human genome is estimated to contain approximately 30 million CpGs [Milosavljevic 2011], 70-90% of which are methylated under normal, healthy conditions [Miranda and Jones 2007]. Methylation of individual CpGs situated outside of the CGI context, particularly those located within DNA sequence repeats, retrotransposons, telomeres and pericentromeric regions, help to maintain genomic stability [Ehrlich 2002; Hoffmann and Schulz 2005; Kulis and Esteller 2010]. In these regions, DNA methylation, in concert with changes in chromatin conformation, represses the expression of transposable elements, preventing them from actively transcribing and reinserting into the genome [Wilson et al. 2007], thus diminishing the possibility of ensuing genetic recombination that could potentially compromise genomic stability or induce untoward mutations through retrotransposition. Methylation can also help to repress latent viral elements that have integrated into the DNA, further contributing to genomic stability [Kulis and Esteller 2010]. Additionally, in stark contrast to promoter methylation, methylation in the gene body can actually lead to increased transcriptional activation [Rivera and Bennett 2010; Baylin and Jones 2011]. The afore-described multiple roles of DNA methylation in gene expression and genomic stability, and corresponding consequences of its dysregulation, make it an intriguing target for molecular epidemiology studies.
Epigenetics, Aging & Human Disease
The contribution of epigenetic alterations to human disease is now widely acknowledged. Epigenetic marks are plastic and can be altered at different points during the life course, which can have varying impact on disease susceptibility and development. This has long been appreciated in cancer research; DNA methylation is among the most common somatic errors involved in carcinogenesis [Brennan and Flanagan 2012] and in fact may account for a higher proportion of tumor suppressor gene inactivation than mutation. An estimated 5-10% of CGIs that are normally unmethylated under non-pathologic conditions are methylated in cancer cells [Baylin and Jones 2011], although this is frequently accompanied by an overall epigenome-wide methylation loss [Kulis and Esteller 2010]. In recent years, momentum has also been building around the study of epigenetic events in the context of aging and other diseases (including autoimmune disorders, cardiovascular disease, diabetes, and neurodegenerative disorders, among others), with the role of alterations in DNA methylation now becoming well recognized as an important component of these processes [Calvanese et al. 2009; Brooks et al. 2010; Ordovas and Smith 2010; Jayaraman 2011; Qureshi and Mehler 2011; Cowley et al. 2012]. While replication of DNA methylation marks occurs with high fidelity and precision during mitosis (on the order of 95-99%) [Bell and Spector 2011], it should be noted that this is not a perfect process and errors do occur. It is these errors, along with interpersonal epigenetic differences including those arising in response to variations in the genome or exposome of individuals, that we as epidemiologists seek to study in the context of health-related states.
Twin studies have demonstrated that epigenetic discordance between identical siblings increases over time; particularly as more time is spent apart [Fraga et al. 2005; Petronis 2006; Ballestar 2010]. It is presently unclear whether such variation stems from stochastic events that occur over time, are preprogrammed, or represent an adaptive response to various exposures encountered during life (or a combination of these events). The latter notion of epigenetic adaptability in response to exogenous or endogenous stimuli represents a resurgence of the old and once dismissed Lamarkian theory of evolution [Handel and Ramagopalan 2010]. However, while such plasticity may convey an immediate or potential survival advantage for the adapted cell or tissue, it may ultimately predispose the individual to or contribute to the genesis of disease later in life.
There is additional mounting evidence that environmental or dietary exposures or maternal signals such as stress that are sustained during the developmental stages, in particular during prenatal development, can result in epigenetic errors or modifications that propagate throughout somatic tissues [Calkins and Devaskar 2011; Gluckman et al. 2011]. Similar as was described for epigenetic adaptations during adulthood, epigenetic alterations sustained early in pre- or neonatal development may confer a growth or survival advantage in the face of the stimulus that may dissipate over time, ceasing to confer a benefit but rather predisposing the individual to disease later in adult life (referred to as fetal origins of adult disease).
Further, newly emerging evidence indicates that disease risk or altered phenotype can derive from constitutional epimutations, which occur during development and propagate soma-wide systemic epigenetic aberrations that potentially predispose the affected individual to later disease [Hitchins 2010; Issa and Garber 2011; Brennan and Flanagan 2012]. From the viewpoint of the epidemiologist, identification and characterization of such constitutional DNA methylation events are of great interest due to their potential importance in etiologic research and implications towards risk stratification and disease prevention. Several types of epimutations have been postulated [Brennan and Flanagan 2012]. This includes those arising from genetic mutations (either cis or trans), such as has been described for with the heritable soma-wide epigenetic silencing of MSH2 (resulting in Lynch syndrome), which occurs due to terminal deletions of the adjacent upstream EPCAM gene causing the transcription machinery to errantly transcribe through the MSH2 gene, leading to its eventual epigenetic silencing [Ligtenberg et al. 2009]. Other sources of epimutation include errors in the setting of methylation marks during embryogenesis, or purely occurring heritable constitutive errors that arise independent of genetics, for which the constitutive silencing of MLH1 is presently a candidate, although it is still not entirely clear that this does not stem from an unknown trans influence [Hesson et al. 2010].
Emergence of Epigenome-Wide Association Studies
Powerful new genome-wide technologies have begun to emerge in recent years, rapidly replacing the candidate gene approach to molecular epidemiology and offering a systematic, unbiased strategy for the study of epigenetics in human subjects. This strategy has been likened to the genome-wide association studies (GWAS), which constituted a large part of the early “omics” revolution, and is credited with uncovering more than 800 genetic variations associated with more than 150 disease states or phenotypes [Hindorff et al. 2009]. However, despite early enthusiasm, the GWAS approach has been disappointing in regard to the identification of high-penetrance genes (most, although not all, have been low penetrance variants with uncertain clinical relevance) [Hindorff et al. 2009], and we are still only able to explain a fraction of familial cancer risk (8% for breast cancer, 20% for prostate cancer and 6% for colorectal cancer [Stadler et al. 2010]). DNA sequence variations identified through GWAS are estimated to account for less than 30% of phenotypic variation in humans [Lander 2011], although this may be somewhat of an underestimate since it assumes independence of genotypes, ignoring intergenic and environmental interactions. Regardless, it can be agreed that whatever the actual phenotypic contribution, genetics is not wholly responsible for our traits, underscoring the notion that our inherited genetic code does not unilaterally determine our health. In other words, our DNA does not fully dictate our destiny – other forces (including epigenetics) are at play.
This has contributed to the current enthusiasm for the post-GWAS era of molecular epidemiology, bringing about a rapid expansion of “omic” technologies, including the emergence of epigenome-wide association studies (EWAS). Such comprehensive studies provide new opportunities for advancing our understanding of epigenetic changes associated with complex disease states and are made possible in large-scale epidemiologic studies by the availability of relatively cost-effective high-throughput technologies, in particular for DNA methylation studies. Table I provides select examples of recent EWAS studies from the epidemiologic literature [Feber et al. 2011; Marsit et al. 2011; Rakyan et al. 2011a; Bell et al. 2012; Hasler et al. 2012; Joubert et al. 2012; Shenker et al. 2012], illustrating the diversity in research question, study design and sample source.
Table I.
Select examples of EWAS studies in the current literature.
| Study | Year | Platform | Study Design | Number of Subjects | Sample Source | Primary Outcome or Exposure | Adjustment for Tissue Heterogeneity |
|---|---|---|---|---|---|---|---|
| Rakyan VK et al. | 2011a | Infinium HumanMethylation27 | Discordant twin-pair | 15 monozygotic twin pairs | CD14+ monocytes | Type-1 diabetes | N/A (homogenous sample) |
| Marsit CJ et al. | 2011 | Infinium HumanMethylation27 | Case-control | 223 cases; 237 controls | Peripheral blood | Bladder cancer | None |
| Feber A et al. | 2011 | MeDIP-seq | Case-control | 10 MPNST; 10 NF; and 6 normal Schwann cell samples | Nerve sheath tumor biopsies; non-neoplastic Schwann cell biopsies (expanded in culture) | Nerve sheath tumors | Nonea |
| Bell JT et al. | 2012 | Infinium HumanMethylation27 | Twin Cohort | 172 healthy female twins (monozygotic, dizygotic, singletons) | Whole blood | Aging | WBC counts (FACS) |
| Hasler R et al. | 2012 | Infinium HumanMethylation27; MeDip-chip (385k tiling array) | Discordant twin-pair | 10 monozygotic twin pairs | Colon biopsy | Ulcerative colitis | None |
| Joubert BR et al. | 2012 | Infinium HumanMethylation450 | Prospective cohort | 1,062 newborns | Cord blood | Maternal smoking | Noneb |
| Shenker NS et al. | 2012 | Infinium HumanMethylation450 | Nested case-control | 374 cases; 374 controls | Peripheral blood | Colon cancer; breast cancer; smoking | None |
Abbreviations:
N/A = not applicable; MPNST = malignant peripheral nerve sheath tumor; NF = neurofibroma; WBC = white blood cells; FACS = fluorescence-activated cell sorting
Although the cultured Schwann cells represent purified samples, the tumor biopsies are still prone to contamination by non-neoplastic and/or connective tissue
450K array was applied to a subset of isolated monocytes and neutrophils from 21 cord blood samples to evaluate differential methylation at the top 26 CpGs identified
Application of EWAS in Human Studies
There are several general ways in which EWAS can be applied to the study of human health and disease:
1. Etiology
Etiologic research using human subjects is crucial to our understanding of the genesis and behavior of human disease, interpersonal epigenetic variation, and epigenetic alterations that occur in response to aging, environmental exposures, or other potential modifiers that may play a role such as genes, hormones or in-utero events. The comprehensive nature of EWAS can greatly facilitate the study of epigenetic drivers of physiologic processes and disease, rapidly advancing our understanding of aging and pathogenesis.
2. Tissue differentiation
To study epigenetic biomarkers and determinants of disease, we first must be able to define “normal” or “healthy” so that we have a frame of reference for determining the normalcy of our observed epigenetic marks. Epigenome-wide interrogation of CpGs allows for the identification of differentially methylated CpG loci or regions that can be used as markers of cell or tissue specific lineage (tDMRs) and provides a baseline from which to determine whether changes have occurred in epigenetic studies of disease.
3. Biomarkers
DNA methylation lends itself nicely for use in diagnostic and prognostic biomarkers of disease, as well as prediction of disease behavior. It is relatively stable, at least as compared with RNA or protein, and can be readily converted into PCR-based locus-specific assays that can potentially be used in a clinical setting. As previously discussed, it is involved in a variety of cellular functions, including transcriptional repression, differentiation and maintenance of genomic stability; and alterations to these marks have been associated with a variety of disease states. Epigenetic biomarker research has been greatly enhanced through the introduction of epigenome-wide platforms, expanding the ability of the researcher to identify relevant loci and capacity to identify clinically-relevant multi-locus methylation profiles.
4. Pharmaco-epigenetics
A subset of the latter category, epigenetic biomarkers may also have utility in the recently emerging field of pharmaco-epigenetics by providing indications of therapeutic response, sensitivity or resistance, or as predictors of potentially adverse reactions. A quintessential example of this may be the association of promoter methylation of MGMT, a gene encoding a direct DNA repair enzyme of the same name that removes alkyl damage, and enhanced survival among glioma patients treated with the alkylating agent, temozolomide [Fukushima et al. 2009].
Special Considerations in EWAS
As was the case with GWAS, properly conducted EWAS require adequate sample-size to achieve the necessary statistical power to address the research question at hand, particularly for detection of small to moderate effect sizes. Power calculations, however, can be challenging in this field, as it is newly emerging and thus less preliminary data and corresponding theory exists than is available for GWAS (this has recently been recently reviewed in more detail by Rakyan et al [Rakyan et al. 2011b]). Epigenome-wide platforms are now capable of interrogating methylation at hundreds of thousands to millions of CpGs. It is therefore critical that multiple comparisons are accounted for to control type I error, and will become even more crucial as the size of the epigenome-wide platforms (and thus the number of data points generated per sample) continues to expand. Even with safeguards to limit multiple comparisons, replication of significant results in an independent dataset remains of paramount importance to mitigate reporting of spurious disease associations. Furthermore, when applying epigenomics for candidate gene discovery, it may be necessary to prioritize candidate loci for efficient replication in single locus studies [Fazzari and Greally 2010]. This can be achieved in a number of ways, including ranking by p-value, effect size, or test statistic, either alone or in conjunction with biological considerations.
Despite the common genomic scale and complementary potential of GWAS and EWAS, there are fundamental differences that must be heeded when considering implementation of a EWAS design in epidemiologic studies. In particular, these differences center on the dynamic nature of DNA methylation as compared to the static existence of the genome, as described below. A side-by-side contrast of GWAS and EWAS is presented in Table II.
Table II.
Side-by-side contrast of genome-wide association studies (GWAS) and epigenome-wide association studies (EWAS).

|

|
|
|---|---|---|
|
| ||
| Scale | Genomic scale | (Epi)genomic scale |
| Cell lineage specificity | Genotype is static soma-wide | DNA methylation profile varies by cell/tissue type [Ohgane et al. 2008] |
| Age | Genotype is stable soma-wide (excepting somatic mutations) |
Changes during the life course [Terry et al. 2011; Lim and Song 2012] |
| Gender | Differ by sex chromosomes (male: XY; female: XX) |
Sex chromosomes can lead to gender bias with array-based approaches due to X-inactivation Differential methylation is reported by gender [Terry et al. 2011; Lim and Song 2012] |
| Race/ethnicity | Population differences in frequency of genetic polymorphisms | Limited evidence of racial variation in methylation profile [Terry et al. 2011; Lim and Song 2012] |
| Diet | Genotype is stable soma-wide (excepting somatic mutations) |
Dietary factors can impact methyl availability [Anderson et al. 2012] Has been associated with altered methylation profile [Lim and Song 2012] |
| Smoking | Genotype is stable soma-wide (excepting somatic mutations) |
Has been associated with altered methylation profile [Terry et al. 2011; Lim and Song 2012] |
| Other exogenous exposures | Genotype is stable soma-wide (excepting somatic mutations) |
A host of environmental factors have been associated with altered methylation profiles [Christensen and Marsit 2011; Terry et al. 2011; Lim and Song 2012] |
Tissue heterogeneity
There are hundreds of distinct cell types in the human body, all possessing an identical genetic code [Slomko et al. 2012]. Whereas genotype is stable across somatic cells, the epigenetic signature varies in a cell and tissue dependent manner [Igarashi et al. 2008; Christensen et al. 2009; Accomando et al. 2012; Houseman et al. 2012; Koestler et al. 2012; Reinius et al. 2012; Wiencke et al. 2012]. This can have profound implications on the interpretation of EWAS data since it means that some or all of the epigenetic variation observed in biospecimens containing more than one cell type may be attributed to differences in cell proportions in the samples. Furthermore, tissues sustain differential levels of exposure to endogenous and exogenous agents and may also vary in their response to such exposures in a tissue or cell-specific manner. Thus biospecimen selection for EWAS must be carefully considered, depending upon the disease or outcome of interest and corresponding study hypothesis.
Blood as a surrogate tissue
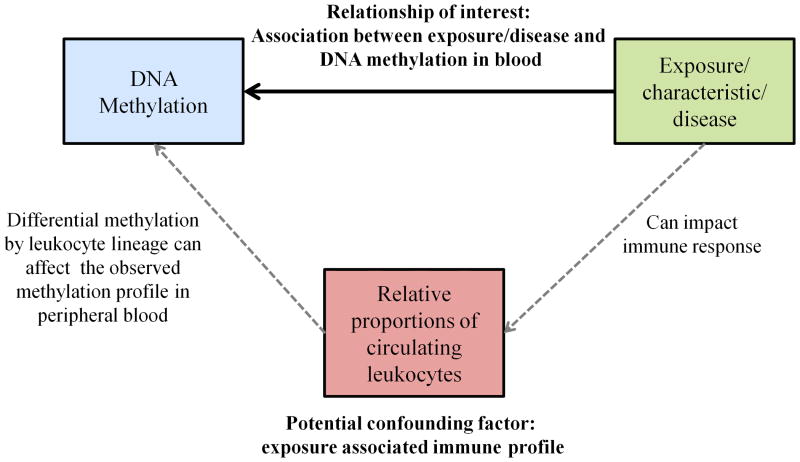
Blood may be an attractive DNA source for conducting EWAS, as it is commonly collected and archived in epidemiologic studies (including neonatal cord blood and Guthrie card samples). It represents a logical target tissue for many cardiovascular or leukocyte-associated disorders (including autoimmune diseases) or may be employed as a surrogate tissue to examine systemic alterations in DNA methylation due to constitutional epimutations or in response to aging, exposures or disease. For these reasons, blood may often be a choice tissue for EWAS, although there are several considerations that must be kept in mind when designing such a study. If using blood as a surrogate, it should be noted that epigenetic alterations that are observed in blood may not necessarily reflect changes occurring in other tissues. Further, blood is a very heterogeneous tissue, comprised of a broad mixture of multiple discrete cell types. Therefore, when blood is used for EWAS, it is imperative that the relative proportions of these constituents be taken into account, as each cell type bears its own specific DNA methylation signature. Since personal characteristics, behaviors, exogenous exposures, or pathologic states can modulate the circulating immune profile [Cheng et al. 2004; Mehta et al. 2008; Szabo and Mandrekar 2009; Federico et al. 2010; Kiecolt-Glaser et al. 2010; Oertelt-Prigione 2012], inter-subject variation in leukocyte distribution can potentially confound the observed relationship between methylation and the disease or exposure of interest (Figure 2). Until now, this has proven a major practical challenge (and is often altogether ignored), requiring additional time-consuming quantification of relative leukocyte proportions in the peripheral blood. Fluorescent activated cell sorting (a.k.a. FACS or flow cytometry) is expensive and requires fresh blood samples (and therefore is not practical for use with most archival specimens collected as part of epidemiologic studies), while manual blood differentials do not distinguish between T, B and NK cells, simply classifying them collectively as “lymphocytes” and thus losing important information. Large-scale international efforts, such as the International Human Epigenome Consortium (IHEC; http://ihec-epigenomes.net/), BLUEPRINT (http://www.blueprint-epigenome.eu/), ENCODE (http://genome.ucsc.edu/ENCODE/) and Roadmap Epigenomics Project (http://www.roadmapepigenomics.org/), are currently underway to establish epigenomic roadmaps for humans, creating reference epigenomes that can be used to identify tDMRs. New statistical methods have recently been described that apply tDMR information to estimate leukocyte fractions using epigenomic profiles of peripheral blood [Houseman et al. 2012], which can in turn be applied to blood-based EWAS analyses to adjust for inter-individual immune variation.
Figure 2.
The relationship between exposure/personal characteristic, relative leukocyte proportions, and DNA methylation profiles in blood.
Findings from blood-based EWAS will be a practical challenge to interpret, in most cases. For example, an EWAS finding may represent constitutional alterations in DNA methylation (akin to ‘epimutations’), reflect the emergence of small but important and heretofore not characterized subsets of immune cells (e.g. rare activated immune cells that are phenotypically important such as dendritic cells, etc.) that signal an immune response (potentially chronic and one that could presage disease but also change over time) associated with the disease of interest or, finally, an EWAS hit may possibly signal the effects of an exposure that acts over time on all cells in the body. Results should be evaluated with caution as these interpretations may be difficult to distinguish in some cases.
Temporality
The characteristic mitotic stability of epigenetic modifications does not necessarily imply a complete static state. Unlike genotype, which is essentially the same for each person at conception as it is at death, the epigenetic profile of an individual changes over time. Epigenetic events occur during the aging process and in response to exogenous exposures, bringing about an increased potential for confounding and necessitating careful consideration of this lurking pitfall when designing studies or interpreting results. It is also another reason why thoughtful selection of a biospecimen source is so critical for EWAS, as cells and tissues may be differentially vulnerable to exposures or may age in divergent ways [Brennan and Flanagan 2012] arousing the possibility of interaction between tissue-specificity and temporality of DNA methylation marks. Aging is a combination of both cellular mitosis and temporal existence in the face of damaging processes, such as oxidative stress; these are quite different situations and may have very different epigenetic consequences. Additionally, heed must be paid to the notion of reverse causality. Not only can epigenetic dysregulation lead to disease, but pathologies can instigate epigenetic dysregulation. Whereas cross-sectional genotyping (i.e. at study enrollment) in case-control studies provides a genetic record of the subject that almost certainly preceded the onset of disease, this is not a valid assumption in the case of DNA methylation. This leaves molecular epidemiogists faced with a paradox reminiscent of the chicken and the egg in establishing whether an observed epigenetic alteration is a cause or consequence of disease, underscoring the critical importance of establishing a temporal relationship between the two. While case-control studies may still be appropriate for initial conduct of EWAS, particularly for studying rare diseases or outcomes, significant findings should be replicated in prospective studies to rule out reverse causality, necessitating the continued establishment of large consortiums for studying associations between epigenetics and rare events.
Platform selection
In a perfect world with infinite resources, high-throughput methylome sequencing (i.e. next-generation sequencing) would be the ideal technology for use in EWAS due to its complete genomic coverage at a single-base resolution and reliable reproducibility [Zhao and Zhang 2011]. However, at the present time this technology is still extremely cost prohibitive for most researchers, although the pricing has rapidly deescalated over the past few years and is anticipated to reach manageable levels for routine epidemiologic use in the relatively near future. Until that day arrives, other powerful technologies are available on the commercial market for interrogation on an epigenome-wide scale that may be used to conduct EWAS. Our personal technology of choice for use in epidemiologic studies is the Infinium 450K BeadArray (Illumina, San Diego, CA) - an array-based platform capable of interrogating methylation status at more than 450,000 bases - due to cost (it can be run for hundreds of dollars per sample rather than thousands) and coverage considerations. However, a drawback of this technology is that use of archival formalin-fixed paraffin-embedded tissue can be challenging due to the damaged and fragmented nature of such samples, although additional ligation steps have been described to mitigate this issue [Thirlwell et al. 2010]. Molecular epidemiologists should select the optimal platform based on the specific needs and resource availability for their study; the advantages and disadvantages of these technologies have been recently reviewed in detail elsewhere [Fazzari and Greally 2010; Rakyan et al. 2011b; Bock 2012].
Summary
It is important to remember that the EWAS is still a relatively nascent approach to molecular epidemiology and is not yet as well established as the GWAS. Consequently, the methodology is still developing, as there are distinct issues and considerations that accompany epigenetic research as compared with genomic analyses, and thus we will encounter bumps in the road as we progress. However, continued efforts involving carefully designed studies by investigators that account for the temporal and histologic plasticity of DNA methylation will continue to drive the field forward allowing it to overcome such growing pains, unlocking the vast potential of epigenomics for use in epidemiologic studies. As time progresses, the eventual integration of epigenomics with other “omic” approaches will critically inform our understanding of phenotypic expression and disease risk and provide for rapid advancements in biomedical science, which in turn will contribute to the ultimate goal of improving public health and well-being.
Footnotes
Author Contributions: SML and KTK both contributed to the writing/content of this manuscript.
Conflicts of Interest and Disclosure: Karl T. Kelsey has applied for a patent covering DNA methylation arrays as biomarkers of immune cell distributions.
References
- Accomando WP, Wiencke JK, Houseman EA, Butler RA, Zheng S, Nelson HH, Kelsey KT. Decreased NK Cells in Patients with Head and Neck Cancer Determined in Archival DNA. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23(8):853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestar E. Epigenetics lessons from twins: prospects for autoimmune disease. Clin Rev Allergy Immunol. 2010;39(1):30–41. doi: 10.1007/s12016-009-8168-4. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27(3):116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, Shin SY, Dempster EL, Murray RM, Grundberg E, Hedman AK, Nica A, Small KS, Dermitzakis ET, McCarthy MI, Mill J, Spector TD, Deloukas P. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8(4):e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Brennan K, Flanagan JM. Epigenetic epidemiology for cancer risk: harnessing germline epigenetic variation. Methods Mol Biol. 2012;863:439–465. doi: 10.1007/978-1-61779-612-8_27. [DOI] [PubMed] [Google Scholar]
- Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. J Autoimmun. 2010;34(3):J207–219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Calkins K, Devaskar SU. Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care. 2011;41(6):158–176. doi: 10.1016/j.cppeds.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8(4):268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Cheng CK, Chan J, Cembrowski GS, van Assendelft OW. Complete blood count reference interval diagrams derived from NHANES III: stratification by age, sex, and race. Lab Hematol. 2004;10(1):42–53. doi: 10.1532/lh96.04010. [DOI] [PubMed] [Google Scholar]
- Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245(3):378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Marsit CJ. Epigenomics in environmental health. Front Genet. 2011;2:84. doi: 10.3389/fgene.2011.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley AW, Jr, Nadeau JH, Baccarelli A, Berecek K, Fornage M, Gibbons GH, Harrison DG, Liang M, Nathanielsz PW, O'Connor DT, Ordovas J, Peng W, Soares MB, Szyf M, Tolunay HE, Wood KC, Zhao K, Galis ZS. Report of the National Heart, Lung, and Blood Institute Working Group on epigenetics and hypertension. Hypertension. 2012;59(5):899–905. doi: 10.1161/HYPERTENSIONAHA.111.190116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr. 2002;132(8 Suppl):2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Introduction to epigenomics and epigenome-wide analysis. Methods Mol Biol. 2010;620:243–265. doi: 10.1007/978-1-60761-580-4_7. [DOI] [PubMed] [Google Scholar]
- Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, Down TA, Rakyan VK, Noon LA, Lloyd AC, Stupka E, Schiza V, Teschendorff AE, Schroth GP, Flanagan A, Beck S. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 2011;21(4):515–524. doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A, D'Aiuto E, Borriello F, Barra G, Gravina AG, Romano M, De Palma R. Fat: a matter of disturbance for the immune system. World J Gastroenterol. 2010;16(38):4762–4772. doi: 10.3748/wjg.v16.i38.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Takeshima H, Kataoka H. Anti-glioma therapy with temozolomide and status of the DNA-repair gene MGMT. Anticancer Res. 2009;29(11):4845–4854. [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Low FM. The role of developmental plasticity and epigenetics in human health. Birth Defects Res C Embryo Today. 2011;93(1):12–18. doi: 10.1002/bdrc.20198. [DOI] [PubMed] [Google Scholar]
- Handel AE, Ramagopalan SV. Is Lamarckian evolution relevant to medicine? BMC Med Genet. 2010;11:73. doi: 10.1186/1471-2350-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler R, Feng Z, Backdahl L, Spehlmann ME, Franke A, Teschendorff A, Rakyan VK, Down TA, Wilson GA, Feber A, Beck S, Schreiber S, Rosenstiel P. A functional methylome map of ulcerative colitis. Genome Res. 2012;22(11):2130–2137. doi: 10.1101/gr.138347.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesson LB, Hitchins MP, Ward RL. Epimutations and cancer predisposition: importance and mechanisms. Curr Opin Genet Dev. 2010;20(3):290–298. doi: 10.1016/j.gde.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP. Inheritance of epigenetic aberrations (constitutional epimutations) in cancer susceptibility. Adv Genet. 2010;70:201–243. doi: 10.1016/B978-0-12-380866-0.60008-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83(3):296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187(4173):226–232. [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi J, Muroi S, Kawashima H, Wang X, Shinojima Y, Kitamura E, Oinuma T, Nemoto N, Song F, Ghosh S, Held WA, Nagase H. Quantitative analysis of human tissue-specific differences in methylation. Biochem Biophys Res Commun. 2008;376(4):658–664. doi: 10.1016/j.bbrc.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6(1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Garber JE. Time to think outside the (genetic) box. Cancer Prev Res (Phila) 2011;4(1):6–8. doi: 10.1158/1940-6207.CAPR-10-0348. [DOI] [PubMed] [Google Scholar]
- Jayaraman S. Epigenetics of autoimmune diabetes. Epigenomics. 2011;3(5):639–648. doi: 10.2217/epi.11.78. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Gouin JP, Hantsoo L. Close relationships, inflammation, and health. Neurosci Biobehav Rev. 2010;35(1):33–38. doi: 10.1016/j.neubiorev.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, Nelson HH, Karagas MR, Wiencke JK, Kelsey KT. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1293–1302. doi: 10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, Tsui WY, Kong CK, Brunner HG, van Kessel AG, Yuen ST, van Krieken JH, Leung SY, Hoogerbrugge N. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–376. doi: 10.1007/978-1-61779-612-8_23. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. J Clin Oncol. 2011;29(9):1133–1139. doi: 10.1200/JCO.2010.31.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn E, Schulz R. Next generation sequencing in epigenetics: insights and challenges. Semin Cell Dev Biol. 2012;23(2):192–199. doi: 10.1016/j.semcdb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Mehta H, Nazzal K, Sadikot RT. Cigarette smoking and innate immunity. Inflamm Res. 2008;57(11):497–503. doi: 10.1007/s00011-008-8078-6. [DOI] [PubMed] [Google Scholar]
- Milosavljevic A. Emerging patterns of epigenomic variation. Trends Genet. 2011;27(6):242–250. doi: 10.1016/j.tig.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213(2):384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Oertelt-Prigione S. The influence of sex and gender on the immune response. Autoimmun Rev. 2012;11(6-7):A479–485. doi: 10.1016/j.autrev.2011.11.022. [DOI] [PubMed] [Google Scholar]
- Ohgane J, Yagi S, Shiota K. Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta. 2008;29(Suppl A):S29–35. doi: 10.1016/j.placenta.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010;7(9):510–519. doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics and twins: three variations on the theme. Trends Genet. 2006;22(7):347–350. doi: 10.1016/j.tig.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Advances in epigenetics and epigenomics for neurodegenerative diseases. Curr Neurol Neurosci Rep. 2011;11(5):464–473. doi: 10.1007/s11910-011-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, Daunay A, Busato F, Mein CA, Manfras B, Dias KR, Bell CG, Tost J, Boehm BO, Beck S, Leslie RD. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011a;7(9):e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011b;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavare S, Beck S. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18(9):1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RM, Bennett LB. Epigenetics in humans: an overview. Curr Opin Endocrinol Diabetes Obes. 2010;17(6):493–499. doi: 10.1097/MED.0b013e3283404f4b. [DOI] [PubMed] [Google Scholar]
- Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90(3):314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- Slomko H, Heo HJ, Einstein FH. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153(3):1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler ZK, Vijai J, Thom P, Kirchhoff T, Hansen NA, Kauff ND, Robson M, Offit K. Genome-wide association studies of cancer predisposition. Hematol Oncol Clin North Am. 2010;24(5):973–996. doi: 10.1016/j.hoc.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Szabo G, Mandrekar P. A recent perspective on alcohol, immunity, and host defense. Alcohol Clin Exp Res. 2009;33(2):220–232. doi: 10.1111/j.1530-0277.2008.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6(7):828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirlwell C, Eymard M, Feber A, Teschendorff A, Pearce K, Lechner M, Widschwendter M, Beck S. Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods. 2010;52(3):248–254. doi: 10.1016/j.ymeth.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44(1):71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Wiencke JK, Accomando WP, Zheng S, Patoka J, Dou X, Phillips JJ, Hsuang G, Christensen BC, Houseman EA, Koestler DC, Bracci P, Wiemels JL, Wrensch M, Nelson HH, Kelsey KT. Epigenetic biomarkers of T-cells in human glioma. Epigenetics. 2012;7(12) doi: 10.4161/epi.22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y. Epigenome sequencing comes of age in development, differentiation and disease mechanism research. Epigenomics. 2011;3(2):207–220. doi: 10.2217/epi.10.78. [DOI] [PubMed] [Google Scholar]